Improved Mixture Cure Model Using Machine Learning Approaches
Abstract
1. Introduction
2. Framework of the Mixture Cure Model
3. Improved Mixture Cure Models Based on Machine Learning
3.1. Improvement of Mixture Cure Models Based on Neural Networks
3.2. Improvement of Mixture Cure Models Based on Bayesian Classifiers
3.3. Improvement of Mixture Cure Models Based on Support Vector Machines
3.4. Improvement of Mixture Cure Models Based on Decision Tree Classifiers
4. Algorithms and R Packages for Implementing Mixture Cure Models
4.1. Overview
| Package | Author | Usage | Advantages | Disadvantages |
|---|---|---|---|---|
| cuRe | Lasse Hjort Jakobsene et al. [35] https://cran.r-project.org/web/packages/cuRe/index.html (accessed on 1 February 2025) | Functions are employed for estimating generalized parametric mixture and non-mixture cure models, lifetime loss, the mean residual life, and crude event probabilities. | (1) Includes functions that calculate a variety of useful post-estimation metrics. (2) Fits both parametric mixture cure models for overall survival functions and relative survival mixture cure models. | (1) Excludes model diagnostics functionality. (2) Excludes functionality for calculating cure proportions on the cumulative incidence scale. |
| mixcure | Yingwei Peng [30] https://cran.r-project.org/web/packages/mixcure/index.html (accessed on 1 February 2025) | Used for mixture cure models. | (1) Applicable to various mixture cure models. (2) Utilizes existing R packages to fit both parametric and semiparametric cure models. | Unable to fit relative survival mixture cure models. |
| npcure | Ignacio López-de-Ullibarri et al. [32] | Non-parametric estimation within mixture cure models, including the performance of non-parametric estimation and conducting significance tests for cure probabilities. | (1) Allows covariates to exert different effects on cured and uncured patients. (2) Selectable bandwidth for non-parametric association estimation. | (1) A non-parametric method that does not estimate parameters. (2) Estimates of cure rates are biased. (3) Due to the nature of non-parametric methods, it cannot provide global survival estimates for uncured individuals. |
| smcure | Chao Cai et al. [31] | Estimation of semiparametric PH and AFT mixture cure models. | The probability component is estimated using a generalized linear model, accommodating various link functions such as logit, probit, and cloglog. The delay component can follow either a PH model or an AFT model. | (1) Fitting performance is poor when covariates follow a binomial distribution with low cure rates and low censoring rates. (2) Computational time is slow, and it is unstable during repeated calculations. |
| intcure | Yingwei Peng [37] | Used for mixture cure models with random effects, as described by Peng and Taylor (2011) [37], pertaining to a mixture treatment model with random effects. | Good initial values for bt, gm, and basepara obtained from mixture cure models without random effects can accelerate the program or help find optimal estimates. | —— |
| GORCure | Jie Zhou et al. [25] | Fitting odds rate mixture cure models with interval-censored data. | (1) Well-suited for handling survival data with low cure rates and low censoring rates. (2) Results are less affected by cure rates and censoring rates, but more influenced by the distribution of covariates. | (1) Not suitable for right-censored data. (2) Bias is relatively large in cases with high cure rates and high censoring rates. |
| Motahareh-Parsa /kmcure | Mahmood-Taghavi et al. [38] | Fitting a semiparametric accelerated failure time (AFT) mixture cure model using the KME-KDE (Kaplan–Meier estimation and kernel density estimation) method. | —— | —— |
| Oswaldogressani /mixcurelps | —— | Laplace approximation and p-splines for rapid approximate Bayesian inference in mixture cure models. | The proportion of uncured subjects (also known as incidence) is modeled using a logit link function, while the survival function for uncured patients (latent period) is approximated using a flexible Cox proportional hazards model, with the baseline risk approximated by penalized cubic B-splines. | —— |
