1. Introduction
A fundamental aspect of graph theory and combinatorial optimization is the study of independent sets in graphs, which offers extensive insights into the structural characteristics of graphs. Fundamentally, it is NP-hard to find an independent set of maximal cardinality [
1]. The independent number, which is the size of the largest subset of vertices, such that no two vertices are adjacent, holds significance for a variety of applications, including route planning [
2], computer graphics [
3,
4], network analysis [
5], computational biology [
6,
7,
8], network architecture, and resource allocation. Enabling sensors to conserve power while maintaining network coverage, it helps wireless sensor network designers create energy-efficient communication protocols [
9,
10]. By preventing conflicting tasks from being scheduled concurrently, it aids in the optimization of timetables and test schedules for graph coloring and scheduling. Independent sets, which are effective for maximizing influence and advertising, are non-connected groups in social network analysis. Additionally, in coding theory, it stands in for distinct codewords in error correction. Also, they assist in locating security rules or filters that do not conflict in network security [
11,
12]. DNA methods are presented by Zhaocai Wang et al. [
13] to solve the maximal independent set problem using biological processes in the Adleman–Lipton model.
Studying the interaction between algebraic structures, such as cofinite submonoids and graph theory, has received more attention in the last few years. The extensively explored concepts within this domain include the zero divisor graph [
14], nilpotent graphs [
15], and Cayley graph [
16]. In [
17], Cameron explored the recent surge in research on graphs defined on algebraic structures, focusing on how these studies benefit both graph theory and algebra, particularly through three types of interactions between graphs and groups, with examples, and raises several open problems. Recently, different mathematicians assign graphs to the numerical semigroups and studied many invariants such as planarity, clique, and metric dimension [
18,
19,
20,
21,
22,
23].
Let be the set of non-negative integers; then, a numerical semigroup is a subset that is closed under addition, contains the zero element, and has a finite complement in . The elements of are called gaps, and their total number is the genus of the semigroup. The Frobenius number F is the largest integer not in , and the multiplicity is the smallest nonzero element of , denoted by . Given a nonempty set , the submonoid generated by A, denoted , is a numerical semigroup if and only if . Every numerical semigroup has a unique minimal system of generators, and the number of these generators defines the embedding dimension . A symmetric numerical semigroup is one where F is odd and for every , belongs to .
The rest of the paper is organized as follows:
Section 2 provides a theoretical approach in the form of a sequence of results to compute the independent number of genus graphs. In
Section 2, we present an algorithm and its implementation to compute the independent number of a genus graph.
2. Independent Number of Genus Graph of Symmetric Cofinite Submonoid
It is well known that the computation of the independent number is an NP-hard problem, and therefore, difficult to solve for general graphs. So, researchers continue to solve this problem by considering the special classes of graphs. We consider the genus graphs of symmetric cofinite submonoid
and give an algorithmic way to compute the independent number of these graphs. Details of the class
can be found in [
24]. The genus graph is a simple, undirected graph denoted by
, having the set of vertices
and the set of edges
Consider the sets
If
is an even integer, then
and define
In the following lemma, we show that
and
are contained in
by showing that
and
C consist on gaps of
. Note that the Frobenius number
F for
is
.
Lemma 1. Let be a genus graph, then .
Proof. For
, we have
and for
(even)
Since
, therefore
. Note that
and
If
, then
, and by symmetric property
. If
, then
Since
, therefore
Also for
,
, this gives
. Hence,
Since
, therefore
. This implies
□
The proof of the following lemma can be found in [
25] (see Lemma 1).
Lemma 2. Let be a numerical semigroup with . Then if and only if with and .
Now, in the following lemma, we show that union of and form an independent set for the genus graph.
Lemma 3. Let be a graph, then is an independent set for the graph .
Proof. To prove and are independent sets, we have the following cases:
Case-I: If
then
Since
, therefore
. This implies
Case-II: If
then
where
. Note that
We can see that
, since
. If
then by Lemma 2, we have
If
then
This implies . Hence, .
Case-III: If
then
where
. Note that
and
; therefore,
. Again by Lemma 2,
. Hence,
.
Case-IV: If
and
then
Note that
Also,
, as
. If
, then
If
, then
As
, therefore
and
This implies
. Consequently,
.
Case-V: If
and
then
Here,
If
, then
. If
, then
This implies
. Consequently,
.
