1. Introduction
In 2022, breast cancer was the most frequently diagnosed cancer in women worldwide, with 2.30 million recorded cases. More than 666 thousand of these resulted in fatalities, representing 15.5% of all cancer-related deaths [
1]. By 2040, this number is expected to reach 3.16 million cases [
2].
Mammography is the most used technique for the early detection of breast cancer [
3,
4], and its use has contributed to reducing the mortality rate of the disease by up to 48% [
5]. Comparisons between different breast imaging techniques have highlighted mammography as the most effective and cost-efficient breast examination option [
6]. Furthermore, mammography is considered the gold standard for breast cancer detection in women over 40 years old [
7].
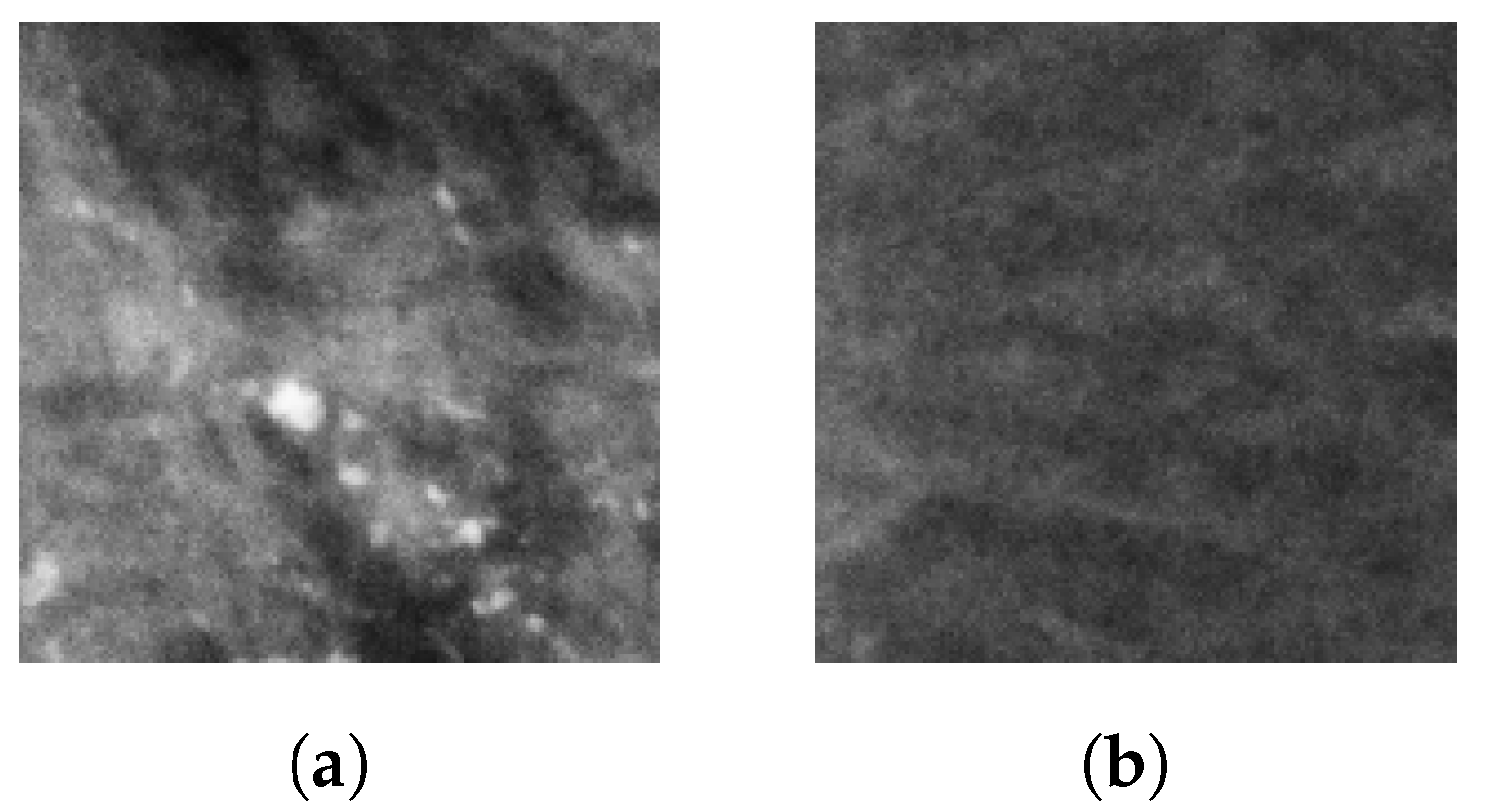
Microcalcifications (MCs) are the most significant indirect indicators of breast cancer. MCs appear as small deposits of calcium dots with diameters ranging from 0.1 mm to 1 mm and can be found either scattered or clustered along the mammary ducts [
3,
8]. In an X-ray image, MCs appear as pixel clusters that vary in brightness and contrast, and they can be dispersed or grouped in different regions of the breast [
6,
9].
MC detection is highly complex due to their size, shape, and distribution. Depending on their characteristics, it is possible to determine whether they are malignant or not at a high degree of accuracy. If diagnosed as malignant, a biopsy is mandatory for confirmation [
10].
Groups of microcalcifications or microcalcification clusters (MCCs) describe situations where a set of MCs occupies a small portion of the breast tissue. This term is applied when at least three MCs are grouped within an area of 1 cm
2 [
11,
12,
13]. MCCs are present in up to 50% of mammograms with confirmed cancer [
12,
13] and have a probability of malignancy ranging from 10% to 50%, which corresponds to the suspicious category with code 4 in the BI-RADS system [
11].
Basile et al. [
3] and Mordang et al. [
14] highlighted that detecting MCCs represents a challenge, as they are imperceptible to touch. Therefore, imaging techniques are necessary for their detection. However, acquiring these images presents additional challenges, such as variability in breast composition, differences in texture, and low contrast.
The characteristics of mammograms vary according to the patient’s age and specific conditions, making MCC detection a complex challenge that can lead to misdiagnoses [
15]. The early detection of MCCs, particularly in the initial stage of cancer, significantly increases survival rates. At this stage, the five-year survival rate reaches 99% [
16].
Given the difficulty in detecting MCCs, caused by their small size, low contrast, variable appearance, and differences in breast tissue composition, researchers have explored artificial intelligence (AI) techniques to improve detection. Some studies have confirmed that using AI in medical imaging is safe and reliable [
17]. Among the various AI techniques, deep learning (DL) has shown particularly promising results [
18,
19]. When trained with a large dataset, DL has achieved high levels of accuracy. DL architectures, such as convolutional neural networks (CNNs), are currently being studied for their effectiveness in detecting MCCs [
3].
