De Novo Single-Cell Biological Analysis of Drug Resistance in Human Melanoma Through a Novel Deep Learning-Powered Approach
Abstract
1. Introduction
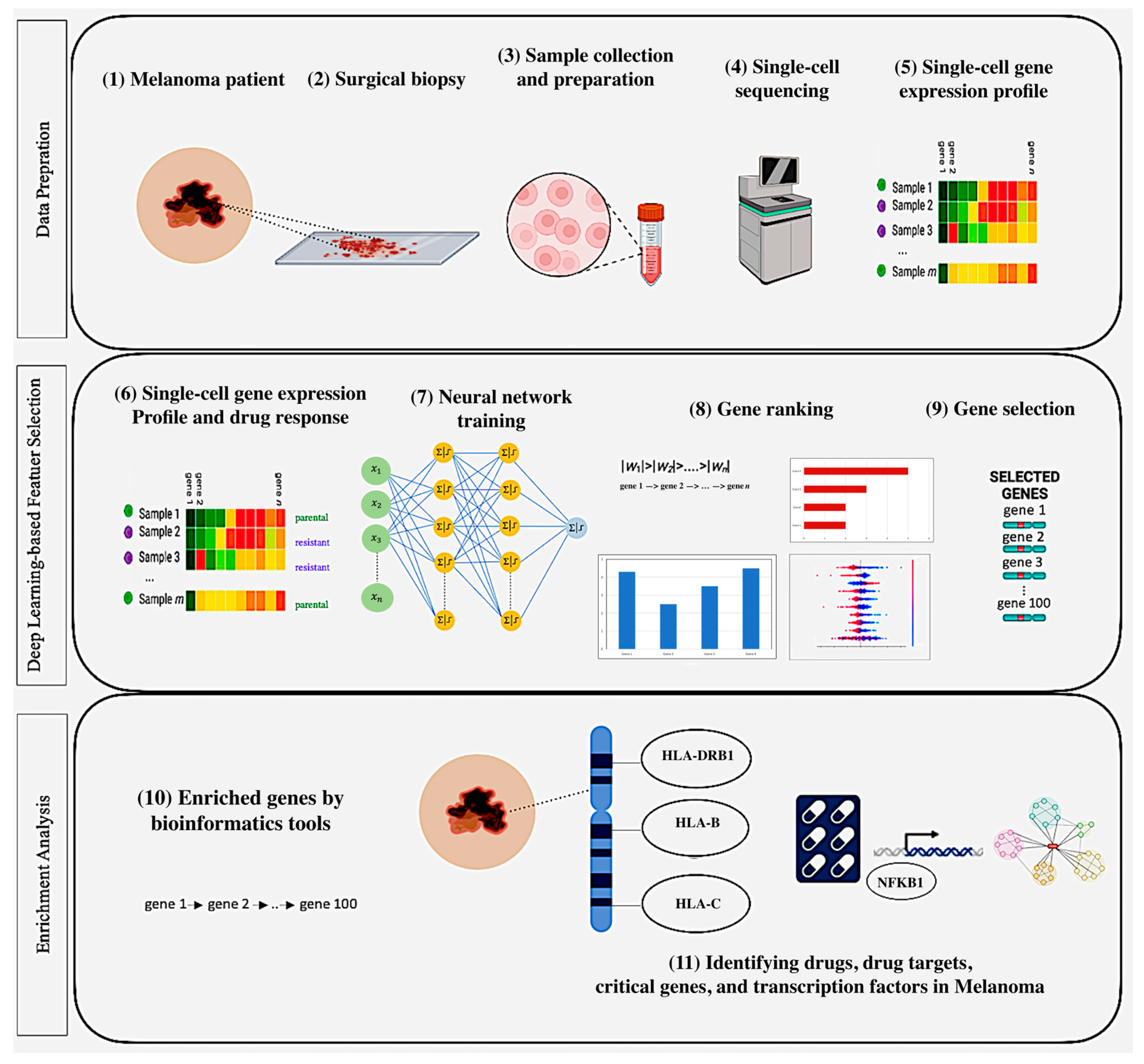
- DL-powered computational approach: We introduce a DL-based approach for gene selection with enrichment analysis to analyze single-cell gene expression data, unveiling the mechanisms underlying melanoma drug response, critical genes, drugs, therapeutic targets, and other critical factors such as key pathways and regulators associated with melanoma treatment outcomes.
- Model training: We downloaded and processed single-cell gene expression datasets related to melanoma treatment response from the GEO database under accession numbers GSE108383_A375 and GSE108383_451Lu, corresponding to the A375 and 451Lu cell lines, respectively. Then, a fully connected deep neural network was trained to learn complex non-linear relationships in the single-cell gene expression data and implicitly reduce dimensionality, enabling more accurate identification of crucial genes for predicting drug response.
- Gene ranking and selection: Adapting DL-based gene selection methods—including L1-Regularization, DeepLIFT, SHAP, Integrated Gradients (IG), and Layer-wise Relevance Propagation (LRP)—to rank genes based on their importance to the model predictions. Subsequently, we selected the top-ranked genes determined by each method for further enrichment analysis, including using Enrichr and Metascape to uncover the underlying biological pathways and processes associated with drug response and resistance in melanoma.