Case-VI: If
and
then
Note that
If
, then
.
Since,
, therefore
. If
, then
This implies
. Consequently,
. □
The following results give us a maximum independent set and the independent number of genus graph .
Proposition 1. Let be a graph associated with symmetric cofinite submonoid of arbitrary multiplicity and embedding dimension.
- 1.
If (odd) then is a maximal independent set.
- 2.
If (even) then is a maximal independent set.
Proof. Let , where and .
Case 1: If
, and
, where
, then for
, we have
because
.
Case 2: If
, and
, where
then
, where
, we have
Since
, also
therefore by Lemma 2,
.
Case 3: If
,
, and
,
then
because
.
Case 4: If
, with
,
, and
, then
because
. □
Theorem 1. Let be a graph associated with symmetric cofinite submonoid of arbitrary multiplicity and embedding dimension. Then the independent number of is:
Proof. To prove the independent number we have to show that and C are disjoint. For this we discuss following cases:
Case-I: Let , then . This is a contradiction, since from the definition of set A, we have . This implies .
Case-II: Let , then . This is again a contradiction, since from the definition of set A, we have . This gives .
Case-III: Let , then . This is a contradiction, since from the definition of set B, we have . This implies .
So, for
(odd), we have
and for
(even)
□
3. Algorithmic Framework and Implementation
In this section, we present the Python-based implementation of a greedy algorithm used to compute the independent number of genus graph associated with cofinite submonoid (see Algorithm 1).
| Algorithm 1 Computation of the Genus Graph and Its Independent Number |
- Input:
, is the minimal system of generator of cofinite submonoid - Output:
Genus Graph and the independent number of
- 1:
, frontier , max_limit - 2:
while frontier not empty do - 3:
current ← frontier.pop(0) - 4:
if current > max_limit then continue - 5:
end if - 6:
if current then - 7:
- 8:
for all do - 9:
frontier.append(current ) - 10:
end for - 11:
end if - 12:
end while - 13:
- 14:
if then - 15:
sum of two largest gaps - 16:
else - 17:
- 18:
end if - 19:
Reset , frontier - 20:
while frontier not empty do - 21:
current ← frontier.pop(0) - 22:
if current > max_check then continue - 23:
end if - 24:
if current then - 25:
- 26:
for all do - 27:
frontier.append(current ) - 28:
end for - 29:
end if - 30:
end while - 31:
- 32:
Create graph G with nodes = gaps - 33:
- 34:
for all distinct pairs from gaps do - 35:
if then - 36:
Add edge to G and - 37:
end if - 38:
end for - 39:
return G, gaps, edge_list - 40:
Use greedy strategy to build independent set - 41:
return independent set and its size - 42:
while True do - 43:
Prompt user: “Enter generators (comma-separated) or ‘q’ to quit:” - 44:
if user enters ‘q’ then break - 45:
end if - 46:
Parse input ⇒ generators - 47:
numerical_semigroup_graph(generators) - 48:
Print number of nodes, edges, and list of gaps - 49:
if edges not empty then - 50:
Print all edge pairs where - 51:
Call find_maximum_independent_set(G) and print result - 52:
end if - 53:
Visualize G using circular layout (optional) - 54:
end while
|
To compute an independent set for the , we employ a greedy approximation algorithm. The algorithm proceeds as follows: Initialization: Create an empty independent set I. Select a vertex v in G with minimum degree, then add to independent set I. Remove the vertex v and all its neighbors from G and repeat this process. The greedy algorithm employs a minimum-degree vertex ordering rule. In each iteratation, it selects the vertex with the smallest number of connections in the graph. This heuristic aims to minimize the number of neighboring vertices removed in the subsequent step, thereby preserving more options for expanding the independent set. In case of ties, the algorithm selects the vertex with smallest numerical value. The algoritthm iterates until the vertex set of the graph is empty.
The space and time complexity of the purposed algorithm is quadratic , where k is the genus. Python has emerged as a versatile computational platform, distinguished by its efficiency, extensibility, and extensive library support, which collectively enable the seamless implementation of complex mathematical algorithms. Its adaptability and scalability render it an indispensable tool for the rigorous verification of theoretical results and for the exploration of large-scale computational structures arising in algebraic and combinatorial research.