In the literature, several studies have demonstrated the high performance of CNNs in MCC detection. For instance, Wang et al. [
13] developed a CNN for MCC detection using 521 screen-film mammography (SFM) images and 188 full-field digital mammography (FFDM) images, all collected by the Department of Radiology at the University of Chicago. The model achieved a 90% sensitivity, 0.69 false positives (FP) per image, and an area under the curve (AUC) of 0.971. Rehman et al. [
15] designed a fully connected deep CNN (FC-DSCNN) for MCC detection and malignancy classification using 2885 SFM images from the Pakistan Institute of Nuclear Medicine and Radiotherapy (PINUM) dataset and 3568 SFM images from the Digital Database for Screening Mammography (DDSM). Although the original resolution is not specified, both datasets provide mammograms with a size of
pixels, which suggests low spatial resolution. The model achieved a 99% sensitivity, 82% specificity, 2.45 FP per image, and 89% accuracy.
Hsieh et al. [
10] used the VGG16 for MCC detection in mammograms, followed by an R-CNN to segment MCs within the detected MCCs and remove background noise. The InceptionV3 model was employed to classify the MCCs into benign or malignant. The model was trained and tested on 1,586 mammograms from a private dataset provided by the Department of Medical Imaging at the affiliated hospital of Chung-Shan Medical University. The study does not specify the image resolution or whether the mammograms are FFDM or SFM. The method achieved 93% and 95% accuracy in detection and segmentation, respectively, and 91% accuracy in malignancy classification. Overall, it reached 87% accuracy, 89% specificity, and 90% sensitivity.
Liu et al. [
20] proposed a deep learning model to predict the malignancy of suspicious MCs, trained exclusively on patches extracted from craniocaudal (CC) and mediolateral oblique (MLO) mammographic views. The architecture consists of two MobileNetV2 networks, each trained on a specific view, sharing weights to extract features that are concatenated for malignancy prediction. The model was evaluated on a private dataset of 824 FFDM mammograms (CC and MLO views) corresponding to 414 BI-RADS 4 microcalcifications from 384 patients, acquired at Xinhua Hospital, Shanghai Jiao Tong University School of Medicine. The resolution and mammography system details were not reported. The image-based model achieved 76.5% sensitivity and 83.8% specificity on the test dataset.
Terrassin et al. [
21] used ResNet-22, the ConvNeXt, and the UNet3+ CNNs to classify and segment malignant MCs in digital mammograms. The models were trained and evaluated on two public datasets: INbreast and the Breast Microcalcifications Dataset (BMCD), both annotated at the pixel level by expert radiologists. Although it is known that INbreast images were acquired at 70
m resolution, this detail was not reported in the article. The total number of mammograms was also not specified. They achieved an AUC of 0.93 for classification and 70% accuracy in segmentation.
Luna et al. [
22] developed a residual shallow CNN for MCC detection. The model was trained and evaluated on 10 FFDM images from the INbreast dataset containing MCCs, achieving an accuracy of 99.71%. Additionally, when evaluated on the MEXBreast dataset at 70
m resolution, the same model reached 99.8% accuracy. In a comparative study using various CNN architectures trained and tested on the same INbreast subset, MobileNetV2 obtained the highest accuracy, with 99.84% [
23].
Most studies, such as the previously mentioned ones, do not explicitly consider or report the mammograms’ resolution, primarily because they rely on publicly available datasets with varying resolutions and different acquisition technologies, implicitly assuming that CNNs can be trained without accounting for differences in image resolution and mammography type [
24]. A summary of these studies and their dataset characteristics is provided in
Table 1. Therefore, selecting an optimal image resolution may improve the performance of CNNs in radiology-based tasks. Hence, diagnostic tasks could benefit from higher image resolutions, as different pathologies may have varying resolution requirements for optimal detection. Whether variations in mammogram resolution across datasets affect the ability of CNNs to generalize effectively constitutes a research gap [
25].
The performance of MCC detection algorithms is highly dependent on the database and the criteria by which the results are evaluated. This further emphasizes the need to carefully consider resolution variability when designing and assessing CNN models for MCC detection, as noted by Karale et al. [
26]. Furthermore, the presence of both FFDM and SFM in available datasets introduces additional variability in resolution, since digital and film-based mammograms may exhibit different spatial characteristics. For instance, the images in the DDSM database were acquired using different scanners [
27].
Table 2 lists available mammography datasets and their corresponding resolutions and imaging modalities.
Due to the variability in mammogram resolution resulting from different acquisition scanners, it is important to assess whether CNN architectures can generalize across multiple resolutions. Although available mammography datasets provide valuable resources for MCC detection research, they exhibit variability in resolution and imaging modality, raising concerns about whether CNNs trained on a specific resolution can generalize effectively across different resolutions. Consequently, we propose a cross domain transfer learning (TL) architecture for MCC detection based on the MobileNetV2 architecture [
35]. In a previous study [
23], several CNN architectures were evaluated using the INbreast dataset at 70
m resolution, and the four best performing models were identified: VGG16, ResNet50, DenseNet121, and MobileNetV2. Among them, MobileNetV2 achieved the highest accuracy. To enable a fair comparison and assess the generalizability of these models, the same four architectures were trained from scratch using patches from our publicly available MEXBreast dataset [
33,
34] at the same resolution of 70
m, as described in
Section 2.3. MobileNetV2 once again obtained the best performance, which supports its use in the present work.
We developed three new models adapted to 50
m, 70
m, and 100
m resolutions based on [
23], following a feature extraction strategy. Their performance was then evaluated on independent mammography datasets to assess robustness across different imaging conditions.
The present study is driven by the need to address existing challenges, build upon previous advancements, and fill current gaps in the area. Specifically, most CNN-based approaches for MCC detection do not explicitly consider mammogram resolution, despite the variability introduced by different acquisition systems and imaging modalities. In addition, model performance often depends heavily on the characteristics of the datasets used, including whether they combine digital and film-based mammograms, which may differ in spatial quality. Finally, there is limited investigation into whether CNN models trained at one resolution can generalize to others, raising concerns about their robustness in heterogeneous clinical scenarios. By developing new methodologies that integrate the strengths of various techniques, current and future studies aim to improve the accuracy and reliability of early detection of breast lesions. Ultimately, these efforts contribute to developing AI models that can be integrated into devices to support more effective healthcare. The contributions of this paper are as follows:
Feature extraction from the pretrained MobileNetV2: Features extracted from MobileNetV2, trained on 70 m resolution mammograms, were used to create three new models at 50 m, 70 m, and 100 m resolutions, improving detection performance on both lower and higher resolution images.
A cross-domain CNN architecture for MCC detection with three different resolutions: A CNN architecture based on MobileNetV2 is proposed and trained from scratch to detect MCCs in mammograms at any of the three resolutions (50 m, 70 m, and 100 m), addressing the generalization limitations of training with single-resolution data.
