All for One or One for All? A Comparative Study of Grouped Data in Mixed-Effects Additive Bayesian Networks
Abstract
1. Introduction
2. Materials and Methods
2.1. Mixed-Effect Additive Bayesian Networks
2.1.1. Modeling Grouped Data with Mixed-Effect Additive Bayesian Networks
2.1.2. Additive Bayesian Networks for Mixed Distribution Data
2.1.3. Model Complexity
2.1.4. Structure Learning and Parameter Estimation in Mixed-Effect ABNs
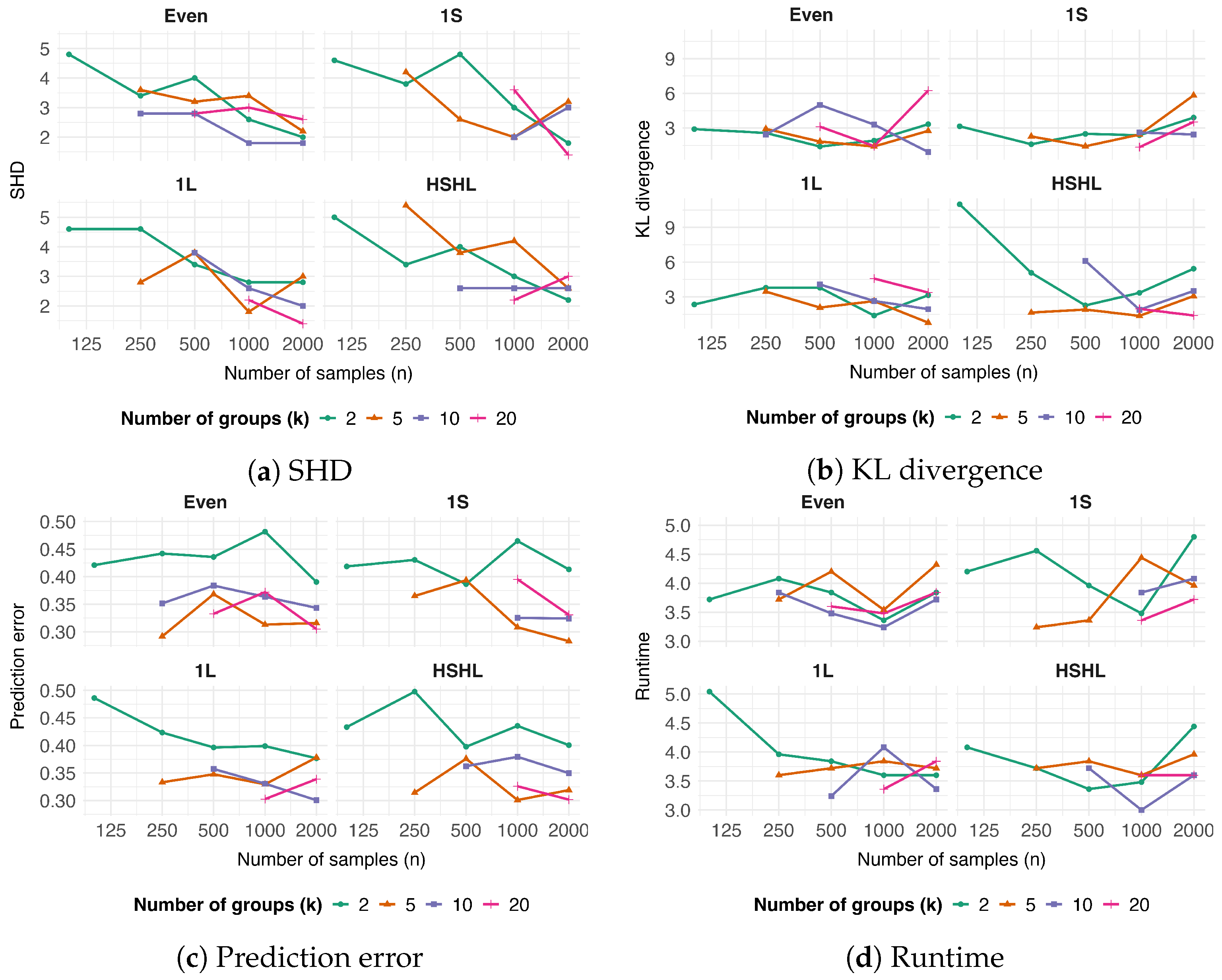
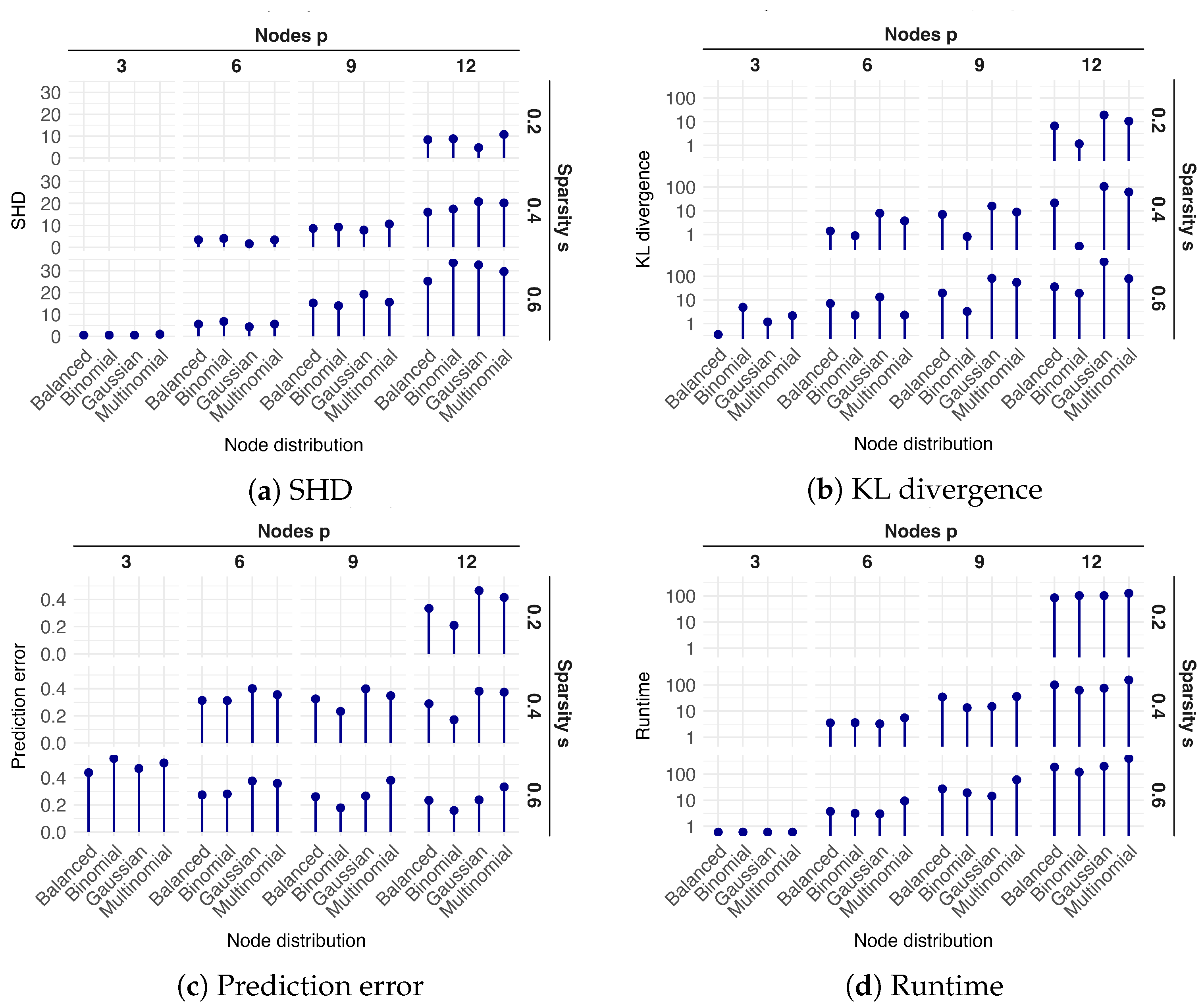
2.2. Simulation Study
2.2.1. DAG Generation
- Number of nodes ;
- Sparsity level , which defines the proportion of possible edges, yielding a number of edges.