To further enhance accessibility, we provide an online interface through which users can directly validate their results, thereby eliminating the necessity of installing Python or auxiliary software environments.
To support the theoretical results, we provide a Python implementation that constructs the graph associated with a cofinite submonoid and computes its independent number. Interested readers can access via the following link:
https://independent-number.onrender.com/ (accessed on 27 July 2025).
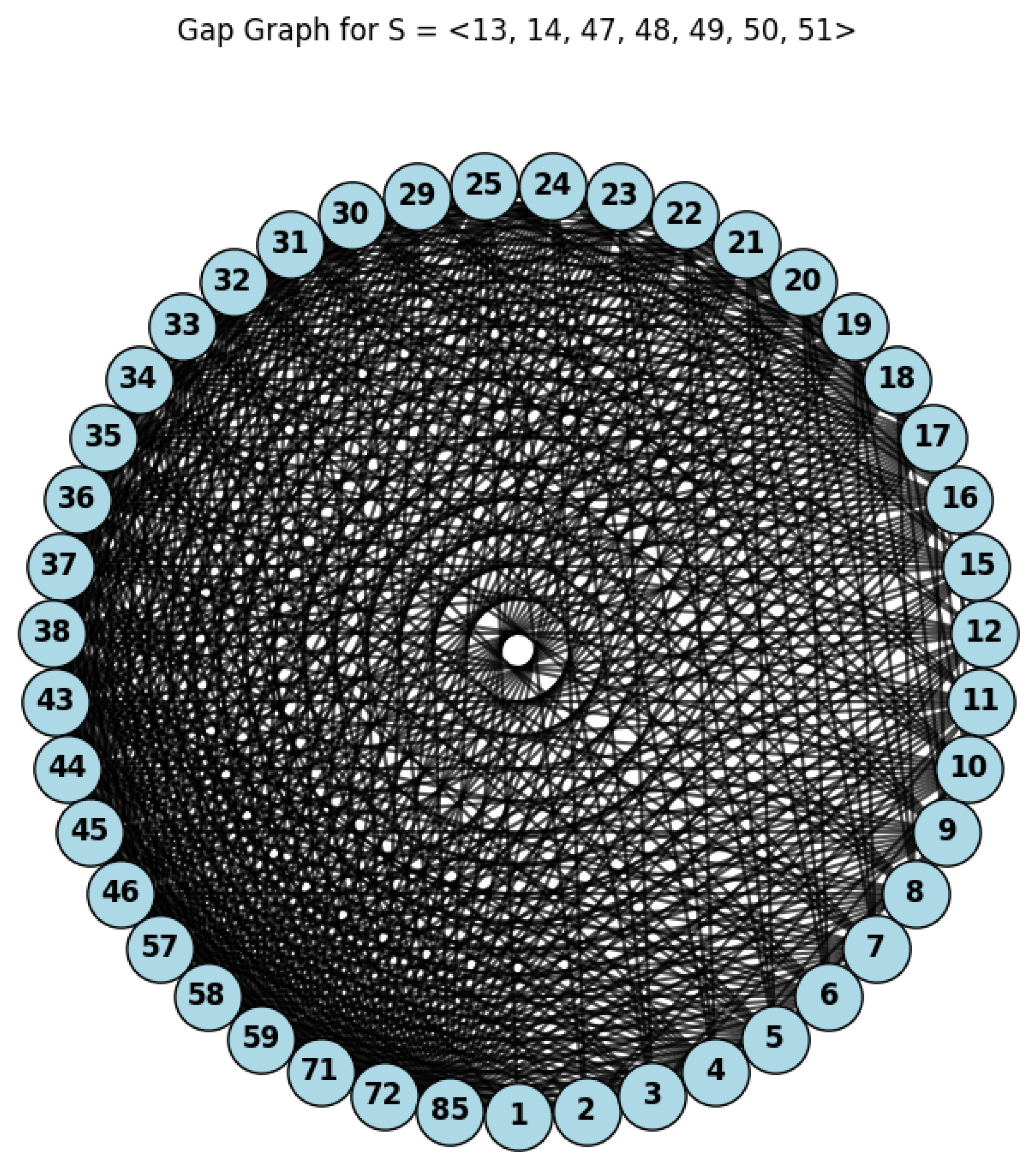
Example 1. Let , then we can use the following Python code (Listing 1) to compute the genus graph and its independent number
By Lemma 1 and Theorem 1, we have
, and
. So,
(see
Figure 1).