Demonstration of the superiority of TL across resolutions: Experimental results show that TL improves MCC detection accuracy compared to models trained from scratch, especially for non-native resolutions (98.32% vs. 96.07% for 50 m, 99.27% vs. 99.20% for 70 m, and 89.17% vs. 83.59% for 100 m).
Efficient patch-based processing: The study introduces a consistent preprocessing method using 1 cm2 patches, enabling scalable and resolution-consistent input handling across datasets.
The article is organized as follows:
Section 2 describes the materials and methods;
Section 3 presents the experimental setup and results;
Section 4 discusses the findings; and
Section 5 provides the conclusions and outlines future work.
3. Experiments and Results
In this section, we present the training, validation, and testing of the CNN architecture introduced in
Section 2. The experiments were conducted on the MEXBreast database [
33,
34] for the 50, 70 and 100
m resolutions, considering two scenarios. In the first, the CNN is trained and evaluated from scratch. In the second, the CNN leverages the knowledge acquired during training with the 70
m resolution patches [
23]. Additionally, for the 70
m resolution, we evaluated the performance of the model pretrained on the INbreast dataset [
23] by testing it directly on the MEXBreast dataset at the same resolution, in order to assess its generalizability to a different dataset.
As a result, we generated seven different models:
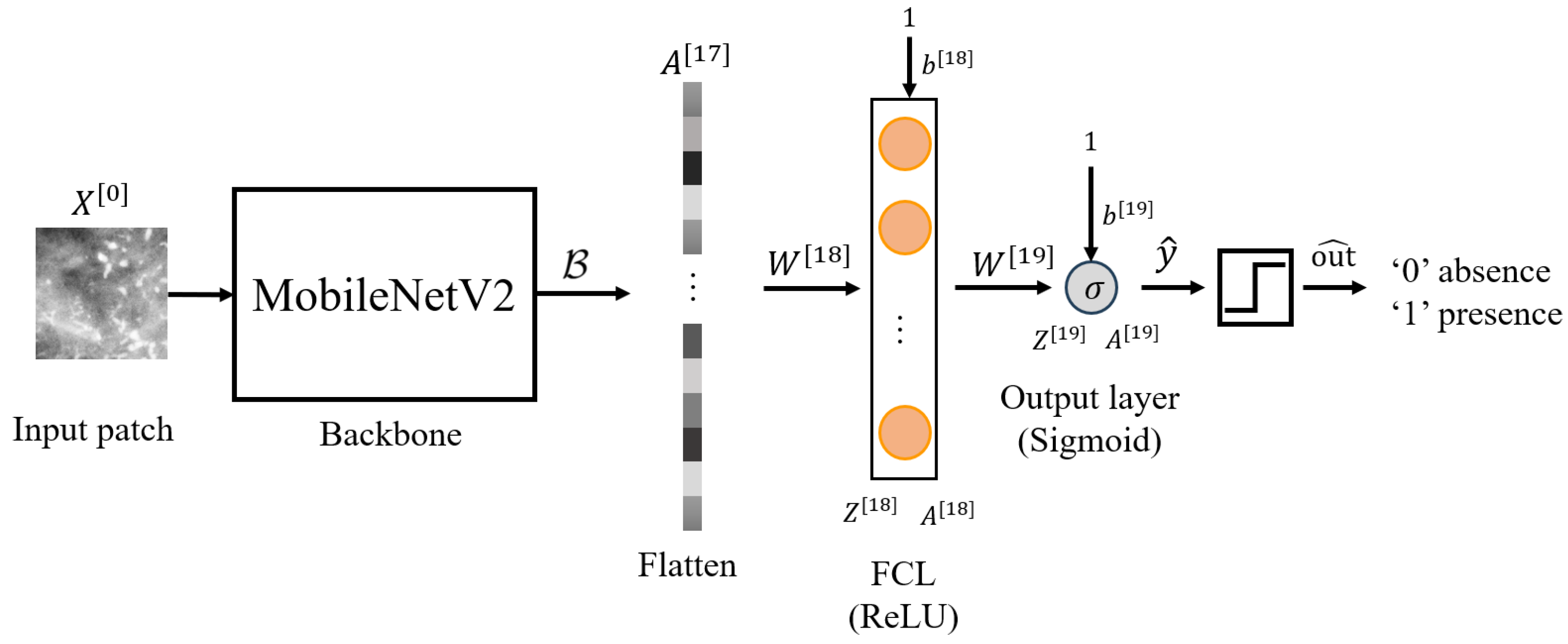
Scratch-50: Model trained from scratch on 50 m resolution mammograms from the MEXBreast dataset.
Transfer-50: Feature extraction model using TL by freezing the backbone () of the pretrained 70 m model (trained on INbreast) and training on 50 m patches from the MEXBreast dataset.
Scratch-70: Model trained from scratch on 70 m resolution mammograms from the MEXBreast dataset.
Transfer-70: Feature extraction model using TL by freezing the backbone () of the pretrained 70 m model (trained on INbreast) and training on 70 m patches from the MEXBreast dataset.
Scratch-100: Model trained from scratch on 100 m resolution mammograms from the MEXBreast dataset.
Transfer-100: Feature extraction model using TL by freezing the backbone () of the pretrained 70 m model (trained on INbreast) and training on 100 m patches from the MEXBreast dataset.
Eval-70: The model pretrained on the INbreast dataset at 70 m, directly evaluated on the MEXBreast dataset at the same resolution.
The MobileNetV2 backbone (
) retains the trained weights in [
23] with the 70
m resolution. Furthermore, we only trained the weights of the additional layers, namely
and
. Therefore, the frozen layers can be expressed as
The results for Scratch-50, Transfer-50, Scratch-70, Transfer-70, Scratch-100, and Transfer-100 models are shown in
Table 8. Training metrics, including accuracy and loss for training, validation, and test, are summarized. The best validation epoch for each model and the total training time is also shown.