2.2.2. Data Simulation
- Even: Equal group sizes ;
- : One small group of size and large groups, with the remaining observations distributed equally among them;
- : One large group of size and small groups, with the remaining observations distributed equally among them;
- : Half of the groups are small (size ) and the other half are large, with the remaining observations distributed equally among them.
- Balanced: Variables are equally distributed among Gaussian, binomial, multinomial;
- Gaussian: One binomial, one multinomial, with all others being Gaussian;
- Binomial: One Gaussian, one multinomial, with all others being binomial;
- Multinomial: One Gaussian, one binomial, with all others being multinomial.
2.2.3. Parameters of the Algorithm
2.2.4. Summary of Simulation Settings
2.3. Model Evaluation
- Structural accuracy, measured by the Structural Hamming Distance (SHD) between the learned DAG and the true data-generating DAG.
- Parametric accuracy, assessed using the Kullback–Leibler (KL) divergence between the learned and true conditional probability distributions for each node.
- Prediction error, evaluated using a 5-fold cross-validation procedure. To specifically isolate the accuracy of parameter estimation, a single consensus DAG was first learned from the full dataset. The model parameters were re-estimated on the training set, and the prediction error was calculated on the held-out test set. The error was then computed as the average of the differences between observed and predicted values across all nodes, more specifically:
- -
- For Gaussian distributed nodes , we use the Mean Squared Error (MSE):
- -
- For binomial distributed nodes , we use the Brier score, which measures the mean squared difference between the predicted probability and the observed binary outcome:
- -
- For multinomial distributed nodes , we use the multi-category Brier score:where and C is the number of categories.
