MT-Tracker: A Phylogeny-Aware Algorithm for Quantifying Microbiome Transitions Across Scales and Habitats
Abstract
1. Introduction
2. Methods
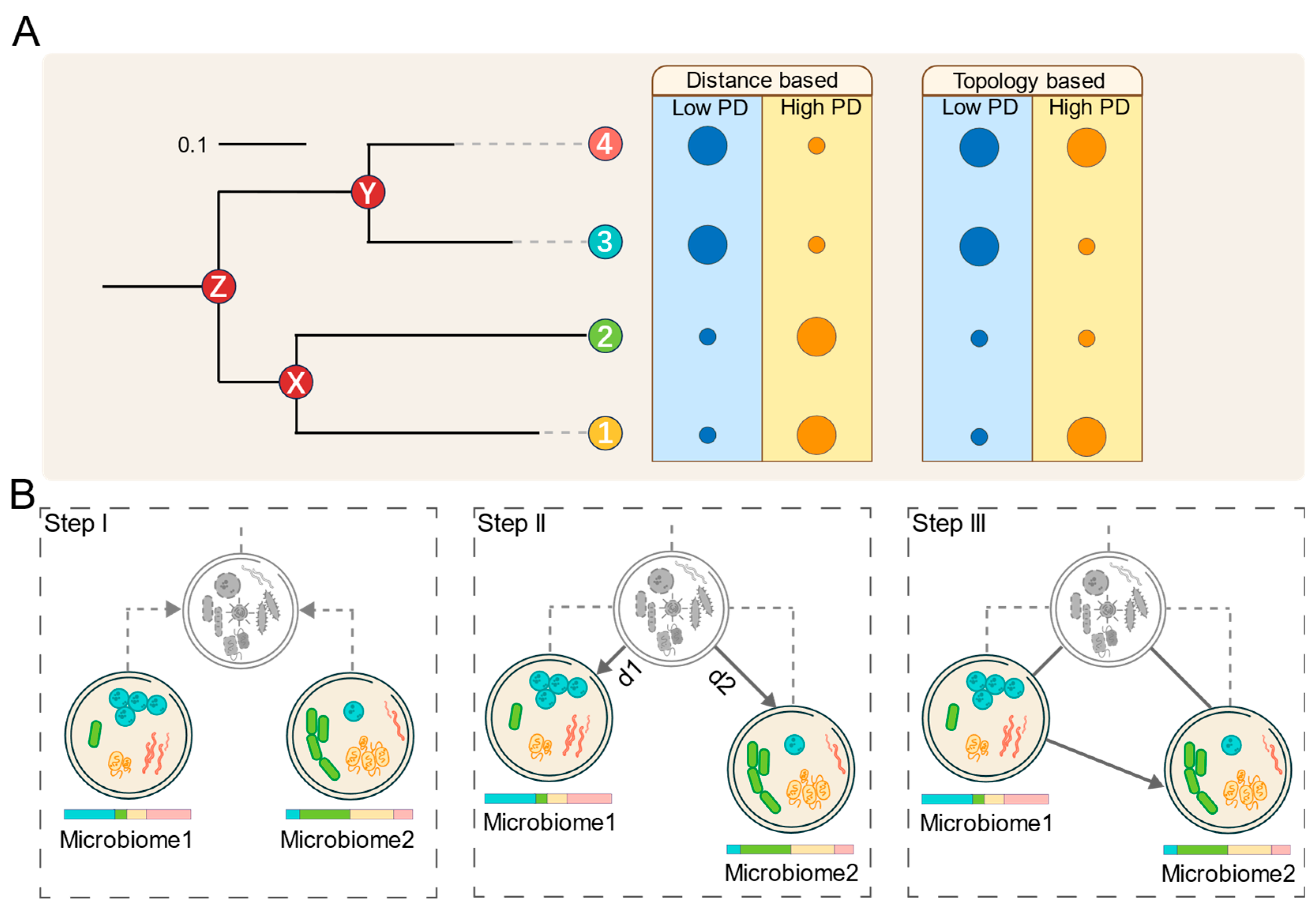
Hypothesis
- a.