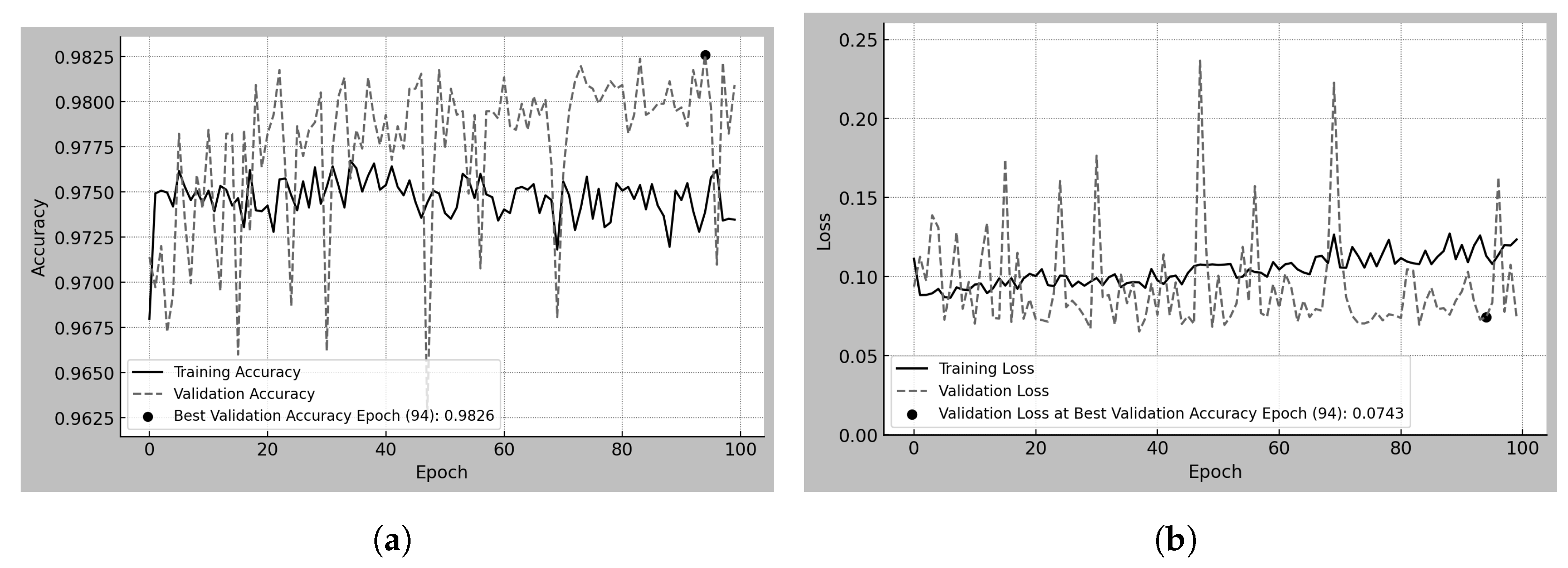
Figure 3a,b present the training and validation accuracy and loss over 100 epochs for the Scratch-50 model, which was trained from scratch using 50
m resolution patches.
Figure 4a,b correspond to the Transfer-50 model, which was trained using feature extraction from a pretrained version at 70
m. Similarly,
Figure 5a,b and
Figure 6a,b depict the same metrics for the Scratch-70 and Transfer-70 models. Finally,
Figure 7a,b and
Figure 8a,b depict the same metrics for the Scratch-100 and Transfer-100 models. In all cases, the model corresponding to the epoch with the highest validation accuracy was saved and later used for evaluation. Each figure includes a visual marker and a legend indicating the epoch at which this maximum validation accuracy was achieved.
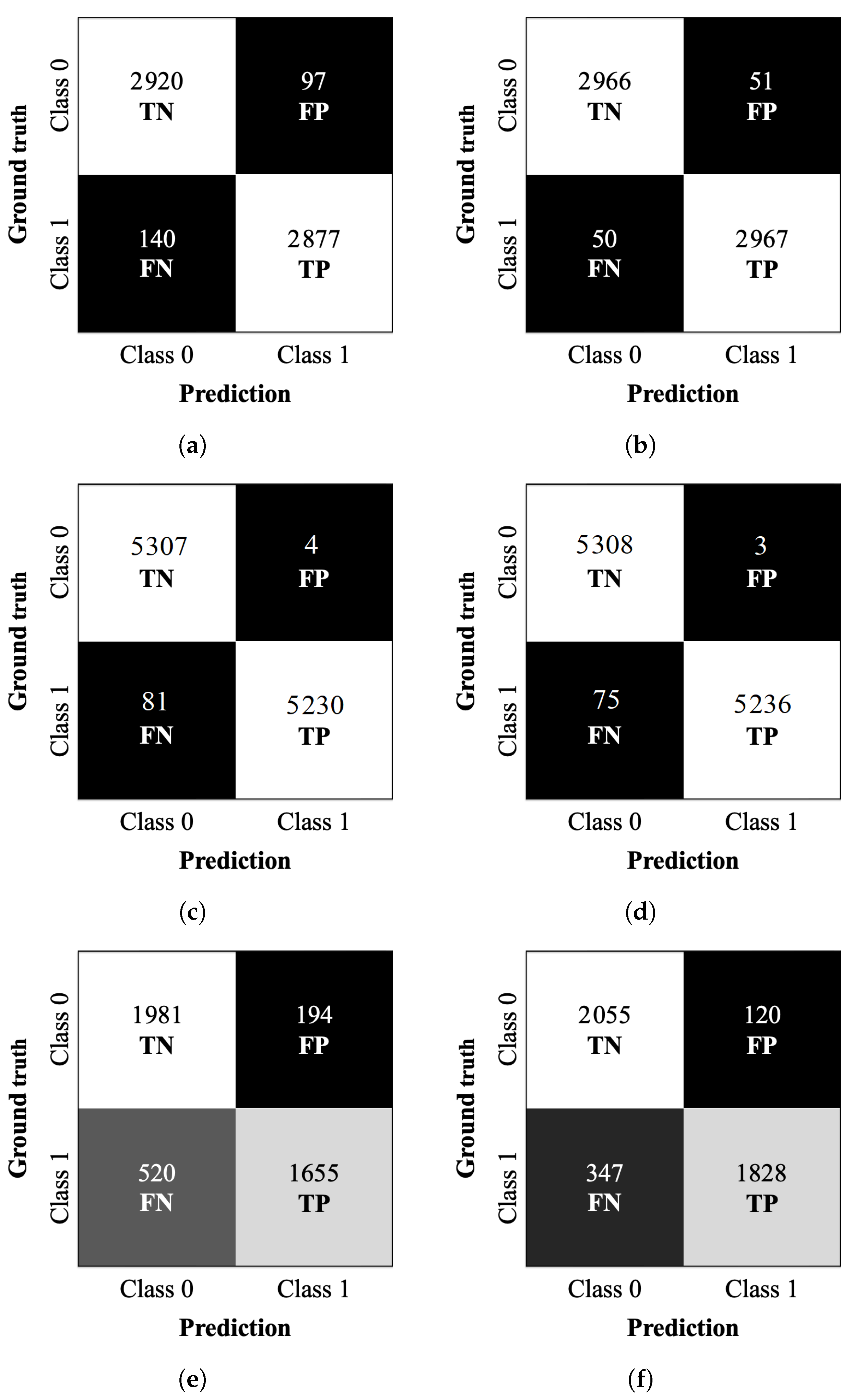
Confusion matrices were calculated for the test datasets corresponding to the Scratch-50, Transfer-50, Scratch-70, Transfer-70, Scratch-100, and Transfer-100 models. The matrices are shown in
Figure 9. The matrices summarize the relationships between the predicted and true classes and provide additional insights into the models’ generalization capabilities. The true negatives (TN) are the number of correctly classified negative samples (in our case, the true class 0). The false positives (FP) are the number of negative samples classified as positive (number of true class 0 classified as class 1). The false negatives (FN) are the number of positive samples classified as negative (number of true class 1 classified as class 0). The true positives (TP) are the number of correctly classified positive samples (in our case, the true class 1).