The final prediction error is the average of these scores across all nodes and all five test folds, yielding a single predictive accuracy score for each simulation. - Computational efficiency, measured by the total runtime (in minutes) of the algorithm from structure learning to parameter estimation.
3. Results
3.1. Mixed-Effect Additive Bayesian Networks and Their Parameter Behavior
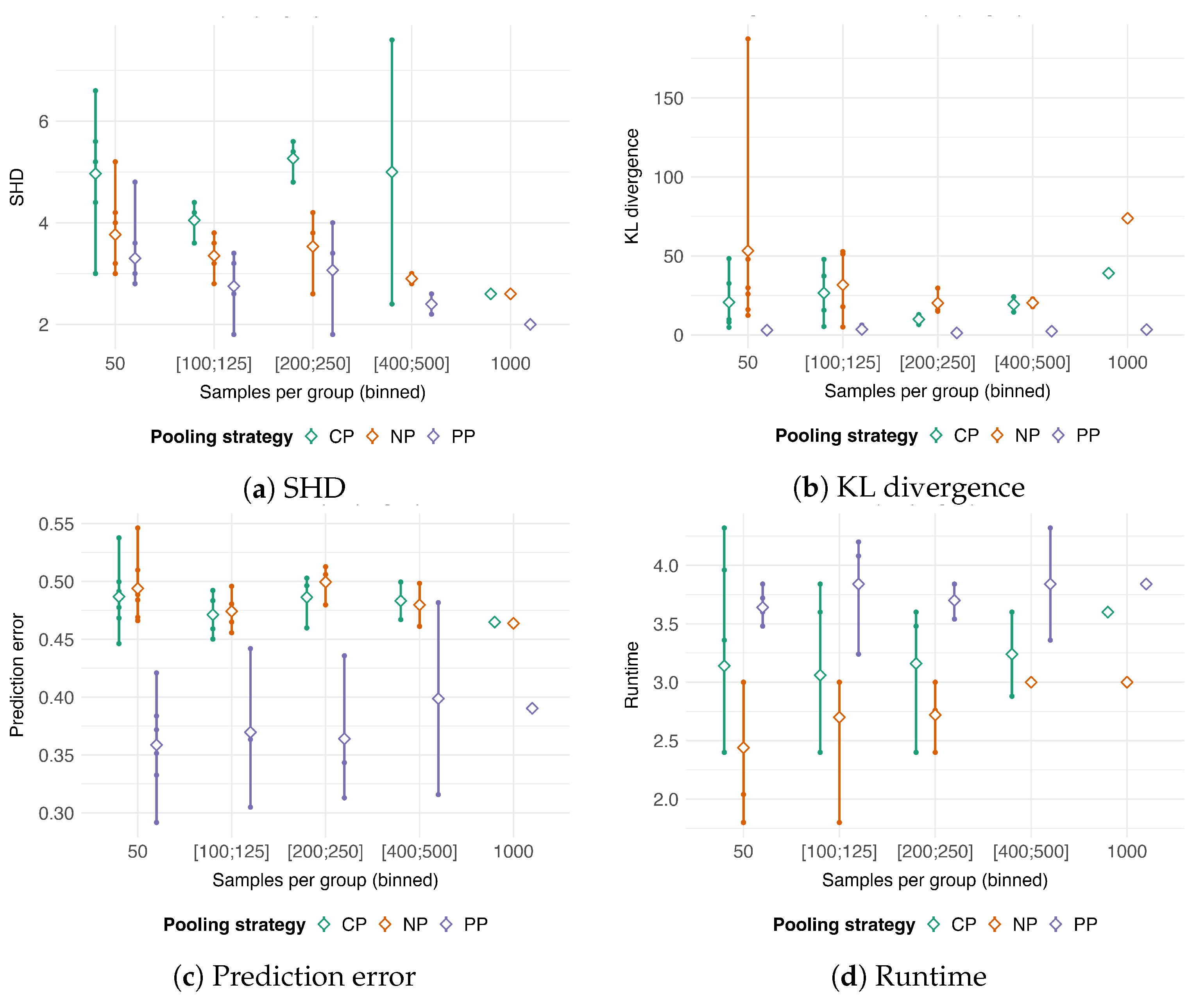
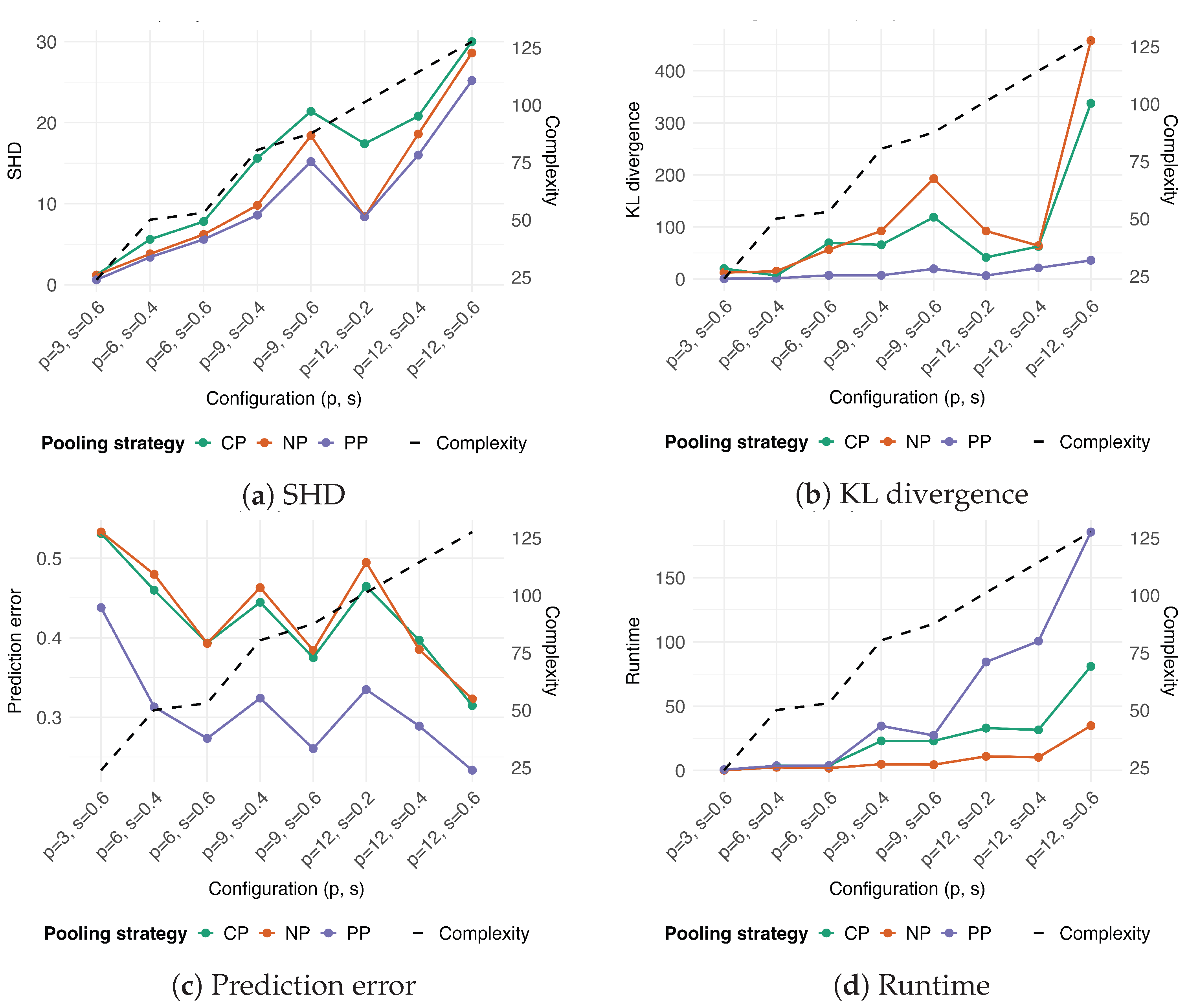
3.2. Comparison with Pooling Strategies
- 50, corresponding to the , , and settings.
- , corresponding to the , , and settings.
- , corresponding to the , and settings.
- , corresponding to the and .
- 1000, corresponding to the setting.
4. Discussion
4.1. Comparative Performance of Pooling Strategies
4.2. Limitations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABN | Additive Bayesian Network |
| GLM | Generalized Linear Model |
| BN | Bayesian Network |
| DAG | Directed Acyclic Graph |
| GLMM | Generalized Linear Mixed Model |
| BIC | Bayesian Information Criterion |
| SHD | Structural Hamming Distance |
| KL | Kullback–Leibler |
| MSE | Mean Squared Error |
| PP | Partial Pooling |
| NP | No Pooling |
| CP | Complete Pooling |
Appendix A. Empirical Validation of Search Constraint
| p | max.parents | SHD | KL | Prediction Error | Runtime (min) |
|---|---|---|---|---|---|
| 3 | 3.4 | 1.41 | 0.313 | 3.45 | |
| 4 | 3.4 | 1.41 | 0.314 | 3.24 | |
| 5 | 3.4 | 1.41 | 0.312 | 3.48 | |
| 3 | 7.6 | 6.62 | 0.327 | 38.0 | |
| 4 | 7.4 | 7.12 | 0.328 | 35.2 | |
| 5 | 6.8 | 5.72 | 0.329 | 44.2 | |
| 8 | 6.8 | 5.73 | 0.328 | 51.1 |
Appendix B. Numerical Stability in Poisson Modeling
| Nodes Distribution | Sparsity (s) | Number of Nodes (p) | |||
|---|---|---|---|---|---|
| 0.2 | - | - | 0.02 | 0.04 | |
| 0.4 | - | 0.02 | 0.04 | 0 | |
| 0.6 | 0.01 | 0.05 | 0.01 | 0 | |
| 0.2 | - | - | 0.02 | 0.02 | |
| 0.4 | - | 0.01 | 0.02 | 0.01 | |
| 0.6 | 0.01 | 0.02 | 0.03 | 0.02 | |
| 0.2 | - | - | 0.02 | 0.02 | |
| 0.4 | - | 0.03 | 0.04 | 0.04 | |
| 0.6 | 0.01 | 0.03 | 0.04 | 0.05 | |