| nltm | Gilda Garibotti et al. [39] | Nonlinear transformation models for analyzing survival data. | The categories of nltm include the following currently supported models: Cox proportional hazards, proportional hazards cure, proportional odds, proportional hazards–proportional hazards cure, proportional hazards–proportional odds cure, gamma frailty, and proportional hazards–proportional odds. | —— |
| geecure | Yi Niu et al. [34] | Used for estimating the marginal proportional hazards mixture cure (PHMC) model using generalized estimating equation (GEE) methods. | (1) Results are stable and accurate. (2) In the delay component and the semiparametric PHMC model, the parametric PHMC model with a Weibull baseline distribution is used to accommodate multivariable survival data with treatment scores. | Due to the need to estimate within-cluster correlation, the computational burden is substantial, resulting in lengthy processing times. |
| penPHcure | A. Beretta et al. [36] | Variable selection for time-dependent covariate proportional hazards (PHs) mixture cure models. | Time-dependent covariates can be incorporated into proportional hazards mixture cure models. | —— |
| CureAuxSP | J. Ding et al. [40] | Estimation of mixture cure models with auxiliary survival probabilities. | External subgroup survival probabilities can be adaptively integrated into the analysis of internal survival data, using subgroup survival probabilities as auxiliary information in survival analysis. | —— |
4.2. Simulation Study
5. The Application of Mixture Cure Models
5.1. Review of Existing Applications in Medicine
5.2. Illustration of Decision Tree Improved Mixture Cure Model Using Cervical Carcinoma Data
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cox, D.R. Regression models and life-tables. J. R. Stat. Soc. Ser. B Methodol. 1972, 34, 187–220. [Google Scholar] [CrossRef]
- Boag, J.W. Maximum likelihood estimates of the proportion of patients cured by cancer therapy. J. R. Stat. Soc. Ser. B Methodol. 1949, 11, 15–53. [Google Scholar] [CrossRef]
- Maller, R.A.; Ghitany, M.E.; Zhou, S. Exponential mixture models with long-term survivors and co-variates. J. Multivar. Anal. 1994, 49, 218–241. [Google Scholar]
- Gage, R.P.; Berkson, J. Survival curve for cancer patients following treatment. J. Am. Stat. Assoc. 1952, 47, 501–515. [Google Scholar]
- Farewell, V.T. A model for a binary variable with time-censored observations. Biometrika 1977, 64, 43–46. [Google Scholar] [CrossRef]
- Farewell, V.T. The use of mixture models for the analysis of survival data with long-term survivors. Biometrics 1982, 38, 1041–1046. [Google Scholar] [CrossRef]
- Chen, C.; Kuk, A.Y.C. A mixture model combining logistic regression with proportional hazards regression. Biometrika 1992, 79, 531–541. [Google Scholar]
- Dear, K.B.; Peng, Y. A nonparametric mixture model for cure rate estimation. Biometrics 2000, 56, 237–243. [Google Scholar]
- Lu, W. Maximum likelihood estimation in the proportional hazards cure model. Ann. Inst. Stat. Math. 2008, 60, 545–574. [Google Scholar] [CrossRef]
- Taylor, J.M.; Li, C.S. A semi-parametric accelerated failure time cure model. Stat. Probab. Lett. 2002, 21, 3235–3247. [Google Scholar]
- Peng, Y.; Zhang, J. A new estimation method for the semiparametric accelerated failure time mixture cure model. Stat. Med. 2007, 26, 3157–3171. [Google Scholar]
- Musta, E.; Valentin, P.; Ingrid, V.K. A two-step estimation procedure for semi-parametric mixture cure models. arXiv 2024, arXiv:2207.08237. [Google Scholar]
- Ortega, E.M.; Cancho, V.G.; Paula, G.A. Generalized log-gamma regression models with cure fraction. Lifetime Data Anal. 2009, 15, 79–106. [Google Scholar] [CrossRef]
- Gordon, N.H. Application of the theory of finite mixtures for the estimation of ’cure’ rates of treat-ed cancer patients. Stat. Med. 1990, 9, 397–407. [Google Scholar] [CrossRef]
- Zhang, J.; Peng, Y. Accelerated hazards mixture cure model. Lifetime Data Anal. 2009, 15, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Engelhard, M.; Henao, R. Disentangling Whether from When in a Neural Mixture Cure Model for Failure Time Data. Proc. Mach. Learn. Res. 2022, 151, 9571–9581. [Google Scholar] [PubMed]