Evaluation metrics such as accuracy, precision, recall (sensitivity), specificity, and F1-score are summarized in
Table 9 for all the models.
Accuracy measures the overall correctness of the model. Precision represents the proportion of correctly identified positive samples among all predicted positives. Recall reflects the model’s ability to detect positive cases, while specificity quantifies its ability to identify negative cases correctly. The F1-score is the harmonic mean of precision and recall, providing a single metric that balances both aspects and penalizes large discrepancies between them.
Table 10 reports the original test results of the Eval-70 model on the INbreast test dataset, obtained in our previous work [
23], and included here only as a reference for comparison, along with the new results obtained on the MEXBreast test dataset in the present study.
Figure 10 shows the confusion matrix obtained from the predictions on the MEXBreast test dataset; the evaluation metrics presented in
Table 11 were calculated based on this matrix.
4. Discussion
In this section, we provide an analysis and interpretation of the results obtained.
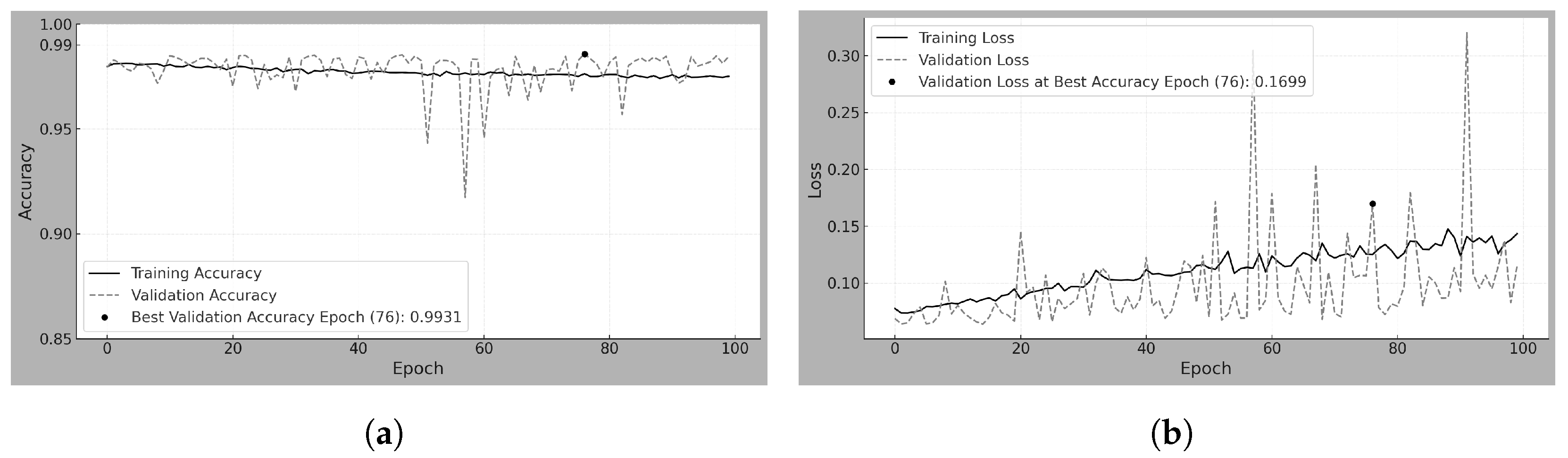
4.1. Scratch-50
Scratch-50 refers to the model trained from scratch using 50
m resolution mammograms from the MEXBreast dataset.
Table 8 and
Figure 3a,b analyze the training and validation behavior over 100 epochs. Regarding accuracy, the training trend shows a steady increase without abrupt changes, reaching 92.56% at the best validation epoch. However, validation accuracy exhibits high variability across epochs, with sudden fluctuations. Despite this, at the best validation epoch (epoch 95 and accuracy of 96.83%), training and validation accuracy exceeded 92%.
The behavior of the loss reflects the fluctuations observed in accuracy. Observe that the loss does not range between 0 and 1 but reaches a maximum value of 350 during training (see
Figure 3b). The high loss is largely due to the 50% decision threshold used for classification. Even when the model correctly classifies a sample as class 1, if the predicted probability is close to 0.5, the loss function penalizes it heavily. Furthermore, the high variability in the patch content contributes to this effect, as the model encounters a wide range of mammographic regions with different levels of complexity.
The BCE loss function (Equation (
14)) evaluates the classification performance. Its behavior is strongly influenced by the relationship between the predicted probabilities and the ground-truth labels, as it penalizes predictions based on their confidence: high-confidence correct predictions result in very small loss values; low-confidence correct predictions incur a noticeably higher loss despite being classified correctly; and high-confidence incorrect predictions are heavily penalized, leading to large loss values. As a result, even with a high overall classification accuracy, the loss can remain elevated due to low-confidence or incorrect predictions. This effect is reflected in
Table 8, where the test accuracy reached 96% (0.96), while the test loss remained high at 4.71.
Table 9 shows the model’s effectiveness in classifying unseen samples, resulting in an accuracy of 96.07%. The model demonstrates high precision (96.74%) and specificity (96.78%), which indicates a low number of FP (97). The recall (sensitivity) of 95.36% tells us that the model correctly identifies most positive cases, with only 140 FN. The F1-score of 96.04% further supports the balance between precision and recall, showing a well-calibrated classification performance.