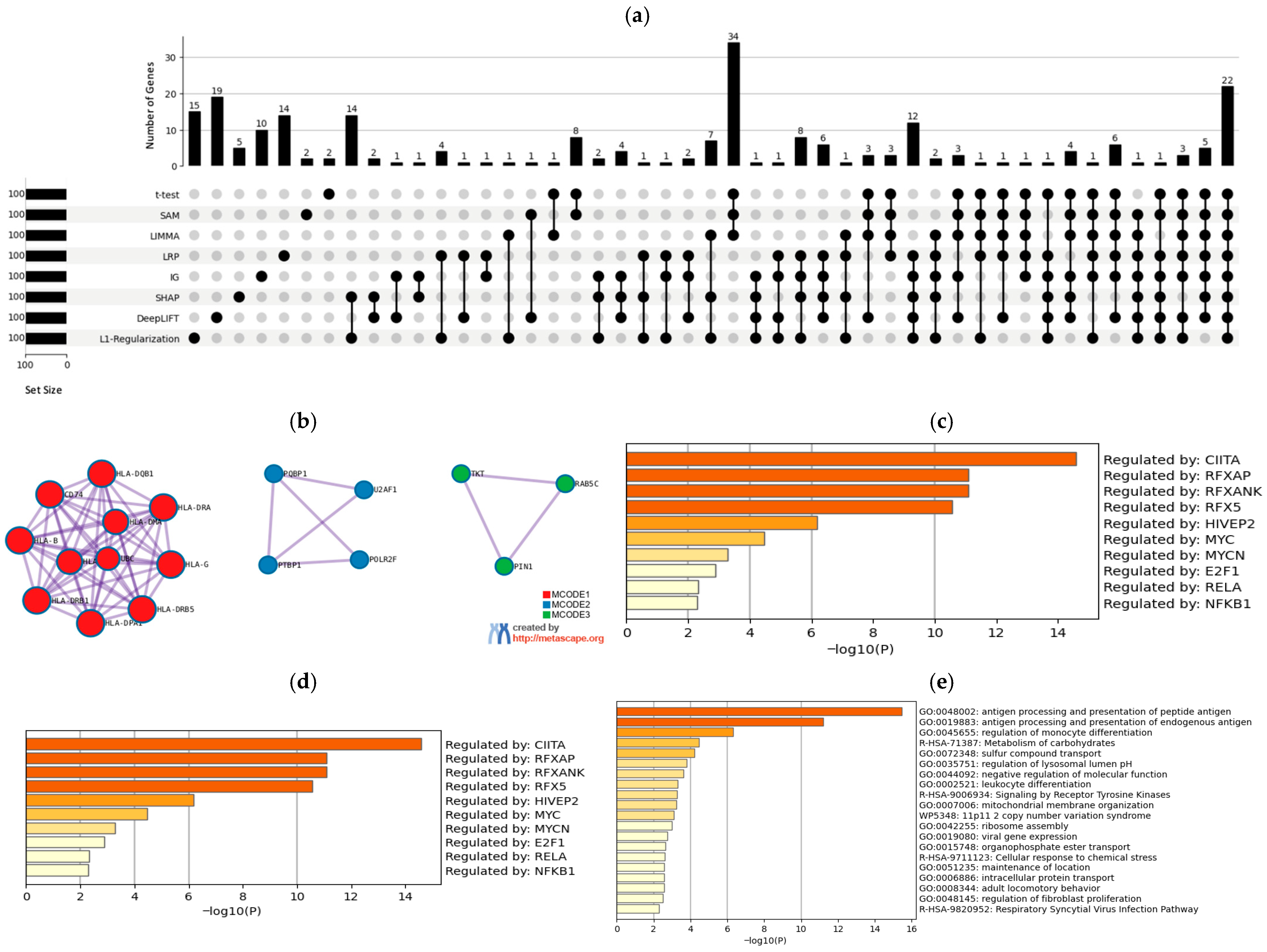
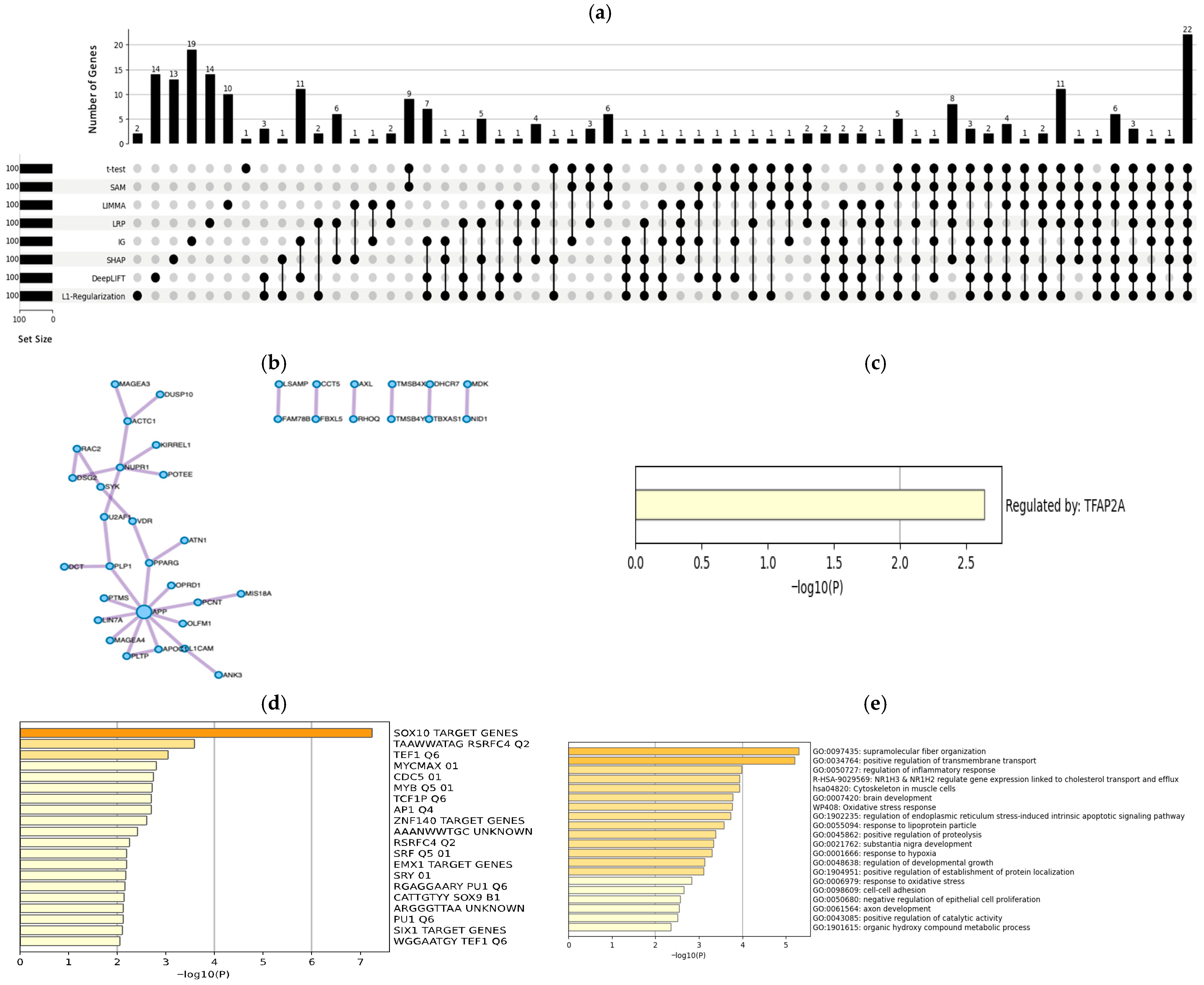
- Results from a biological perspective: Our developed DL-based approach demonstrated superior performance when compared to baseline methods across the two datasets. Notably, DeepLIFT and IG produced the highest performance on the first dataset, whereas both DeepLIFT and IG were most effective for the second, with higher numbers of expressed genes in established cell lines (i.e., MALME-3M, MDA-MB435, SKMEL28, and SKMEL5). Additionally, our methods successfully identified several drugs, including FDA-approved melanoma treatments like Vemurafenib and Dabrafenib, significant genes (e.g., ARAF, SOX10, SLC7A5, DCT, and AXL), and TFs (such as MITF, RELA, E2F1, and TFAP2A).