- Xie, Y.; Yu, Z. Promotion time cure rate model with a neural network estimated nonparametric component. Stat. Med. 2021, 40, 3516–3532. [Google Scholar] [CrossRef]
- Gressani, O.; Faes, C.; Hens, N. Laplacian-P-splines for Bayesian inference in the mixture cure model. Stat. Med. 2022, 41, 2602–2626. [Google Scholar] [CrossRef] [PubMed]
- Hurtado Rúa, S.M.; Dey, D.K. A Bayesian piecewise survival cure rate model for spatially clustered data. Spat. Spatio-Temporal Epidemiol. 2019, 29, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, W.; Tang, Y. A Bayesian semiparametric accelerate failure time mixture cure model. Int. J. Biostat. 2022, 18, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Cai, B.; Sui, X. A Bayesian proportional hazards mixture cure model for interval-censored data. Lifetime Data Anal. 2024, 30, 327–344. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, J.; Lu, W. Computationally Efficient Estimation for the Generalized Odds Rate Mixture Cure Model with Interval-Censored Data. J. Comput. Graph. Stat. 2018, 27, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Peng, Y.; Jiang, P.; Dong, Q. A support vector machine based semiparametric mixture cure model. Comput. Stat. 2020, 35, 931–945. [Google Scholar] [CrossRef]
- Pal, S.; Peng, Y.; Aselisewine, W.; Barui, S. A support vector machine-based cure rate model for interval censored data. Stat. Methods Med. Res. 2023, 32, 2405–2422. [Google Scholar] [CrossRef] [PubMed]
- Aselisewine, W.; Pal, S. On the integration of decision trees with mixture cure model. Stat. Med. 2023, 42, 4111–4127. [Google Scholar] [CrossRef]
- Peng, Y.; Dear, K.B.G.; Denham, J.W. A generalized F mixture model for cure rate estimation. Stat. Med. 1998, 17, 813–830. [Google Scholar] [CrossRef]
- Yuan, S.; Zheng, P.; Wu, X. SAFE: A Neural Survival Analysis Model for Fraud Early Detection. Proc. AAAI Conf. Artif. Intell. 2019, 33, 1278–1285. [Google Scholar]
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.; Kluger, Y. DeepSurv: Personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef]
- Kvamme, H.; Borgan, Ø.; Ida, S. Time-to-Event Prediction with Neural Networks and Cox Regression. J. Mach. Learn. Res. 2019, 20, 1–30. [Google Scholar]
- Peng, Y.W. Fitting semiparametric cure models. Comput. Stat. Data Anal. 2003, 41, 481–490. [Google Scholar] [CrossRef]
- Cai, C.; Zou, Y.; Peng, Y.; Zhang, J. smcure: An R-package for estimating semiparametric mixture cure models. Comput. Methods Programs Biomed. 2012, 108, 1255–1260. [Google Scholar] [CrossRef]
- Cao, R.; López-Cheda, A.; Jácome, M.A. Nonparametric incidence estimation and bootstrap band-width selection in mixture cure models. Comput. Stat. Data Anal. 2017, 105, 144–165. [Google Scholar]
- Mukhopadhyay, P.; Ouwens, M.J.N.M.; Zhang, Y.D. Estimating lifetime benefits associated with immunooncology therapies: Challenges and approaches for overall survival extrapolations. Pharm. Econ. 2019, 37, 1129–1138. [Google Scholar]
- Niu, Y.; Wang, X.; Peng, Y. geecure: An R-package for marginal proportional hazards mixture cure models. Comput. Methods Programs Biomed. 2018, 161, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.K.; Clements, M.; Gjærde, L.K.; Jakobsen, L.H. Fitting parametric cure models in R using the packages cuRe and rstpm2. Comput. Methods Programs Biomed. 2022, 226, 107125. [Google Scholar] [CrossRef]
- Beretta, A.; Heuchenne, C. penPHcure: Variable Selection in Proportional Hazards Cure Model with Time-Varying Covariates. R J. 2021, 13, 53–66. [Google Scholar] [CrossRef]
- Peng, Y.; Taylor, J.M. Mixture cure model with random effects for the analysis of a multi-center tonsil cancer study. Stat. Med. 2011, 30, 211–223. [Google Scholar] [CrossRef]
- Parsa, M.; Taghavi-Shahri, S.M.; Van Keilegom, I. kmcure: Fits AFT Semiparametric Mixture Cure Model Using the KME-KDE Method. 2022. Available online: https://github.com/Motahareh-Parsa/kmcure/ (accessed on 1 February 2025).
- Garibotti, G.; Tsodikov, A.; Clements, M. nltm: Non-Linear Transformation Models. 2023. Available online: https://cran.r-project.org/web/packages/nltm/index.html (accessed on 1 February 2025).