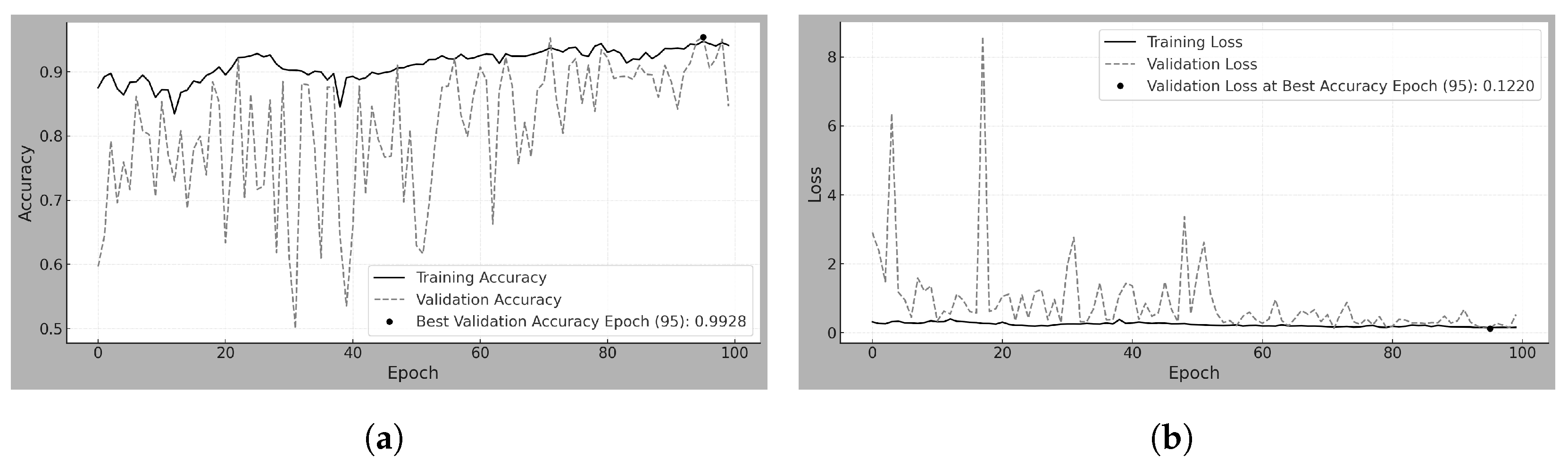
4.2. Transfer-50
Transfer-50 refers to the model using TL by freezing the backbone (
) of the pretrained 70
m model [
23] (trained on INbreast) and training on 50
m patches from the MEXBreast dataset.
Table 8 and
Figure 4a,b show that, from the first epoch, the accuracy exceeds 95% due to the transfer of knowledge from the 70
m model. Training and validation accuracies remain stable throughout the process. The range between 96.25% and 98.25%, indicates the absence of abrupt fluctuations.
Similarly, the loss remains stable for validation and training. The values ranging between 0.05 and 0.25, confirming the consistency of the model’s learning process. The narrow range in accuracy and loss suggests that TL approach enables rapid adaptation without significant variability. The confusion matrix in
Figure 9b, provides further insight into the model’s classification performance on the testing dataset. The strong contrast between the white and black regions indicates a well-defined separation between correctly and incorrectly classified samples. The completely white diagonal, corresponding to correctly classified cases (2966 TN and 2967 TP), suggests high classification performance.
Likewise, the black diagonal, representing misclassified cases, shows 51 FP and 50 FN, reinforcing the low error rate. The clarity in the separation of these regions visually confirms the model’s ability to distinguish between high accuracy classes. The metrics in
Table 9 show the model’s high classification effectiveness. The accuracy of 98.32% depicts the strong separation observed in
Figure 9b, where correctly classified cases dominate.
Precision, recall, and specificity remain balanced, all within 98.31–98.34%, indicating that the model maintains consistent performance across both classes. The F1-score of 98.33% further confirms this balance, highlighting the agreement between precision and recall.
4.3. Scratch-70
Scratch-70 is the model trained from scratch on 70
m resolution mammograms from the MEXBreast dataset.
Table 8 and
Figure 5a,b show the training and validation behavior over 100 epochs. Regarding accuracy, the training curve increases steadily without abrupt changes, reaching 99.28% at the epoch with the best validation result. In contrast, the validation accuracy exhibits high variability across epochs, with noticeable fluctuations. Nonetheless, at the best validation epoch (epoch 95), both training and validation accuracy exceeded 92%. Concerning the loss metric, although the overall loss ranges between 0 and 8, as depicted in
Figure 5b, it remains close to zero for most of the epochs.
Table 9 shows a test accuracy of 99.20%, consistent with the rest of the evaluation metrics. The only metric showing a slight deviation is recall, with a value of 98.62%, which suggests the presence of a few FN. In practical terms, this means that out of 100 truly positive cases, the model might fail to detect fewer than two.
4.4. Transfer-70
Transfer-70 refers to the model trained using TL by freezing the backbone (
) of a pretrained model [
23] (originally trained on 70
m resolution patches from the INbreast dataset) and retraining on 70
m patches from the MEXBreast dataset.
Table 8 and
Figure 6a,b show that, from the very first epoch, both training and validation accuracies exceeded 97% due to the transfer of knowledge from the pretrained 70
m model. Accuracy values remained stable throughout the training process, without abrupt fluctuations, except for a few noticeable variations in validation accuracy between epochs 55 and 65.
Regarding the loss metric, training loss remained within the 0 to 0.15 range, while validation loss mostly stayed in that same range, with occasional spikes reaching up to 0.30. Nevertheless, the overall variation remained small. Although the trend shows a gradual increase in validation loss over the epochs, the fluctuation remains within a narrow band.
The confusion matrix in
Figure 9d clearly shows a strong contrast between correct and incorrect classifications, with only 3 FP among 5308 TN. This visual distinction highlights the model’s reliability in accurately identifying negative cases.
As summarized in
Table 9, accuracy, precision, specificity, and F1-score all exceed 99%, with the exception of recall, which reaches 98.59%. This means that, out of every 100 positive cases, more than 98 are correctly identified as TP, with fewer than two misclassified as FN.
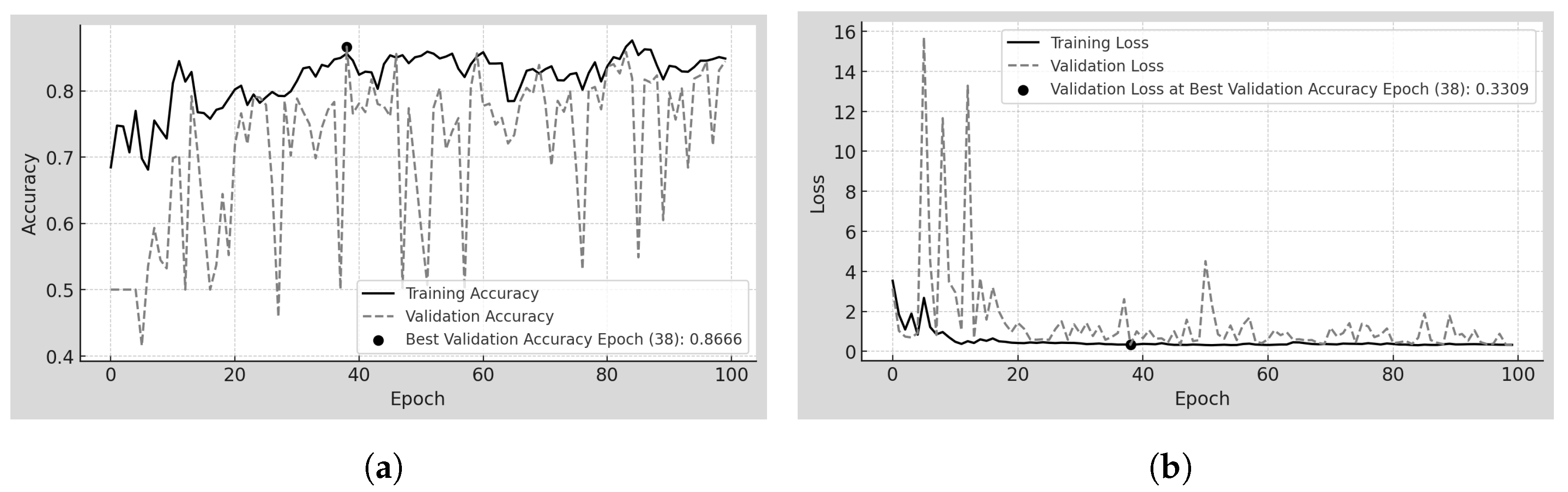
4.5. Scratch-100
Scratch-100 is the model trained from scratch on 100
m resolution mammograms from the MEXBreast dataset.