| 0.2 | - | - | 0.04 | 0 | |
| 0.4 | - | 0.04 | 0 | 0 | |
| 0.6 | 0.01 | 0.02 | 0 | 0 | |
Appendix C. Disaggregated Prediction Error Results
| Node | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | Averaged |
|---|---|---|---|---|---|---|
| b1 | 0.111 | 0.097 | 0.075 | 0.087 | 0.088 | 0.092 |
| b2 | 0.132 | 0.148 | 0.143 | 0.146 | 0.146 | 0.143 |
| b3 | 0.179 | 0.146 | 0.156 | 0.157 | 0.166 | 0.161 |
| b4 | 0.207 | 0.205 | 0.182 | 0.177 | 0.210 | 0.196 |
| averaged_b | - | - | - | - | - | 0.148 |
| c1 | 0.106 | 0.142 | 0.125 | 0.115 | 0.111 | 0.120 |
| c2 | 0.147 | 0.136 | 0.134 | 0.152 | 0.160 | 0.147 |
| c3 | 0.102 | 0.112 | 0.087 | 0.108 | 0.129 | 0.108 |
| c4 | 0.121 | 0.142 | 0.147 | 0.112 | 0.138 | 0.132 |
| averaged_c | - | - | - | - | - | 0.126 |
| n1 | 0.116 | 0.100 | 0.142 | 0.123 | 0.102 | 0.116 |
| n2 | 0.096 | 0.072 | 0.086 | 0.081 | 0.065 | 0.079 |
| n3 | 0.086 | 0.096 | 0.089 | 0.101 | 0.080 | 0.091 |
| n4 | 0.068 | 0.084 | 0.091 | 0.084 | 0.071 | 0.079 |
| averaged_n | - | - | - | - | - | 0.086 |
References
- Spiegelhalter, D.; Abrams, K.; Myles, J. Bayesian Approaches to Clinical Trials and Health-Care Evaluation; Wiley: Hoboken, NJ, USA, 2004. [Google Scholar]
- Gelman, A.; Hill, J. Data Analysis Using Regression and Multilevel/Hierarchical Models; Analytical Methods for Social Research; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- McCulloch, C.; Searle, S.R.; Neuhaus, J. Generalized, Linear and Mixed Models, 2nd ed.; Wiley Series in Probability and Statistics; Wiley: Hoboken, NJ, USA, 2008. [Google Scholar]
- Scutari, M.; Marquis, C.; Azzimonti, L. Using Mixed-Effects Models to Learn Bayesian Networks from Related Data Sets. In Proceedings of the 11th International Conference on Probabilistic Graphical Models, Almeria, Spain, 5–7 October 2022; pp. 73–84. [Google Scholar]
- Delucchi, M.; Spinner, G.R.; Scutari, M.; Bijlenga, P.; Morel, S.; Friedrich, C.M.; Furrer, R.; Hirsch, S. Bayesian network analysis reveals the interplay of intracranial aneurysm rupture risk factors. Comput. Biol. Med. 2022, 147, 105740. [Google Scholar] [CrossRef] [PubMed]