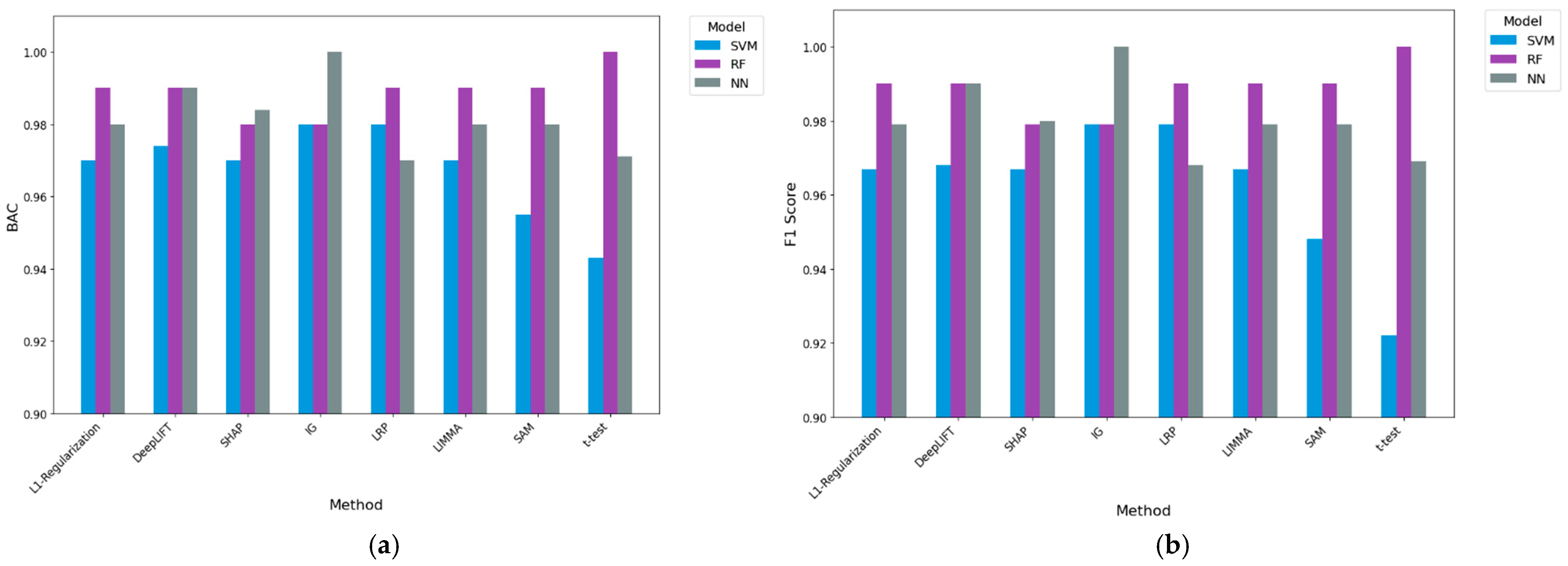
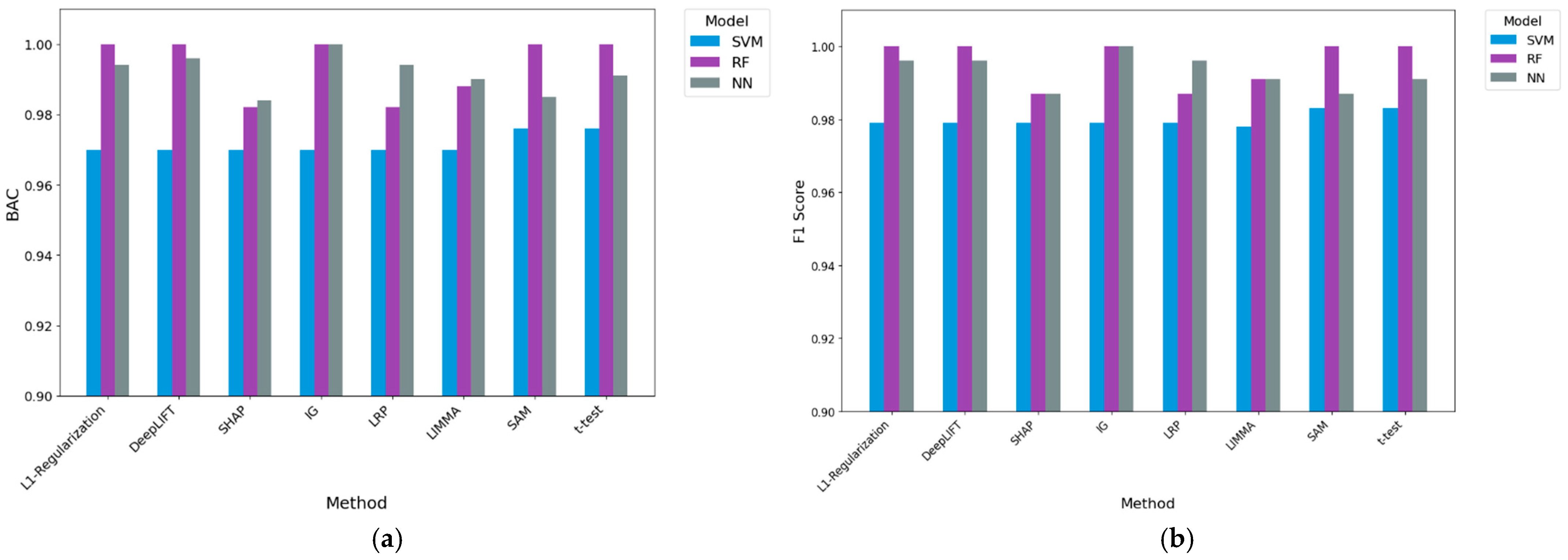
- Results from a classification perspective: Providing the gene sets delivered by our DL-based methods and existing baseline methods to three machine learning algorithms, we evaluated using the average balanced accuracy (BAC) and F1. Compared to the best-performing baseline (LIMMA) in the A375 cell line dataset, IG coupled with machine learning algorithms had performance improvements of 2.2% and 2.0% in F1 and BAC, respectively. For the 451Lu cell line dataset, IG coupled with machine learning algorithms resulted in 0.5% and 0.3% performance improvements when employing F1 and BAC performance measures, respectively, over the best-performing baseline (t-test). Further experimental work against no gene selection demonstrated that IG was the only method to generate statistically significant results, with 14.4% and 11.7% overall performance improvements. These results demonstrate the predictive potential of genes produced via IG in our approach.