Table 8 and
Figure 7a,b show the training and validation behavior over 100 epochs analyzed. Regarding accuracy, a separation between training and validation accuracy is observed throughout most epochs, though this gap remains relatively small despite being visually noticeable. Training accuracy ranges from just below 0.7 to 0.9, while validation accuracy fluctuates between 0.4 and 0.9. However, both metrics converge at the optimal epoch (epoch 38), with the validation accuracy slightly exceeding the training accuracy, reaching 86.66%.
The limited dataset size, consisting of only 22,130 patches, including augmented samples, may have influenced the early stabilization of the model, as the best epoch occurred at 38 rather than later in training. Regarding loss, both training and validation follow a similar trend. However, between epochs 5 and 15, validation loss exhibits sharp peaks ranging from 1 to 16, suggesting higher variability in performance during this phase.
The confusion matrix of
Figure 9e provides further insight into the model’s classification performance on the testing dataset. Unlike previous models, the contrast between the white and black regions is less pronounced, with intermediate gray tones appearing, indicating a higher number of misclassified samples. The model correctly classifies 1981 TN and 1655 TP. However, it also yields 194 FP and 520 FN. The lighter shading in the FN cell suggests a higher number of missed positive cases than previous models, indicating greater difficulty detecting class 1 instances.
The overall pattern in the confusion matrix shows a tendency of the model to misclassify positive samples as negative more frequently than vice versa. This imbalance is influenced by the limited dataset size, as previously noted in the training behavior. The final accuracy and loss values for the Scratch-100 model, presented in
Table 8, provide an overview of the model’s classification performance across training, validation, and test datasets. The accuracy remains relatively stable, with values of 85.60% in training, 86.66% in validation, and 83.59% in testing, showing a slight decrease in the latter. The loss values remain close across datasets, with 0.3311 in training, 0.3309 in validation, and 0.3792 in testing. The increase in test loss compared to training and validation suggests performance degradation when evaluating previously unseen data.
Unlike previous models, where the loss function heavily penalizes predictions close to the 50% classification threshold, the less pronounced effect observed in Scratch-100 may be attributed to the limited number of samples, which could influence the overall distribution of predicted probabilities.
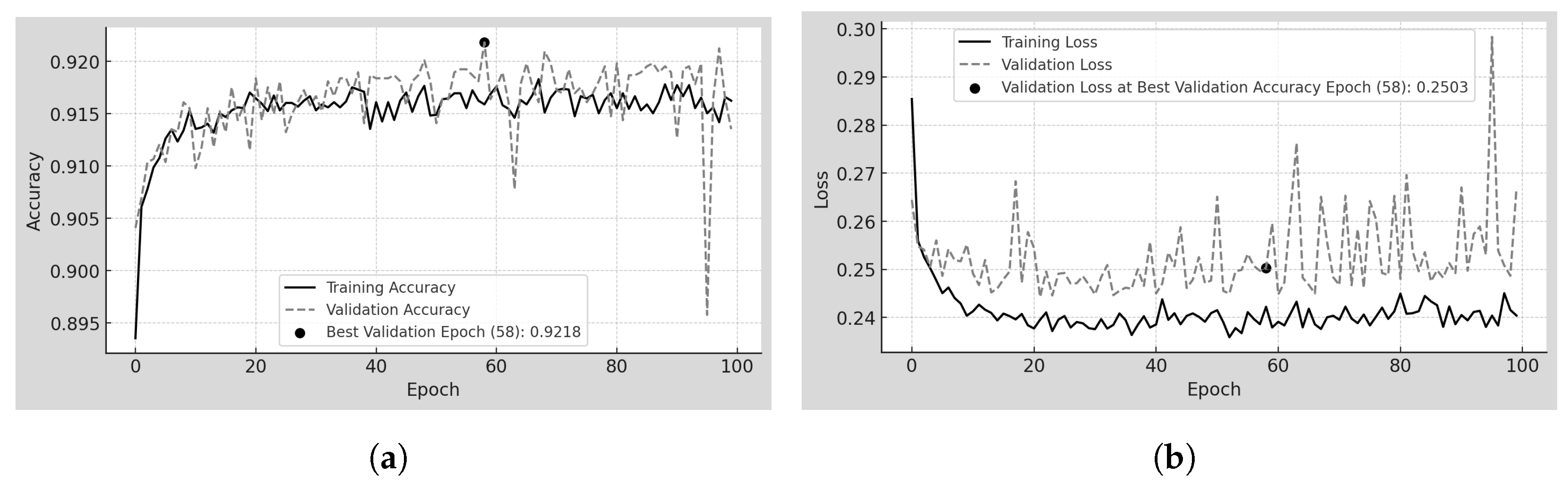
4.6. Transfer-100
Transfer-100 is the model using TL by freezing the backbone (
) of the pretrained 70
m model [
23] (trained on INbreast) and training on 100
m patches from the MEXBreast dataset. In
Table 8 and
Figure 8a,b the training and validation behavior over 100 epochs is analyzed. Unlike Scratch-100, the accuracy trends in training and validation follow a similar pattern, except for a few prominent peaks, particularly near epoch 100 in validation. However, accuracy remains within a narrow range, from 89.5% to 92.0%, indicating minimal variability. The model achieves its best validation accuracy early in training, reaching 92.18% at epoch 58.
Regarding loss, the trends in training and validation do not fully converge, but the separation between them is minimal. Training loss remains between 0.24 and 0.25, while validation loss fluctuates slightly between 0.25 and 0.26. Additionally, the overall loss values range only from 0.24 to 0.30, confirming the low variability in error throughout training.
The confusion matrix in
Figure 9f provides further insight into the model’s classification performance on the testing dataset. The correctly classified cases, 2055 TN and 1828 TP form a diagonal that, although not completely white, appears in a very light gray tone, indicating a high proportion of correctly classified samples.