- Short branch lengths, indicating minimal cumulative genetic variation.
- b.
- A compact topological structure, suggesting high kinship overlap.
- a.
- Long branch lengths, reflecting extensive cumulative genetic variation.
- b.
- A divergent topological structure, indicating low kinship overlap.
3. Algorithm
3.1. Microbiome Virtual Ancestor
| Algorithm 1. Reconstructing the microbiome virtual ancestor | ||
| Input: Output: | Relative abundance of 2 microbiomes: ABD 1 and ABD 2 Phylogeny tree with evolutionary distances: distp (child_node, ancestor_internal_node) Relative abundance of the virtual ancestor VA | |
| 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 | VA [node] ← 0 REM 1 [node], REM 2 [node] ← 0 #Initialization of the remaining abundance all nodes Function PostOrderTraversal (node): if node is tip node then Shared ← min (ABD 1 [node], ABD 2 [node]) VA [node] ← Shared REM 1 [node] ← ABD 1 [node]—Shared REM 2 [node] ← ABD 2 [node]—Shared end if if node is ancestor internal node then ANC 1, ANC 2 ← 0 #Initialize the abundance assigned to the ancestor internal node for child_node in node.children do PostOrderTraversal (child_node) ANC 1 ←ANC 1 + REM 1 [child_node] · (1—distp (child_node, node)) ANC 2 ←ANC 2 + REM 2 [child_node] · (1—distp (child_node, node)) end for Shared ← min (ANC 1, ANC 2) VA [node] ← Shared REM 1 [node] ←ANC 1—Shared REM 2 [node] ←ANC 1—Shared end if return VA | |
3.2. Transition Direction and Probability Between Two Microbiomes
3.3. Calibration of Transition Probability Among Multiple Microbiomes
4. Results
4.1. Evaluating Computational Efficiency of MT-Tracker and Phylogeny Tree-Based Approach
4.2. Validation of MT-Tracker by a Phylosymbiosis Analysis of Animal-Associated Habitat Microbiomes
4.3. A Transitional Perspective on the Global Microbiome Network
4.4. Uncovering Subtle Dynamic Trends in Short-Term Longitudinal Surveys
5. Conclusions and Discussion
6. Materials and Methods
6.1. Microbiome Sample Collection
6.2. Microbiome Profiling
6.3. Construction of Phylogeny Tree for Microbiomes
6.4. Inferring Transitional Direction Using Phylogeny Tree
6.5. Transition Network of the Global Microbiome
6.6. The Transition Trend of Oral Microbiome Transition over Time
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, J.; Kang, S.; Schadt, C.W.; Garten, C.T., Jr. Spatial scaling of functional gene diversity across various microbial taxa. Proc. Natl. Acad. Sci. USA 2008, 105, 7768–7773. [Google Scholar] [CrossRef] [PubMed]
- Chase, J.M.; Myers, J.A. Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 2351–2363. [Google Scholar] [CrossRef] [PubMed]
- Chase, J.M. Stochastic community assembly causes higher biodiversity in more productive environments. Science 2010, 328, 1388–1391. [Google Scholar] [CrossRef] [PubMed]
- Ofiţeru, I.D.; Lunn, M.; Curtis, T.P.; Wells, G.F.; Criddle, C.S.; Francis, C.A.; Sloan, W.T. Combined niche and neutral effects in a microbial wastewater treatment community. Proc. Natl. Acad. Sci. USA 2010, 107, 15345–15350. [Google Scholar] [CrossRef]
- Stegen, J.C.; Fredrickson, J.K.; Wilkins, M.J.; Konopka, A.E.; Nelson, W.C.; Arntzen, E.V.; Chrisler, W.B.; Chu, R.K.; Danczak, R.E.; Fansler, S.J. Groundwater–surface water mixing shifts ecological assembly processes and stimulates organic carbon turnover. Nat. Commun. 2016, 7, 11237. [Google Scholar] [CrossRef]