| Listing 1. Python Console Output. |
![Mathematics 13 02951 i001 Mathematics 13 02951 i001]() |
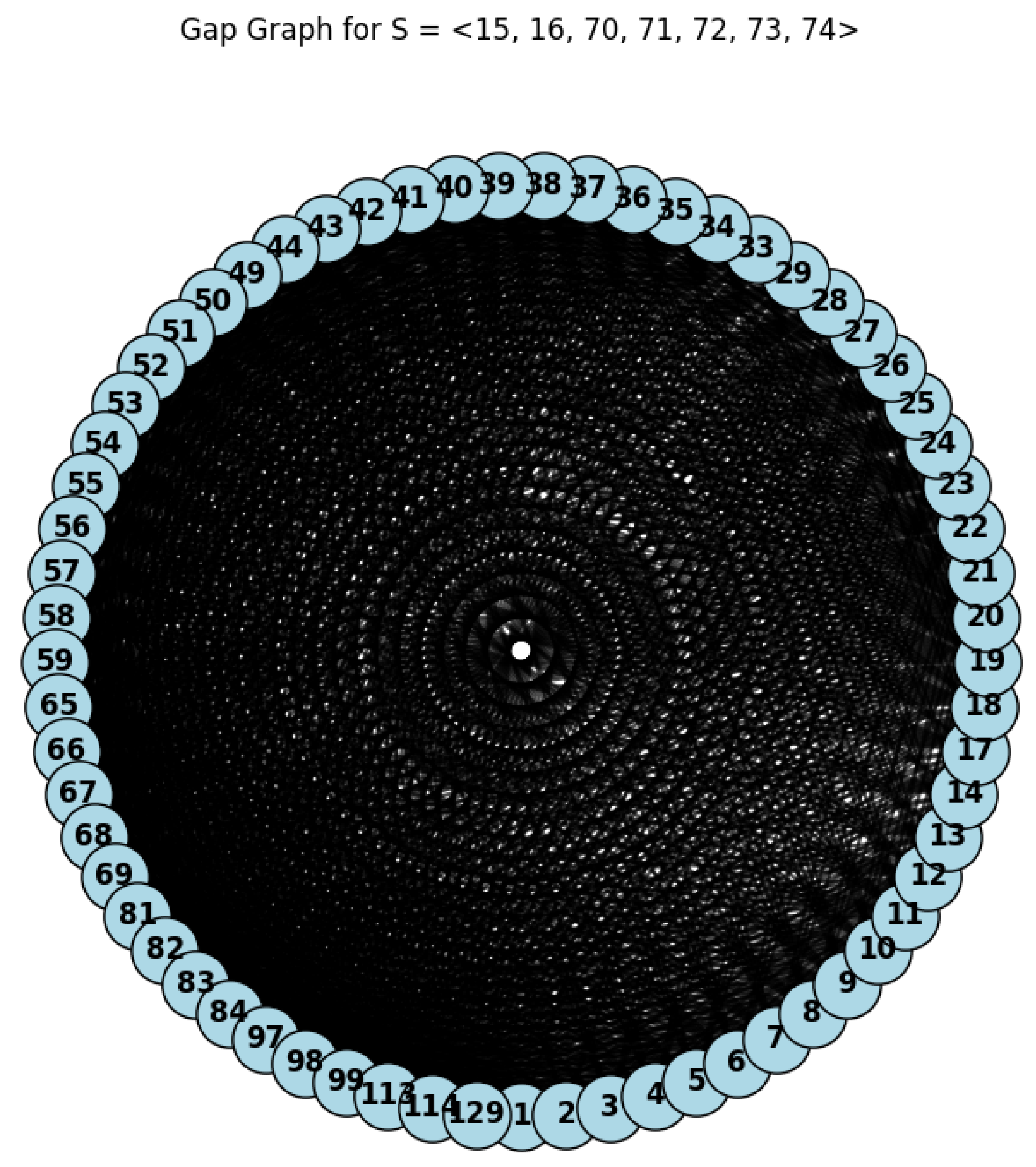
Example 2. Let be a graph associated with , then we can use the following Python code (Listing 2) to compute the genus graph and its independent number .
| Listing 2. Python Console Output. |
![Mathematics 13 02951 i002 Mathematics 13 02951 i002]() |
By Lemma 1 and Theorem 1, we have
, and
. So,
(see
Figure 2).