- Ding, J.; Li, J.; Zhang, M.; Wang, X. CureAuxSP: An R package for estimating mixture cure models with auxiliary survival probabilities. Comput. Methods Programs Biomed. 2024, 251, 108212. [Google Scholar] [CrossRef] [PubMed]
- Amanpour, F.; Akbari, S.; Looha, M.A.; Abdehagh, M.; Pourhoseingholi, M.A. Mixture cure model for estimating short-term and long-term colorectal cancer survival. Gastroenterol. Hepatol. Bed Bench 2019, 12 (Suppl. S1), s37–s43. [Google Scholar]
- Cai, C.; Love, B.L.; Yunusa, I.; Reeder, C.E. Applying mixture cure survival modeling to medication persistence analysis. Pharmacoepidemiol. Drug Saf. 2022, 31, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, M.; Ghamsari, F.S.H.; Rasekhi, A.; Hajifathali, A. Factors affecting survival in bone marrow transplantation using mixture cure model. Health Sci. Monit. 2024, 3, 19–28. [Google Scholar] [CrossRef]
- Ou, X.; You, J.; Liang, B.; Li, X.; Zhou, J.; Wen, F.; Wang, J.; Dong, Z.; Zhang, Y. Prognostic Factors Analysis of Metastatic Recurrence in Cervical Carcinoma Patients Treated with Definitive Radiotherapy: A Retrospective Study Using Mixture Cure Model. Cancers 2023, 15, 2913. [Google Scholar] [CrossRef] [PubMed]
- Maller, R.; Resnick, S.; Shemehsavar, S.; Zhao, M. Mixture cure model methodology in survival analysis: Some recent results for the one-sample case. Stat. Surv. 2024, 18, 82–138. [Google Scholar] [CrossRef]
- Kaplan, E.L.; Paul, M. Nonparametric Estimation from Incomplete Observations. In Break-Throughs in Statistics; Springer: New York, NY, USA, 1992; pp. 319–337. [Google Scholar]
| Incidence | π(z) | Su(∙; x) | S(∙; x, z) | |||
|---|---|---|---|---|---|---|
| Submodel | Bias | MSE | Bias | MSE | Bias | MSE |
| Logistic | 0.274 | 0.138 | 0.345 | 0.225 | 0.152 | 0.048 |
| DT | 0.234 | 0.114 | 0.345 | 0.225 | 0.135 | 0.037 |