- Vellend, M. Conceptual synthesis in community ecology. Q. Rev. Biol. 2010, 85, 183–206. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Sloan, W.T.; Lunn, M.; Woodcock, S.; Head, I.M.; Nee, S.; Curtis, T.P. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 2006, 8, 732–740. [Google Scholar] [CrossRef]
- Martino, C.; Morton, J.T.; Marotz, C.A.; Thompson, L.R.; Tripathi, A.; Knight, R.; Zengler, K. A novel sparse compositional technique reveals microbial perturbations. mSystems 2019, 4, e00016-19. [Google Scholar] [CrossRef]
- Rizvi, A.H.; Camara, P.G.; Kandror, E.K.; Roberts, T.J.; Schieren, I.; Maniatis, T.; Rabadan, R. Single-cell topological RNA-seq analysis reveals insights into cellular differentiation and development. Nat. Biotechnol. 2017, 35, 551–560. [Google Scholar] [CrossRef]
- Lum, P.Y.; Singh, G.; Lehman, A.; Ishkanov, T.; Vejdemo-Johansson, M.; Alagappan, M.; Carlsson, J.; Carlsson, G. Extracting insights from the shape of complex data using topology. Sci. Rep. 2013, 3, 1236. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Deng, Y.; Zhang, P.; Xue, K.; Liang, Y.; Van Nostrand, J.D.; Yang, Y.; He, Z.; Wu, L.; Stahl, D.A.; et al. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc. Natl. Acad. Sci. USA 2014, 111, E836–E845. [Google Scholar] [CrossRef]
- Jing, G.; Zhang, Y.; Liu, L.; Wang, Z.; Sun, Z.; Knight, R.; Su, X.; Xu, J. A Scale-Free, Fully Connected Global Transition Network Underlies Known Microbiome Diversity. mSystems 2021, 6, e0039421. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef]
- Zhou, J.; He, Z.; Yang, Y.; Deng, Y.; Tringe, S.G.; Alvarez-Cohen, L. High-throughput metagenomic technologies for complex microbial community analysis: Open and closed formats. mBio 2015, 6, 02288-14. [Google Scholar] [CrossRef]
- Jeraldo, P.; Sipos, M.; Chia, N.; Brulc, J.M.; Dhillon, A.S.; Konkel, M.E.; Larson, C.L.; Nelson, K.E.; Qu, A.; Schook, L.B.; et al. Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proc. Natl. Acad. Sci. USA 2012, 109, 9692–9698. [Google Scholar] [CrossRef]
- Schweiger, O.; Klotz, S.; Durka, W.; Kühn, I. A comparative test of phylogenetic diversity indices. Oecologia 2008, 157, 485–495. [Google Scholar] [CrossRef]
- Dickerson, R.E. The structure of cytochrome c and the rates of molecular evolution. J. Mol. Evol. 1971, 1, 26–45. [Google Scholar] [CrossRef]
- Guénard, G.; Legendre, P.; Peres-Neto, P. Phylogenetic eigenvector maps: A framework to model and predict species traits. Methods Ecol. Evol. 2013, 4, 1120–1131. [Google Scholar] [CrossRef]
- Cadotte, M.; Albert, C.H.; Walker, S.C. The ecology of differences: Assessing community assembly with trait and evolutionary distances. Ecol. Lett. 2013, 16, 1234–1244. [Google Scholar] [CrossRef]
- Tucker, C.M.; Davies, T.J.; Cadotte, M.W.; Pearse, W.D. On the relationship between phylogenetic diversity and trait diversity. Ecology 2018, 99, 1473–1479. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Xu, J.; Ning, K. Meta-Storms: Efficient search for similar microbial communities based on a novel indexing scheme and similarity score for metagenomic data. Bioinformatics 2012, 28, 2493–2501. [Google Scholar] [CrossRef] [PubMed]