2. Materials and Methods
2.1. Single-Cell Gene Expression Profiles
2.2. Computational Approach
2.2.1. Data Preparation
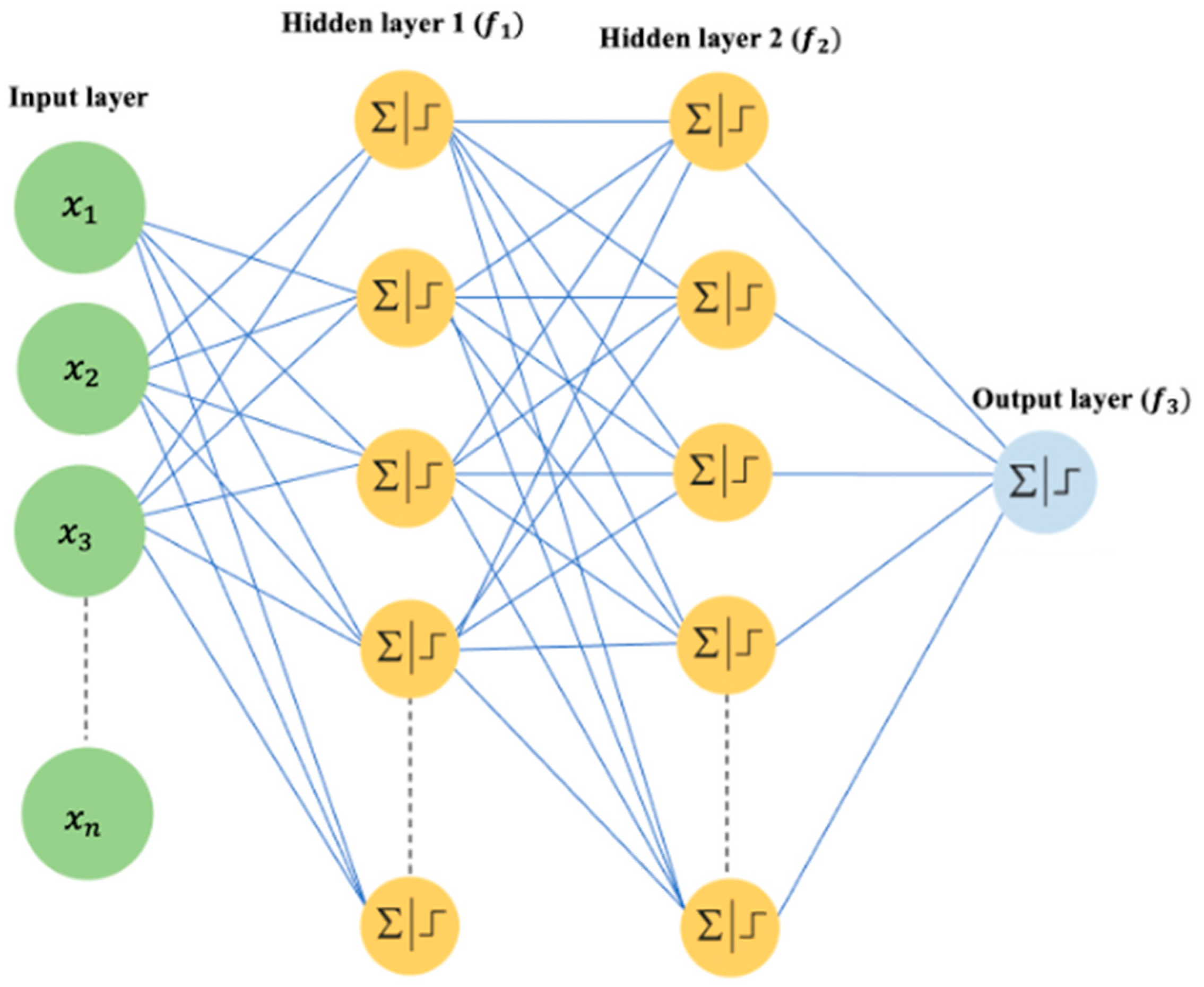
2.2.2. Deep Learning-Based Gene Selection
Neural Network Model
L1-Regularization for Gene Selection
Deep Learning Important Features (DeepLIFT)
SHapley Additive exPlanations (SHAP)
Integrated Gradients (IG)
Layer-Wise Relevance Propagation (LRP)
2.2.3. Enrichment Analysis
3. Experiments and Results
3.1. Experimental Methodology
3.2. Biological Results
3.2.1. A375 Cell Line
3.2.2. 451Lu Cell Line
3.3. Classification Results
4. Discussion
5. Conclusions and Future Work
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tawbi, H.; Nimmagadda, N. Targeted therapy in melanoma. Biol. Targets Ther. 2009, 3, 475–484. [Google Scholar] [CrossRef]
- Su, Y.; Wei, W.; Robert, L.; Xue, M.; Tsoi, J.; Garcia-Diaz, A.; Moreno, B.H.; Kim, J.; Ng, R.H.; Lee, J.W.; et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc. Natl. Acad. Sci. USA 2017, 114, 13679–13684. [Google Scholar] [CrossRef]
- Ho, Y.-J.; Anaparthy, N.; Molik, D.; Mathew, G.; Aicher, T.; Patel, A.; Hicks, J.; Hammell, M.G. Single-cell RNA-seq analysis identifies markers of resistance to targeted BRAF inhibitors in melanoma cell populations. Genome Res. 2018, 28, 1353–1363. [Google Scholar] [CrossRef]
- Schmidt, M.; Mortensen, L.S.; Loeffler-Wirth, H.; Kosnopfel, C.; Krohn, K.; Binder, H.; Kunz, M. Single-cell trajectories of melanoma cell resistance to targeted treatment. Cancer Biol. Med. 2022, 19, 56. [Google Scholar] [CrossRef]
- Zhang, C.; Shen, H.; Yang, T.; Li, T.; Liu, X.; Wang, J.; Liao, Z.; Wei, J.; Lu, J.; Liu, H.; et al. A single-cell analysis reveals tumor heterogeneity and immune environment of acral melanoma. Nat. Commun. 2022, 13, 7250. [Google Scholar] [CrossRef]
- Egan, D.; Kreileder, M.; Nabhan, M.; Iglesias-Martinez, L.F.; Dovedi, S.J.; Valge-Archer, V.; Grover, A.; Wilkinson, R.J.; Slidel, T.; Bendtsen, C.; et al. Small gene networks can delineate immune cell states and characterize immunotherapy response in melanoma. bioRxiv 2022, 11, 1125–1136. [Google Scholar] [CrossRef]
- Li, J.; Smalley, I.; Chen, Z.; Wu, J.-Y.; Phadke, M.S.; Teer, J.K.; Nguyen, T.; Karreth, F.A.; Koomen, J.M.; Sarnaik, A.A.; et al. Single-cell Characterization of the Cellular Landscape of Acral Melanoma Identifies Novel Targets for Immunotherapy. Clin. Cancer Res. 2022, 28, 2131–2146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, D.; Wang, F.; Liu, J.; Jiang, X.; Anuchapreeda, S.; Tima, S.; Xiao, Z.; Duangmano, S. CD3ζ as a novel predictive biomarker of PD-1 inhibitor resistance in melanoma. Mol. Cell. Probes 2023, 72, 101925. [Google Scholar] [CrossRef] [PubMed]
- Bakr, M.N.; Takahashi, H.; Kikuchi, Y. CHRNA1 and its correlated-myogenesis/cell cycle genes are prognosis-related markers of metastatic melanoma. Biochem. Biophys. Rep. 2023, 33, 101425. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, W.; Zhou, H.; Lai, W.; Hu, C.; Dai, Y.; Li, G.; Zhang, R.; Zhao, Y. Tozasertib activates anti-tumor immunity through decreasing regulatory T cells in melanoma. Neoplasia 2024, 48, 100966. [Google Scholar] [CrossRef]
- Chen, S.; Tang, Z.; Wan, Q.; Huang, W.; Li, X.; Huang, X.; Zheng, S.; Lu, C.; Wu, J.; Li, Z.; et al. Machine learning and single-cell RNA sequencing reveal relationship between intratumor CD8+ T cells and uveal melanoma metastasis. Cancer Cell Int. 2024, 24, 359. [Google Scholar] [CrossRef]
- Pinhasi, A.; Yizhak, K. Uncovering gene and cellular signatures of immune checkpoint response via machine learning and single-cell RNA-seq. NPJ Precis. Oncol. 2025, 9, 95. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. GEO Accession Viewer: SAKE (Single-Cell RNA-Seq Analysis and Klustering Evaluation) Identifies Markers of Resistance to Targeted BRAF Inhibitors in Melanoma Cell Populations [Fluidigm scRNA-Seq]. National Library of Medicine. 2018. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108383 (accessed on 10 September 2024).