| SVM | 0.225 | 0.109 | 0.343 | 0.225 | 0.129 | 0.036 |
| NN | 0.229 | 0.136 | 0.344 | 0.224 | 0.127 | 0.042 |
| Variable | Overall | Non-Metastatic Recurrence | Metastatic Recurrence | p-Value † |
|---|---|---|---|---|
| All patients | 462 | 353 (76%) | 109 (24%) | |
| Differentiation | 0.643 | |||
| High | 288 | 218 (76%) | 70 (24%) | |
| Low | 174 | 135 (78%) | 39 (22%) | |
| Stage [n (%)] | <0.001 | |||
| Early | 97 | 84 (87%) | 13 (13%) | |
| Local adva. | 269 | 208 (77%) | 61 (23%) | |
| Advanced | 96 | 61 (36%) | 109 (64%) | |
| Num. of treat. | 0.03 | |||
| 2 | 8 | 6 (75%) | 2 (25%) | |
| 3 | 13 | 8 (62%) | 5 (38%) | |
| 4 | 240 | 191 (80%) | 49 (20%) | |
| 5 | 181 | 138 (76%) | 58 (24%) | |
| 6 | 20 | 10 (50%) | 10 (50%) | |
| CoChemo | 0.724 | |||
| 0 | 223 | 172 (77%) | 52 (23%) | |
| 1 | 239 | 181 (76%) | 58 (24%) | |
| Age (in years) * | 53 (47, 59) | 53 (47, 59) | 52 (45, 57) | 0.3 |
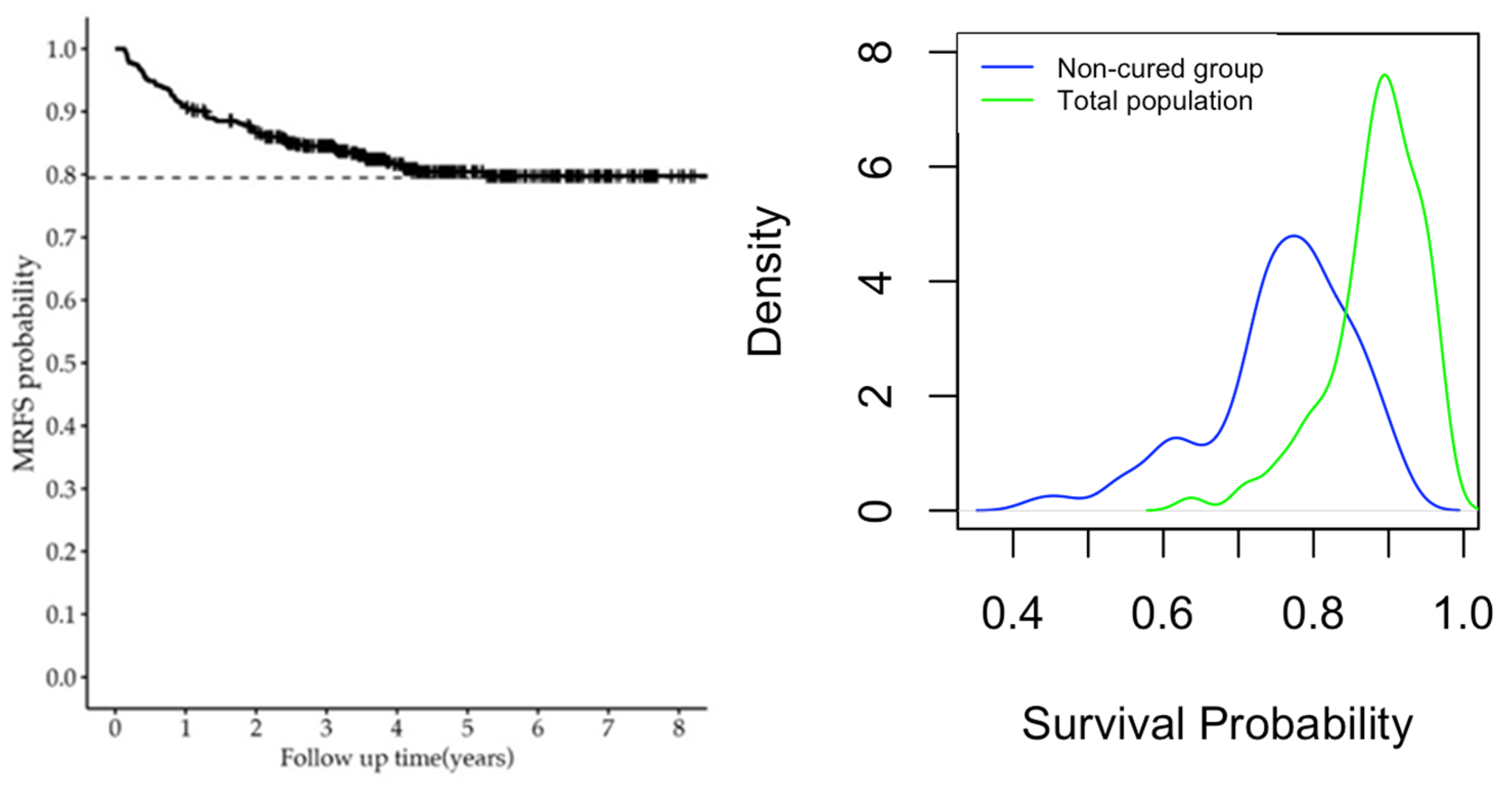
| Survival Probability | Min. | Median | Max. |
|---|---|---|---|
| Non-cured | 0.4338 | 0.7724 | 0.9120 |
| Overall | 0.6371 | 0.8824 | 0.9679 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Feng, T.; Liang, B. Improved Mixture Cure Model Using Machine Learning Approaches. Mathematics 2025, 13, 557. https://doi.org/10.3390/math13040557
Wang H, Feng T, Liang B. Improved Mixture Cure Model Using Machine Learning Approaches. Mathematics. 2025; 13(4):557. https://doi.org/10.3390/math13040557
Chicago/Turabian StyleWang, Huina, Tian Feng, and Baosheng Liang. 2025. "Improved Mixture Cure Model Using Machine Learning Approaches" Mathematics 13, no. 4: 557. https://doi.org/10.3390/math13040557
APA StyleWang, H., Feng, T., & Liang, B. (2025). Improved Mixture Cure Model Using Machine Learning Approaches. Mathematics, 13(4), 557. https://doi.org/10.3390/math13040557