- Groussin, M.; Mazel, F.; Sanders, J.G.; Smillie, C.S.; Lavergne, S.; Thuiller, W.; Alm, E.J. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 2017, 8, 14319. [Google Scholar] [CrossRef]
- Revell, L.J.; Chamberlain, S.A. Rphylip: An R interface for PHYLIP. Methods Ecol. Evol. 2014, 5, 976–981. [Google Scholar] [CrossRef]
- Jing, G.; Liu, L.; Wang, Z.; Zhang, Y.; Qian, L.; Gao, C.; Zhang, M.; Li, M.; Zhang, Z.; Liu, X.; et al. Microbiome Search Engine 2: A Platform for Taxonomic and Functional Search of Global Microbiomes on the Whole-Microbiome Level. mSystems 2021, 6, 00943-20. [Google Scholar] [CrossRef]
- Brooks, A.W.; Kohl, K.D.; Brucker, R.M.; van Opstal, E.J.; Bordenstein, S.R. Phylosymbiosis: Relationships and Functional Effects of Microbial Communities across Host Evolutionary History. PLoS Biol. 2016, 14, e2000225. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N.; et al. Moving pictures of the human microbiome. Genome Biol. 2011, 12, R50. [Google Scholar] [CrossRef]
- Youngblut, N.D.; Reischer, G.H.; Walters, W.; Schuster, N.; Walzer, C.; Stalder, G.; Ley, R.E.; Farnleitner, A.H. Host diet and evolutionary history explain different aspects of gut microbiome diversity among vertebrate clades. Nat. Commun. 2019, 10, 2200. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Andrei, A.S.; Salcher, M.M.; Mehrshad, M.; Rychtecky, P.; Znachor, P.; Ghai, R. Niche-directed evolution modulates genome architecture in freshwater Planctomycetes. ISME J. 2019, 13, 1056–1071. [Google Scholar] [CrossRef] [PubMed]
- Battistuzzi, F.U.; Hedges, S.B. A major clade of prokaryotes with ancient adaptations to life on land. Mol. Biol. Evol. 2009, 26, 335–343. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Hayatsu, M.; Hosokawa, T.; Nagayama, A.; Tago, K.; Fukatsu, T. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 8618–8622. [Google Scholar] [CrossRef]
- Méheust, R.; Castelle, C.J.; Matheus Carnevali, P.B.; Farag, I.F.; He, C.; Chen, L.-X.; Amano, Y.; Hug, L.A.; Banfield, J.F. Groundwater Elusimicrobia are metabolically diverse compared to gut microbiome Elusimicrobia and some have a novel nitrogenase paralog. ISME J. 2020, 14, 2907–2922. [Google Scholar] [CrossRef]
- Banerjee, S.; van der Heijden, M.G.A. Soil microbiomes and one health. Nat. Rev. Microbiol. 2023, 21, 6–20. [Google Scholar] [CrossRef]
- Dodds, W.K.; Zeglin, L.H.; Ramos, R.J.; Platt, T.G.; Pandey, A.; Michaels, T.; Masigol, M.; Klompen, A.M.; Kelly, M.C.; Jumpponen, A. Connections and feedback: Aquatic, plant, and soil microbiomes in heterogeneous and changing environments. BioScience 2020, 70, 548–562. [Google Scholar] [CrossRef]
- Veach, A.M.; Dodds, W.K.; Jumpponen, A. Woody plant encroachment, and its removal, impact bacterial and fungal communities across stream and terrestrial habitats in a tallgrass prairie ecosystem. FEMS Microbiol. Ecol. 2015, 91, fiv109. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-González, C.; Niño-García, J.P.; Del Giorgio, P.A. Terrestrial origin of bacterial communities in complex boreal freshwater networks. Ecol. Lett. 2015, 18, 1198–1206. [Google Scholar] [CrossRef]