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep sparse rectifier neural networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; JMLR Workshop and Conference Proceedings. pp. 315–323. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Shajalal, M.; Boden, A.; Stevens, G. ForecastExplainer: Explainable household energy demand forecasting by approximating shapley values using DeepLIFT. Technol. Forecast. Soc. Change 2024, 206, 123588. [Google Scholar] [CrossRef]
- Rynazal, R.; Fujisawa, K.; Shiroma, H.; Salim, F.; Mizutani, S.; Shiba, S.; Yachida, S.; Yamada, T. Leveraging explainable AI for gut microbiome-based colorectal cancer classification. Genome Biol. 2023, 24, 21. [Google Scholar] [CrossRef]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 3319–3328. [Google Scholar]
- Bach, S.; Binder, A.; Montavon, G.; Klauschen, F.; Müller, K.R.; Samek, W. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 2015, 10, e0130140. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef]
- Team, R. C.; Team, M. R. C; Suggests, M.A.S.S.; Matrix, S. Package Stats. The R Stats Package. 2018, pp. 1–3. Available online: https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html (accessed on 10 October 2024).
- Python Software Foundation. Python Language Reference, Version 3.x. 2023. Available online: https://www.python.org. (accessed on 5 October 2024).
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32, 8026–8037. [Google Scholar]
- Kokhlikyan, N.; Miglani, V.; Martin, M.; Wang, E.; Alsallakh, B.; Reynolds, J.; Melnikov, A.; Kliushkina, N.; Araya, C.; Yan, S.; et al. Captum: A unified and generic model interpretability library for PyTorch. arXiv 2020, arXiv:2009.07896. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Contributors. SHAP (SHapley Additive exPlanations) [Python Library]. GitHub. 2017. Available online: https://github.com/slundberg/shap (accessed on 10 October 2024).
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2008; Volume 25. [Google Scholar]
- Schwender, H. Siggenes: Multiple Testing Using SAM and Efron’s Empirical Bayes Approaches; R Package Version 1; R Core Team: Vienna, Austria, 2012; pp. 1–70. [Google Scholar]
- Martins, I.; Deshayes, F.; Baton, F.; Forget, A.; Ciechomska, I.; Sylla, K.; Aoudjit, F.; Charron, D.; Al-Daccak, R.; Alcaide-Loridan, C. Pathologic expression of MHC class II is driven by mitogen-activated protein kinases. Eur. J. Immunol. 2007, 37, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Singleton, K.R.; Crawford, L.; Tsui, E.; Manchester, H.E.; Maertens, O.; Liu, X.; Liberti, M.V.; Magpusao, A.N.; Stein, E.M.; Tingley, J.P.; et al. Melanoma therapeutic strategies that select against resistance by exploiting MYC-driven evolutionary convergence. Cell Rep. 2017, 21, 2796–2812. [Google Scholar] [CrossRef] [PubMed]
- Bucci, B.; D’AGnano, I.; Amendola, D.; Citti, A.; Raza, G.H.; Miceli, R.; De Paula, U.; Marchese, R.; Albini, S.; Felsani, A.; et al. Myc down-regulation sensitizes melanoma cells to radiotherapy by inhibiting MLH1 and MSH2 mismatch repair proteins. Clin. Cancer Res. 2005, 11, 2756–2767. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, D.; Lu, Y.; Tan, L. E2F transcription factor 1 as a potential prognostic biomarker and promotes tumor proliferation in skin cutaneous melanoma. Pathol. Res. Pract. 2025, 269, 155875. [Google Scholar] [CrossRef]
- Sheen, Y.; Syu, Y.; Chang, Y.; Hsieh, P.; Liao, Y.; Lin, M.; Chen, C.; Chu, C. Insulin-like growth factor 2 mRNA—binding protein 3 enhanced melanoma migration through regulation of AKT1 and RELA expression. Exp. Dermatol. 2024, 33, e15015. [Google Scholar] [CrossRef]
- Ueda, Y.; Richmond, A. NF-κB activation in melanoma. Pigment. Cell Res. 2006, 19, 112–124. [Google Scholar] [CrossRef]
- Capparelli, C.; Purwin, T.J.; Glasheen, M.; Caksa, S.; Tiago, M.; Wilski, N.; Pomante, D.; Rosenbaum, S.; Nguyen, M.Q.; Cai, W.; et al. Targeting SOX10-deficient cells to reduce the dormant-invasive phenotype state in melanoma. Nat. Commun. 2022, 13, 1381. [Google Scholar] [CrossRef]
- Zhou, S.; Li, P.; Qin, L.; Huang, S.; Dang, N. Transcription factor YY1 contributes to human melanoma cell growth through modulating the p53 signalling pathway. Exp. Dermatol. 2022, 31, 1563–1578. [Google Scholar] [CrossRef] [PubMed]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer immune evasion through loss of MHC class I antigen presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Chang, T.; Zhang, T.; Gu, H.; Wang, J. Genomic analyses identify signaling pathways and biological processes in small and large melanoma. Res. Sq. Prepr. 2022. [Google Scholar] [CrossRef]