- Hartnack, S.; Springer, S.; Pittavino, M.; Grimm, H. Attitudes of Austrian Veterinarians towards Euthanasia in Small Animal Practice: Impacts of Age and Gender on Views on Euthanasia. BMC Vet. Res. 2016, 12, 26. [Google Scholar] [CrossRef] [PubMed]
- McCormick, B.J.J.; Sanchez-Vazquez, M.J.; Lewis, F.I. Using Bayesian Networks to Explore the Role of Weather as a Potential Determinant of Disease in Pigs. Prev. Vet. Med. 2013, 110, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Guinat, C.; Comin, A.; Kratzer, G.; Durand, B.; Delesalle, L.; Delpont, M.; Guérin, J.L.; Paul, M.C. Biosecurity risk factors for highly pathogenic avian influenza (H5N8) virus infection in duck farms, France. Transbound. Emerg. Dis. 2020, 67, 2961–2970. [Google Scholar] [CrossRef] [PubMed]
- Aggrey, S.; Egeru, A.; Kalule, J.B.; Lukwa, A.T.; Mutai, N.; Hartnack, S. Household Satisfaction with Health Services and Response Strategies to Malaria in Mountain Communities of Uganda. Trans. R. Soc. Trop. Med. Hyg. 2025, 119, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, M.E.; Shukla, A.K.; Arteaga, K.; Crasta, M.; Dixon, C.; Famose, F.; Hartnack, S.; Pot, S.A. Evaluation of Risk Factors for Treatment Failure in Canine Patients Undergoing Photoactivated Chromophore for Keratitis—Corneal Cross-Linking (PACK-CXL): A Retrospective Study Using Additive Bayesian Network Analysis. BMC Vet. Res. 2023, 19, 227. [Google Scholar] [CrossRef] [PubMed]
- Kratzer, G.; Lewis, F.; Comin, A.; Pittavino, M.; Furrer, R. Additive Bayesian Network Modeling with the R Package abn. J. Stat. Softw. 2023, 105, 1–41. [Google Scholar] [CrossRef]
- Scutari, M. Learning Bayesian Networks with the bnlearn R Package. J. Stat. Softw. 2010, 35, 1–22. [Google Scholar] [CrossRef]
- Gelman, A.; Carlin, J.; Stern, H.; Dunson, D.; Vehtari, A.; Rubin, D. Bayesian Data Analysis, 3rd ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2013. [Google Scholar]