The misclassified cases, 120 FP and 347 FN, are represented by darker shades, though not entirely black. The FN cell, in particular, appears in a dark gray tone, suggesting that the model still misclassified some positive cases as negative, but at a lower rate compared to other misclassifications scenarios. The overall distribution of values in the confusion matrix reflects a relatively stable classification performance, with a clear distinction between correctly and incorrectly classified samples.
The metrics in
Table 9 show the quantitative evaluation of the model’s classification effectiveness. The accuracy of 89.17% indicates that most of the samples were correctly classified. The model achieves a precision of 93.84%, meaning that in most of the cases class 1 is predicted correctly. The specificity of 94.48% further supports this. It indicates a strong ability to identify negative cases correctly (class 0).
The recall (sensitivity) of 84.05% suggests that the model correctly identifies most positive cases, though the presence of 347 FN in the confusion matrix indicates that some positive cases were misclassified as negative. The F1-score of 88.67% reflects a balance between precision and recall, confirming that the model maintains a stable trade-off between these two metrics.
4.7. Eval-70
Eval-70 is the model pretrained on the INbreast dataset at 70
m [
23], directly evaluated on the MEXBreast dataset at the same resolution.
Table 10 presents the accuracy and loss values for the Eval-70 model when tested on the INbreast and MEXBreast test datasets. Since this model was previously trained on INbreast, its performance on this dataset is a reference for evaluating its generalization to MEXBreast.
On the INbreast test dataset, the model achieves an accuracy of 99.84% with a loss of 0.00844. However, when tested on the MEXBreast test dataset, the accuracy decreases to 98.46%, and the loss increases to 0.17292. While the model still performs well, the increase in loss suggests a higher uncertainty in predictions on MEXBreast compared to INbreast.
The variation in loss values may be related to differences in the size and characteristics of the test datasets. The pretrained model on INbreast was trained using only 10,088 patches (including augmented data), while it was evaluated on 10,622 non-augmented patches from MEXBreast. Notably, the test set is even larger than the original training set. The increased size and diversity of the MEXBreast test set may contribute to the observed variability in prediction confidence.
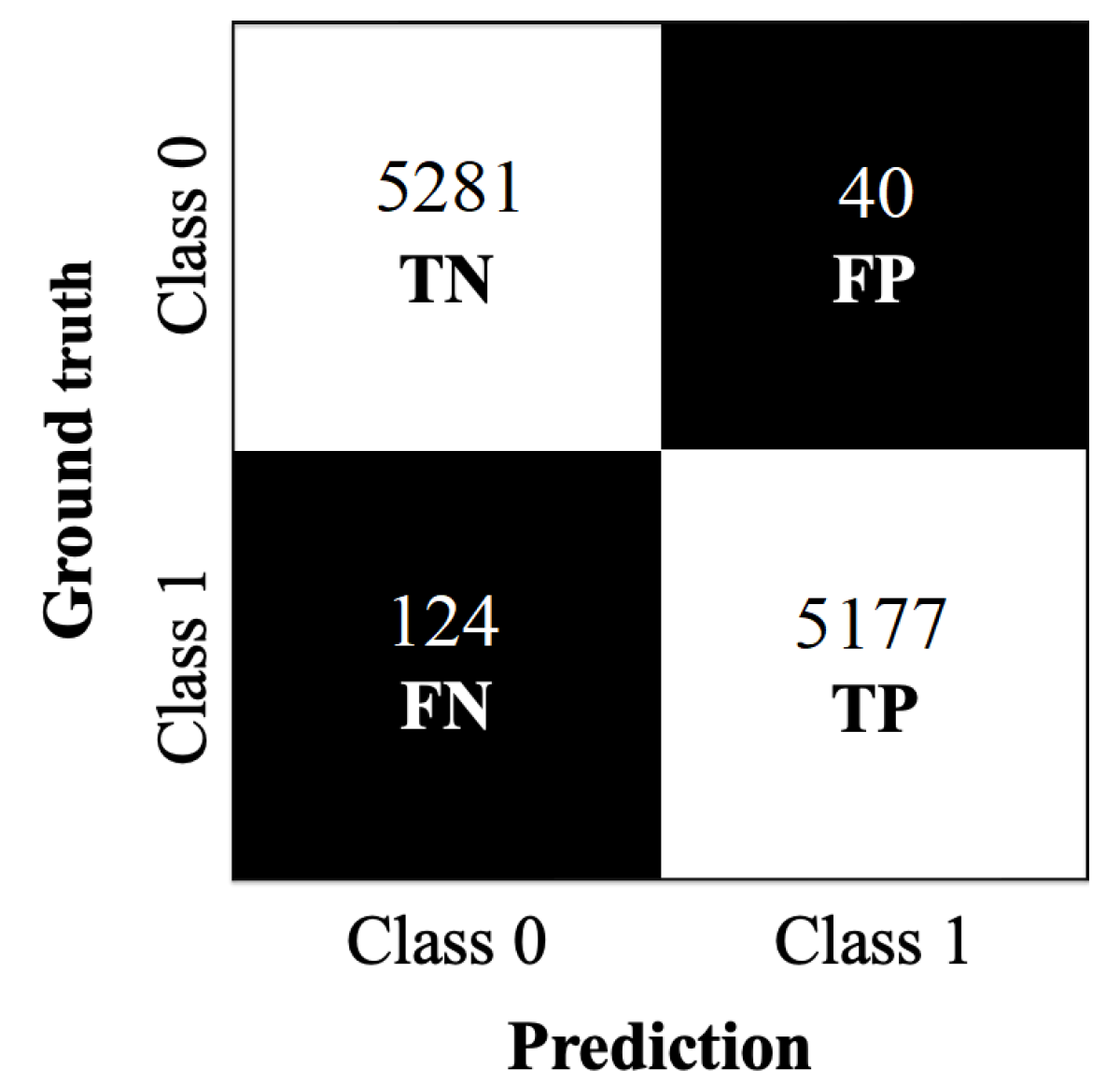
The confusion matrix in
Figure 10 provides further insight into the classification performance of the Eval-70 model on the MEXBreast test dataset. The correctly classified cases, 5281 TN and 5177 TP, form an entirely white diagonal, reflecting the large number of correct predictions in both classes.
The misclassified cases, 40 FP and 124 FN, appear in completely black regions, meaning that these errors represent only a small fraction of the total samples. The overall distribution of values in the confusion matrix visually confirms the model’s strong classification performance, with a well-defined separation between correctly and incorrectly classified samples.
The performance metrics in
Table 11 show the strong classification performance of the Eval-70 model on the MEXBreast test dataset with an accuracy of 98.46%. The model precision is 99.24%. The specificity of 99.25% shows that the model is highly reliable in identifying negative cases (class 0).
The recall (sensitivity) of 97.66% and the F1-score of 98.44% suggest that the model correctly identifies nearly all positive cases, and consequently, precision and recall are closely aligned.