- Oliver, J.; Onieva, J.L.; Garrido-Barros, M.; Berciano-Guerrero, M.; Sánchez-Muñoz, A.; Lozano, M.J.; Farngren, A.; Álvarez, M.; Martínez-Gálvez, B.; Pérez-Ruiz, E.; et al. Association of circular RNA and long non-coding RNA dysregulation with the clinical response to immune checkpoint blockade in cutaneous metastatic melanoma. Biomedicines 2022, 10, 2419. [Google Scholar] [CrossRef]
- Wolf, J.; Boneva, S.; Rosmus, D.-D.; Agostini, H.; Schlunck, G.; Wieghofer, P.; Schlecht, A.; Lange, C. Deciphering the molecular signature of human hyalocytes in relation to other innate immune cell populations. Investig. Ophthalmol. Vis. Sci. 2022, 63, 9. [Google Scholar] [CrossRef]
- Nazarian, R.; Shi, H.; Wang, Q.; Kong, X.; Koya, R.C.; Lee, H.; Chen, Z.; Lee, M.-K.; Attar, N.; Sazegar, H.; et al. Melanomas acquire resistance to B-RAF (V600E) inhibition by RTK or N-RAS upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Cohen, M.S. The discovery of vemurafenib for the treatment of BRAF-mutated metastatic melanoma. Expert Opin. Drug Discov. 2016, 11, 907–916. [Google Scholar] [CrossRef]
- Kim, G.; McKee, A.E.; Ning, Y.-M.; Hazarika, M.; Theoret, M.; Johnson, J.R.; Xu, Q.C.; Tang, S.; Sridhara, R.; Jiang, X.; et al. DA approval summary: Vemurafenib for treatment of unresectable or metastatic melanoma with the BRAFV600E mutation. Clin. Cancer Res. 2014, 20, 4994–5000. [Google Scholar] [CrossRef]
- Robert, C.; Grob, J.J.; Stroyakovskiy, D.; Karaszewska, B.; Hauschild, A.; Levchenko, E.; Chiarion Sileni, V.; Schachter, J.; Garbe, C.; Bondarenko, I.; et al. Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N. Engl. J. Med. 2019, 381, 626–636. [Google Scholar] [CrossRef]
- Liu, L.; Cao, Y.; Chen, C.; Zhang, X.; McNabola, A.; Wilkie, D.; Wilhelm, S.; Lynch, M.; Carter, C. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006, 66, 11851–11858. [Google Scholar] [CrossRef]
- Shi, Z.; Kaneda-Nakashima, K.; Ohgaki, R.; Xu, M.; Okanishi, H.; Endou, H.; Nagamori, S.; Kanai, Y. Inhibition of cancer-type amino acid transporter LAT1 suppresses B16-F10 melanoma metastasis in mouse models. Sci. Rep. 2023, 13, 13943. [Google Scholar] [CrossRef]
- Augustin, R.C.; Huang, Z.; Ding, F.; Zhai, S.; McArdle, J.; Santisi, A.; Davis, M.; Sander, C.; Davar, D.; Kirkwood, J.M.; et al. Metformin is associated with improved clinical outcomes in patients with melanoma: A retrospective, multi-institutional study. Front. Oncol. 2023, 13, 1075823. [Google Scholar] [CrossRef] [PubMed]
- Seberg, H.E.; Van Otterloo, E.; Cornell, R.A. Beyond MITF: Multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment. Cell Melanoma Res. 2017, 30, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Han, W.; Shen, Y.; Shen, G. Comprehensive analysis of aberrantly methylated differentially expressed genes and validation of CDC6 in melanoma. J. Cancer Res. Clin. Oncol. 2024, 150, 362. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, S.; Cheng, S.; Gajendran, N.; Shekoohi, S.; Chesnokova, L.; Yu, X.; Witt, S.N. Transcriptomic analysis of melanoma cells reveals an association of α-synuclein with regulation of the inflammatory response. Sci. Rep. 2024, 14, 27140. [Google Scholar] [CrossRef]
- Goff, P.H.; Baker, K.K.; Lee, S.M.; Tykodi, S.S.; Bhatia, S.; Zeng, J.; Redman, M.; Rengan, R. 610 ImmunoRad: A stratified phase II trial of image guided hypofractionated radiotherapy with concurrent nelfinavir & PD-1 inhibition in advanced melanoma, lung cancer & renal cell carcinoma. J. Immunother. Cancer 2023, 11. [Google Scholar] [CrossRef]
- Kim, K.B.; Eton, O.; Davis, D.W.; Frazier, M.L.; McConkey, D.J.; Diwan, A.H.; Papadopoulos, N.E.; Bedikian, A.Y.; Camacho, L.H.; Ross, M.I.; et al. Phase II trial of imatinib mesylate in patients with metastatic melanoma. Br. J. Cancer 2008, 99, 734–740. [Google Scholar] [CrossRef]
- Carvajal, R.D.; Antonescu, C.R.; Wolchok, J.D.; Chapman, P.B.; Roman, R.A.; Teitcher, J.; Panageas, K.S.; Busam, K.J.; Chmielowski, B.; Lutzky, J.; et al. KIT as a therapeutic target in metastatic melanoma. JAMA 2011, 305, 2327–2334. [Google Scholar] [CrossRef]
- Daud, A.; Kluger, H.M.; Kurzrock, R.; Schimmoller, F.; Weitzman, A.L.; Samuel, T.A.; Moussa, A.H.; Gordon, M.S.; Shapiro, G.I. Phase II randomised discontinuation trial of the MET/VEGF receptor inhibitor cabozantinib in metastatic melanoma. Br. J. Cancer 2017, 116, 432–440. [Google Scholar] [CrossRef]