- Delucchi, M.; Bijlenga, P.; Morel, S.; Furrer, R.; Hostettler, I.; Bakker, M.; Bourcier, R.; Lindgren, A.; Maschke, S.; Bozinov, O.; et al. Mixed-effects Additive Bayesian Networks for the Assessment of Ruptured Intracranial Aneurysms: Insights from Multicenter Data. Comput. Biol. Med. 2025. submitted. [Google Scholar]
- Azzimonti, L.; Corani, G.; Scutari, M. A Bayesian hierarchical score for structure learning from related data sets. Int. J. Approx. Reason. 2022, 142, 248–265. [Google Scholar] [CrossRef]
- Koivisto, M.; Sood, K. Exact Bayesian Structure Discovery in Bayesian Networks. J. Mach. Learn. Res. 2004, 5, 549–573. [Google Scholar]
- Delucchi, M.; Liechti, J.; Spinner, G.; Furrer, R. Additive Bayesian Networks. J. Open Source Softw. 2024, 9, 6822. [Google Scholar] [CrossRef]
- Bolker, B.M.; Brooks, M.E.; Clark, C.J.; Geange, S.W.; Poulsen, J.R.; Stevens, M.H.H.; White, J.S.S. Generalized Linear Mixed Models: A Practical Guide for Ecology and Evolution. Trends Ecol. Evol. 2009, 24, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Lunn, D.; Spiegelhalter, D.; Thomas, A.; Best, N. The BUGS project: Evolution, critique and future directions. Stat. Med. 2009, 28, 3049–3067. [Google Scholar] [CrossRef] [PubMed]
- Plummer, M.; Best, N.; Cowles, K.; Vines, K. CODA: Convergence Diagnosis and Output Analysis for MCMC. R News 2006, 6, 7–11. [Google Scholar]
- Sagan, A. Sample Size in Multilevel Structural Equation Modeling—The Monte Carlo Approach. Econometrics 2019, 23, 63–79. [Google Scholar] [CrossRef]
- Maas, C.; Hox, J. Sufficient sample sizes for multilevel modeling. Methodol. Eur. J. Res. Methods Behav. Soc. Sci. 2005, 1, 86–92. [Google Scholar] [CrossRef]
| Sample Size (n) | ||||||
|---|---|---|---|---|---|---|
| Groups () | 100 | 250 | 500 | 1000 | 2000 | |
| 2 | 50 | 125 | 250 | 500 | 1000 | |
| 5 | 20 | 50 | 100 | 200 | 400 | |
| 10 | 10 | 25 | 50 | 100 | 200 | |
| 20 | 5 | 13 | 25 | 50 | 100 | |
| 2 | (25, 75) | (62, 188) | (125, 375) | (250, 750) | (500, 1500) | |