Example 3. Let , then . From Lemma 1, we haveFor an odd integer, Theorem 1 gives maximum independent set for is . This implies , which is not correct for the numerical semigroup shown as follows (Listing 3). | Listing 3. Python Console Output. |
![Mathematics 13 02951 i003 Mathematics 13 02951 i003]() |
4. Application and Experiments
The application of an independent set: Cancer develops when multiple genes become misregulated, causing disruptions in the pathways that normally regulate healthy cell growth and leading to the uncontrolled accumulation of abnormal cells, while some abnormal gene expressions can be naturally compensated by other genes, many remain uncompensated and act as persistent drivers of tumor progression. In the framework of numerical semigroups, these abnormal gene states are represented by the gaps, and the uncompensated ones are captured by the independent set. This connection allows us to identify precisely which gene expressions must be targeted in therapy. Tumors with larger independent sets are biologically more aggressive, since they contain multiple uncompensated drivers that require combination therapy to suppress. By focusing on the members of the independent set, treatment strategies can be designed to block all critical pathways simultaneously, thereby reducing the risk of cancer survival and drug resistance. Moreover, the independent number itself serves as a measure of tumor complexity and can act as a prognostic marker, guiding both therapeutic design and clinical decision making. Thus, the concept of the independent set in numerical semigroup theory offers a powerful mathematical lens to model tumor robustness and optimize cancer treatment (see
Table 1).
5. Conclusions
In this research work, we find a maximum independent set and the independent number of graphs associated with symmetric cofinite submonoid of arbitrary multiplicity and embedding dimension. This work opens up new avenues for future study, such as generalizing our methods to non-symmetric cofinite submonoid, exploring other graph invariants, and applying these results to more extensive context in combinatorics and algebra. The problem of figuring out the independent number, in general, remains unsolved. Also, we give a Python code to find the independent number of the genus graph .
Author Contributions
Conceptualization, S.A.; methodology, S.A. and A.S.A.; software, A.S.A.; formal analysis, A.S.A.; investigation, S.A.; writing—original draft, S.A. and A.S.A.; project administration, S.A.; funding acquisition, A.S.A. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Deanship of Scientific Research and Libraries in Princess Nourah bint Abdulrahman University for funding this research work through the Program for Supporting Publication in Top-Impact Journals, Grant No. (SPTIF-2025-10).
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research and Libraries in Princess Nourah bint Abdulrahman University for funding this research work through the Program for Supporting Publication in Top-Impact Journals, Grant No. (SPTIF-2025-10).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Garey, M.R.; Johnson, D.S.; Stockmeyer, L. Some simplified NP-complete problems. In Proceedings of the Sixth Annual ACM Symposium on Theory of Computing, Seattle, WA, USA, 30 April–2 May 1974; pp. 47–63. [Google Scholar]
- Kieritz, T.; Luxen, D.; Sanders, P.; Vetter, C. Distributed time-dependent contraction hierarchies. In Proceedings of the International Symposium on Experimental Algorithms, Naples, Italy, 20–22 May 2010; Springer: Berlin/Heidelberg, Germany; pp. 83–93. [Google Scholar]
- Sander, P.V.; Nehab, D.; Chlamtac, E.; Hoppe, H. Efficient traversal of mesh edges using adjacency primitives. ACM Trans. Graph. (TOG) 2008, 27, 1–9. [Google Scholar] [CrossRef]
- Shirinivas, S.G.; Vetrivel, S.; Elango, N.M. Applications of graph theory in computer science an overview. Int. J. Eng. Sci. Technol. 2010, 2, 4610–4621. [Google Scholar]