- Moro-Sibilot, D.; Cozic, N.; Pérol, M.; Mazières, J.; Otto, J.; Souquet, P.; Bahleda, R.; Wislez, M.; Zalcman, G.; Guibert, S.; et al. Crizotinib in c-MET-or ROS1-positive NSCLC: Results of the AcSé phase II trial. Ann. Oncol. 2019, 30, 1985–1991. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Zhu, Y.; Zhang, L.; Hou, W. Gefitinib inhibits malignant melanoma cells through the VEGF/AKT signaling pathway. Mol. Med. Rep. 2018, 17, 7351–7355. [Google Scholar] [CrossRef] [PubMed]
- ZZhang, X.; Fang, X.; Gao, Z.; Chen, W.; Tao, F.; Cai, P.; Yuan, H.; Shu, Y.; Xu, Q.; Sun, Y.; et al. Axitinib, a selective inhibitor of vascular endothelial growth factor receptor, exerts an anticancer effect in melanoma through promoting antitumor immunity. Anti-Cancer Drugs 2014, 25, 204–211. [Google Scholar] [CrossRef]
- Skoko, J.; Rožanc, J.; Charles, E.M.; Alexopoulos, L.G.; Rehm, M. Post-treatment de-phosphorylation of p53 correlates with dasatinib responsiveness in malignant melanoma. BMC Cell Biol. 2018, 19, 28. [Google Scholar] [CrossRef] [PubMed]
| Ref. | Year | Objective | Data Source | Main Findings |
|---|---|---|---|---|
| [3] | 2018 | Identifying resistance markers to BRAF inhibitors in melanoma. | scRNA-seq | Known markers (DCT, AXL, NRG1). |
| [4] | 2022 | Analyzing resistance in BRAF V600E melanoma cells. | scRNA-seq | KDM5B as marker; distinct sensitivity/resistance states. |
| [5] | 2022 | Investigate tumor heterogeneity and immune environment in AM and CM. | scRNA-seq | Five cell clusters (e.g., TGF-β, cell cycle) associated with good melanoma prognosis. |
| [6] | 2022 | Characterizing immune cell states and predicting responses to ICIs. | scRNA-seq | Four immune states; patients clustering into response groups. |
| [7] | 2022 | Characterizing the cellular and immune landscape in AM. | scRNA-seq | AM has fewer immune cells; immune checkpoints (PD-1) as therapeutic targets. |
| [8] | 2023 | Identifying biomarkers predicting resistance to PD-1 inhibitors. | scRNA-seq | CD3ζ predicts PD-1 resistance. |
| [9] | 2023 | Investigating CHRNA1’s role in melanoma metastasis. | scRNA-seq/ bulk | CHRNA1 is highly expressed in metastasis. |
| [10] | 2024 | Assessing the impact of AURKB inhibition on melanoma immune response. | scRNA-seq/ bulk | AURKB inhibition enhances anti-tumor immunity; Tozasertib reduces tumor growth. |
| [11] | 2024 | Identifying prognostic genes for UM metastasis using ML. | scRNA-seq/ bulk | Three prognostic genes identified (SLC25A38, EDNRB, LURAP1). |
| [12] | 2025 | Enhancing the prediction of treatment outcomes in melanoma using ML. | scRNA-seq | Eleven-gene signature (e.g., GAPDH, STAT1); |
| Proposed | 2025 | Unveiling various molecular mechanisms underlying melanoma drug resistance; performance validation from classification and biological perspectives against baseline methods. | scRNA-seq | -Adapting five DL-based methods: L1-Regularization, DeepLIFT, SHAP, IG, LRP. -Biological Perspective: More expressed genes, approved FDA drugs (Vemurafenib and Dabrafenib), TFs such as MITF and TFAP2A. -Classification Results: 2.2% and 0.5% performance improvements over baseline methods |
| Dataset | Cell Line | Genes | Samples | Parental | Resistant | Platform | Organism | Experiment Type |
|---|---|---|---|---|---|---|---|---|
| GSE108383 | A375 | 27,001 | 130 | 79 | 51 | GPL18573 | Homo sapiens | Expression profiling via high-throughput sequencing |
| GSE108383 | 451Lu | 27,001 | 197 | 84 | 113 | GPL18573 | Homo sapiens | Expression profiling via high-throughput sequencing |
| Method | Rank | Term | Overlap | p-Value | Adjusted p-Value |
|---|---|---|---|---|---|
| L1-Regularization | 6 | MALME-3M | 5/366 | 3.67 × 10−2 | 4.90 × 10−1 |
| 3 | MDA-MB435 | 8/621 | 1.26 × 10−2 | 3.37 × 10−1 | |
| 31 | SKMEL28 | 3/436 | 3.73 × 10−1 | 8.96 × 10−1 | |
| 2 | SKMEL5 | 6/354 | 8.73 × 10−3 | 3.37 × 10−1 | |
| DeepLIFT | 4 | MALME-3M | 7/366 | 2.42 × 10−3 | 3.68 × 10−2 |
| 1 | MDA-MB435 | 14/621 | 2.53 × 10−6 | 1.67 × 10−4 | |
| 7 | SKMEL28 | 7/436 | 6.28 × 10−3 | 5.92 × 10−2 | |
| 9 | SKMEL5 | 6/354 | 8.73 × 10−3 | 6.40 × 10−2 | |
| SHAP | 5 | MALME-3M | 6/366 | 1.02 × 10−2 | 1.55 × 10−1 |
| 1 | MDA-MB435 | 11/621 | 2.74 × 10−4 | 2.09 × 10−2 | |
| 7 | SKMEL28 | 6/436 | 2.22 × 10−2 | 2.00 × 10−1 | |
| 4 | SKMEL5 | 6/354 | 8.73 × 10−3 | 1.55 × 10−1 | |
| IG | 4 | MALME-3M | 7/366 | 2.42 × 10−3 | 3.91 × 10−2 |
| 1 | MDA-MB435 | 14/621 | 2.53 × 10−6 | 1.77 × 10−4 | |
| 9 | SKMEL28 | 5/436 | 6.77 × 10−2 | 4.32 × 10−1 | |
| 6 | SKMEL5 | 6/354 | 8.73 × 10−3 | 1.02 × 10−1 | |
| LRP | 3 | MALME-3M | 7/366 | 2.42 × 10−3 | 5.25 × 10−2 |
| 1 | MDA-MB435 | 13/621 | 1.31 × 10−5 | 8.54 × 10−4 | |
| 13 | SKMEL28 | 4/436 | 1.75 × 10−1 | 7.90 × 10−1 | |
| 6 | SKMEL5 | 5/354 | 3.25 × 10−2 | 3.52 × 10−1 | |
| LIMMA | 7 | MALME-3M | 6/366 | 1.02 × 10−2 | 9.17 × 10−2 |