| 5 | (10, 23) | (25, 56) | (50, 113) | (100, 225) | (200, 450) | |
| 10 | (5, 11) | (13, 26) | (24, 53) | (50, 106) | (100, 211) | |
| 20 | (3, 5) | (6, 13) | (13, 26) | (25, 51) | (50, 103) | |
| 2 | (25, 75) | (62, 188) | (125, 375) | (250, 750) | (500, 1500) | |
| 5 | (18, 30) | (44, 75) | (88, 150) | (175, 300) | (350, 600) | |
| 10 | (9, 15) | (24, 38) | (47, 75) | (94, 150) | (189, 300) | |
| 20 | (5,8) | (12, 19) | (24, 38) | (49, 75) | (97, 150) | |
| 2 | (25, 75) | (62, 188) | (125, 375) | (250, 750) | (500, 1500) | |
| 5 | (13, 25) | (31, 63) | (63, 125) | (125, 250) | (250, 500) | |
| 10 | (5, 15) | (13, 38) | (25, 75) | (50, 150) | (100, 300) | |
| 20 | (3, 8) | (6, 19) | (13, 38) | (25, 75) | (50, 150) | |
| Number of Nodes (p) | ||||
|---|---|---|---|---|
| Sparsity () | 3 | 6 | 9 | 12 |
| Average edges | ||||
| 0.2 | 1 | 3 | 7 | 13 |
| 0.4 | 1 | 6 | 14 | 26 |
| 0.6 | 2 | 9 | 22 | 40 |
| max.parents | ||||
| 0.2 | - | - | - | 3 |
| 0.4 | - | 3 | 3 | 3 |
| 0.6 | 2 | 3 | 3 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Champion, M.; Delucchi, M.; Furrer, R. All for One or One for All? A Comparative Study of Grouped Data in Mixed-Effects Additive Bayesian Networks. Mathematics 2025, 13, 3649. https://doi.org/10.3390/math13223649
Champion M, Delucchi M, Furrer R. All for One or One for All? A Comparative Study of Grouped Data in Mixed-Effects Additive Bayesian Networks. Mathematics. 2025; 13(22):3649. https://doi.org/10.3390/math13223649
Chicago/Turabian StyleChampion, Magali, Matteo Delucchi, and Reinhard Furrer. 2025. "All for One or One for All? A Comparative Study of Grouped Data in Mixed-Effects Additive Bayesian Networks" Mathematics 13, no. 22: 3649. https://doi.org/10.3390/math13223649
APA StyleChampion, M., Delucchi, M., & Furrer, R. (2025). All for One or One for All? A Comparative Study of Grouped Data in Mixed-Effects Additive Bayesian Networks. Mathematics, 13(22), 3649. https://doi.org/10.3390/math13223649