- Puthal, D.; Nepal, S.; Paris, C.; Ranjan, R.; Chen, J. Efficient algorithms for social network coverage and reach. In Proceedings of the IEEE International Congress on Big Data, New York, NY, USA, 27 June–2 July 2015; pp. 467–474. [Google Scholar]
- Butenko, S.; Wilhelm, W.E. Clique-detection models in computational biochemistry and genomics. Eur. J. Oper. Res. 2006, 173, 1–17. [Google Scholar] [CrossRef]
- Cheng, T.M.; Lu, Y.E.; Vendruscolo, M.; Lio’, P.; Blundell, T.L. Prediction by graph theoretic measures of structural effects in proteins arising from non-synonymous single nucleotide polymorphisms. PLoS Comput. Biol. 2008, 4, 1000135. [Google Scholar] [CrossRef] [PubMed]
- Mason, O.; Verwoerd, M. Graph theory and networks in biology. IET Syst. Biol. 2007, 1, 89–119. [Google Scholar] [CrossRef] [PubMed]
- Hespe, D.; Lamm, S.; Schorr, C. Targeted branching for the maximum independent set problem. In Proceedings of the 19th International Symposium on Experimental Algorithms, Nice, France, 7–9 June 2021; Volume 190, pp. 1–17. [Google Scholar]
- Balaban, A.T. Applications of graph theory in chemistry. J. Chem. Inf. Comput. Sci. 1985, 25, 334–343. [Google Scholar] [CrossRef]
- Hespe, D.; Schulz, C.; Strash, D. Scalable kernelization for maximum independent sets. J. Exp. Algorithmics (JEA) 2019, 24, 1–22. [Google Scholar] [CrossRef]
- Strash, D. On the Power of Simple Reductions for the Maximum Independent Set Problem; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Wang, Z.; Tan, J.; Zhu, L.; Huang, W. Solving the maximum independent set problem based on molecule parallel supercomputing. Appl. Math. Inf. Sci. 2014, 8, 2361–2366. [Google Scholar] [CrossRef][Green Version]
- Anderson, D.F.; Livingston, P.S. The Zero-Divisor Graph of a Commutative Ring. J. Algebra 1999, 217, 434–447. [Google Scholar] [CrossRef]
- Kumar, B.D.; Ajay, S.; Rahul, D. Nilpotent Graph. Theory Appl. Graphs 2021, 8, 2. [Google Scholar] [CrossRef]
- Meier, J. Groups Graphs and Trees: An Introduction to the Geometry of Infinite Groups; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Cameron, P.J. What can graphs and algebraic structures say to each other? AKCE Int. J. Graphs Comb. 2023, 21, 249–254. [Google Scholar] [CrossRef]
- Chen, R.; Fazal, S.; Aslam, A.; Tchier, F.; Binyamin, M.A. On the metric dimension of graphs associated with irreducible and Arf numerical semigroups. AKCE Int. J. Graphs Comb. 2024, 21, 286–293. [Google Scholar] [CrossRef]
- Rao, Y.; Binyamin, M.A.; Aslam, A.; Mehtab, M.; Fazal, S. On the planarity of graphs associated with symmetric and pseudo symmetric numerical semigroups. Mathematics 2023, 11, 1681. [Google Scholar] [CrossRef]
- Binyamin, M.A.; Adeel; Alali, A.S.; Mahmood, H. On Connectedness, Girth and Diameter of Gap Poset Graphs. Proc. Bulg. Acad. Sci. 2025, 78, 813–820. [Google Scholar] [CrossRef]
- Mehtab, M.; Binyamin, M.A.; Asghar, S.S.; Alali, A.S.; Mehmood, K. On Structural Characterization and Computation of the Diameter and Girth of Bipartite Gap Poset Graphs with Python Application. Axioms 2025, 14, 669. [Google Scholar] [CrossRef]
- Alali, A.S.; Binyamin, M.A.; Mehtab, M. A Conjecture for the Clique Number of Graphs Associated with Symmetric Numerical Semigroups of Arbitrary Multiplicity and Embedding Dimension. Symmetry 2024, 16, 854. [Google Scholar] [CrossRef]
- Binyamin, M.A.; Siddiqui, H.M.A.; Khan, N.M.; Aslam, A.; Rao, Y. Characterization of graphs associated with numerical semigroups. Mathematics 2019, 7, 557. [Google Scholar] [CrossRef]
- Rosales, J.C. Symmetric numerical semigroups with arbitrary multiplicity and embedding dimension. Proc. Am. Math. Soc. 2001, 129, 2197–2203. [Google Scholar] [CrossRef]
- García-Sánchez, P.A.; Rosales, J.C. Numerical semigroups generated by intervals. Pac. J. Math. 1999, 191, 75–83. [Google Scholar] [CrossRef]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).