| 2 | MDA-MB435 | 12/621 | 6.28 × 10−5 | 1.98 × 10−3 | |
| 16 | SKMEL28 | 4/436 | 1.75 × 10−1 | 6.87 × 10−1 | |
| 4 | SKMEL5 | 7/354 | 2.00 × 10−3 | 3.16 × 10−2 | |
| SAM | 8 | MALME-3M | 6/366 | 1.02 × 10−2 | 7.00 × 10−2 |
| 1 | MDA-MB435 | 13/621 | 1.31 × 10−5 | 7.22 × 10−4 | |
| 14 | SKMEL28 | 4/436 | 1.75 × 10−1 | 6.86 × 10−1 | |
| 5 | SKMEL5 | 7/354 | 2.00 × 10−3 | 2.21 × 10−2 | |
| t-test | 8 | MALME-3M | 6/366 | 1.02 × 10−2 | 7.13 × 10−2 |
| 1 | MDA-MB435 | 13/621 | 1.31 × 10−5 | 7.36 × 10−4 | |
| 14 | SKMEL28 | 4/436 | 1.75 × 10−1 | 6.98 × 10−1 | |
| 4 | SKMEL5 | 7/354 | 2.01 × 10−3 | 2.81 × 10−2 |
| Rank | Term | Class | Genes | Approval Status/Study Phase | References |
|---|---|---|---|---|---|
| 5 | Vemurafenib | BRAF/MEK Pathway Inhibitor | ARAF | FDA-Approved | [47] |
| 8 | Dabrafenib | BRAF/MEK Pathway Inhibitor | ARAF | FDA-Approved | [48,49] |
| 15 | Sorafenib | VEGFR, BRAF, and c-KIT | ARAF | Clinical Trial | [50] |
| 1 | Phenylalanine | LAT1 Inhibition | SLC7A5 | Preclinical Studies | [51] |
| 14 | Metformin | Mitochondrial Complex I Inhibition | NDUFA11 | Clinical Trials | [52] |
| Method | Rank | Term | Overlap | p-Value | Adjusted p-Value |
|---|---|---|---|---|---|
| L1-Regularization | 3 | MALME-3M | 7/366 | 2.42 × 10−3 | 6.30 × 10−2 |
| 23 | MDA-MB435 | 5/621 | 1.99 × 10−1 | 6.78 × 10−1 | |
| 7 | SKMEL28 | 6/436 | 2.22 × 10−2 | 2.48 × 10−1 | |
| 1 | SKMEL5 | 9/354 | 7.11 × 10−5 | 5.55 × 10−3 | |
| DeepLIFT | 6 | MALME-3M | 7/366 | 2.42 × 10−3 | 3.07 × 10−2 |
| 10 | MDA-MB435 | 7/621 | 3.62 × 10−2 | 2.74 × 10−1 | |
| 5 | SKMEL28 | 8/436 | 1.55 × 10−3 | 2.35 × 10−2 | |
| 1 | SKMEL5 | 9/354 | 7.11 × 10−5 | 5.41 × 10−3 | |
| SHAP | 3 | MALME-3M | 7/366 | 2.42 × 10−3 | 5.97 × 10−2 |
| 32 | MDA-MB435 | 4/621 | 3.77 × 10−1 | 8.59 × 10−1 | |
| 5 | SKMEL28 | 7/436 | 6.28 × 10−3 | 9.29 × 10−2 | |
| 2 | SKMEL5 | 7/354 | 2.01 × 10−3 | 5.97 × 10−2 | |
| IG | 4 | MALME-3M | 8/366 | 5.02 × 10−4 | 8.78 × 10−3 |
| 35 | MDA-MB435 | 4/621 | 3.77 × 10−1 | 8.18 × 10−1 | |
| 3 | SKMEL28 | 9/436 | 3.38 × 10−4 | 8.56 × 10−3 | |
| 1 | SKMEL5 | 10/354 | 1.12 × 10−5 | 8.52 × 10−4 | |
| LRP | 6 | MALME-3M | 6/366 | 1.02 × 10−2 | 1.19 × 10−1 |
| 13 | MDA-MB435 | 6/621 | 9.11 × 10−2 | 4.91 × 10−1 | |
| 1 | SKMEL28 | 8/436 | 1.55 × 10−3 | 1.08 × 10−1 | |
| 4 | SKMEL5 | 6/354 | 8.73 × 10−3 | 1.19 × 10−1 | |
| LIMMA | 5 | MALME-3M | 6/366 | 9.26 × 10−3 | 1.37 × 10−1 |
| 13 | MDA-MB435 | 5/621 | 1.89 × 10−1 | 7.00 × 10−1 | |
| 2 | SKMEL28 | 8/436 | 1.36 × 10−3 | 5.03 × 10−2 | |
| 7 | SKMEL5 | 5/354 | 3.01 × 10−2 | 3.02 × 10−1 | |
| SAM | 3 | MALME-3M | 8/366 | 4.08 × 10−4 | 8.96 × 10−3 |
| 9 | MDA-MB435 | 6/621 | 8.14 × 10−2 | 6.60 × 10−1 | |
| 6 | SKMEL28 | 7/436 | 5.32 × 10−3 | 6.47 × 10−2 | |
| 2 | SKMEL5 | 8/354 | 4.02 × 10−4 | 1.05 × 10−2 | |
| t-test | 4 | MALME-3M | 7/366 | 2.03 × 10−3 | 3.71 × 10−2 |
| 10 | MDA-MB435 | 6/621 | 8.14 × 10−2 | 5.66 × 10−1 | |
| 7 | SKMEL28 | 6/436 | 1.94 × 10−2 | 1.91 × 10−1 | |
| 2 | SKMEL5 | 8/354 | 3.27 × 10−4 | 1.19 × 10−2 |
| Rank | Term | Class | Genes | Approval Status/Study Phase | References |
|---|---|---|---|---|---|
| 11 | Nelfinavir | ER stress/PI3K-Akt inhibition | OPRD1; TBXAS1 | Clinical trials | [56] |
| 54 | Imatinib | KIT (c-KIT) | SYK; TBXAS1 | Clinical trials | [57,58] |
| 69 | Cabozantinib | AXL, MET, VEGFR2 | AXL | Clinical trials | [59] |
| 100 | Crizotinib | ALK, MET, AXL, RON | SYK; AXL | Clinical trials | [60] |
| 112 | Gefitinib | EGFR | AXL | Clinical trials | [61] |
| 114 | Axitinib | VEGFR1–3 | AXL | Clinical trials | [62] |
| 120 | Dasatinib | BCR-ABL/Src (SYK modulator) | SYK | Clinical trials | [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alghamdi, S.; Turki, T.; Taguchi, Y.-h. De Novo Single-Cell Biological Analysis of Drug Resistance in Human Melanoma Through a Novel Deep Learning-Powered Approach. Mathematics 2025, 13, 3334. https://doi.org/10.3390/math13203334
Alghamdi S, Turki T, Taguchi Y-h. De Novo Single-Cell Biological Analysis of Drug Resistance in Human Melanoma Through a Novel Deep Learning-Powered Approach. Mathematics. 2025; 13(20):3334. https://doi.org/10.3390/math13203334
Chicago/Turabian StyleAlghamdi, Sumaya, Turki Turki, and Y-h. Taguchi. 2025. "De Novo Single-Cell Biological Analysis of Drug Resistance in Human Melanoma Through a Novel Deep Learning-Powered Approach" Mathematics 13, no. 20: 3334. https://doi.org/10.3390/math13203334
APA StyleAlghamdi, S., Turki, T., & Taguchi, Y.-h. (2025). De Novo Single-Cell Biological Analysis of Drug Resistance in Human Melanoma Through a Novel Deep Learning-Powered Approach. Mathematics, 13(20), 3334. https://doi.org/10.3390/math13203334