- Lv, H.; Yang, M.; Cheng, Y.; Li, K.; Ji, G.; Huang, T.; Wen, G. Disentangling the assembly patterns and drivers of microbial communities during thermal stratification and mixed periods in a deep-water reservoir. Sci. Total Environ. 2024, 946, 174398. [Google Scholar] [CrossRef]
- Morlon, H. Phylogenetic approaches for studying diversification. Ecol. Lett. 2014, 17, 508–525. [Google Scholar] [CrossRef] [PubMed]
- Kyrpides, N.C.; Eloe-Fadrosh, E.A.; Ivanova, N.N. Microbiome data science: Understanding our microbial planet. Trends Microbiol. 2016, 24, 425–427. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wu, Z.; Zhang, S.; Zhao, J. Cov-trans: An efficient algorithm for discontinuous transcript assembly in coronaviruses. BMC Genom. 2024, 25, 1257. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.R.; Kahn, S.; Kashyap, P.; Laine, L.; Rubin, D.; Atreja, A.; Moore, T.; Wu, G. Update on Fecal Microbiota Transplantation 2015: Indications, Methodologies, Mechanisms, and Outlook. Gastroenterology 2015, 149, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Ivanova, N.N.; Szeto, E.; Palaniappan, K.; Chu, K.; Dalevi, D.; Chen, I.M.; Grechkin, Y.; Dubchak, I.; Anderson, I.; et al. IMG/M: A data management and analysis system for metagenomes. Nucleic Acids Res. 2008, 36, D534–D538. [Google Scholar] [CrossRef]
- Chen, Y.; Li, J.; Zhang, Y.; Zhang, M.; Sun, Z.; Jing, G.; Huang, S.; Su, X. Parallel-Meta Suite: Interactive and rapid microbiome data analysis on multiple platforms. IMeta 2022, 1, e1. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Warshall, S. A theorem on boolean matrices. J. ACM 1962, 9, 11–12. [Google Scholar] [CrossRef]
- Kruskal, J.B. On the shortest spanning subtree of a graph and the traveling salesman problem. Proc. Am. Math. Soc. 1956, 7, 48–50. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]





| Datasets | No. of Samples | Description |
|---|---|---|
| MSE | 456,702 | Microbiome samples from the Microbiome Search Engine database (http://mse.ac.cn, accessed on 9 September 2024), encompassing diverse habitats including soil, water, plant rhizospheres, and animal intestines. Refer to Table 2 for details. |
| MAG | 280 | Mammal animal gut sample is from the dataset provided by Groussin et al. [24]. A dataset of 28 mammalian species containing deduplicated and non-chimeric 16S rRNA gene amplicons (V2 region). |
| SAG | 319 | Small animal gut samples come from the dataset provided by Brooks et al. [27]. Microbial community data from 24 species of four small animal types (white-footed mice, fruit flies, mosquitoes, and golden wasps), spanning multiple populations. |
| TS | 373 | The time series samples come from the dataset provided in the study by Caporaso et al. [28]. Longitudinal dataset of human microbiota collected across 396 time points from four body sites, providing detailed temporal dynamics of microbial communities. Qiita project number 550. |
| Sample Type | Habitat | Source | No. of Samples |
|---|---|---|---|
| Human-associated | Gut | Feces, etc. | 156,156 |
| Skin | Hand, arm, head, leg, etc. | 32,622 | |
| Oral | Tongue, saliva, plaque, etc. | 29,025 | |
| Other human body-site | Hair, lung, blood, eye, etc. | 16,188 | |
| Urogenital | Vagina, urine, etc. | 9741 | |
| Nose | Nostril | 8519 | |
| Animal-associated | Mammal animal | Mouse, rabbit, dog, deer, etc. | 96,024 |
| Nonmammal animal | Sponge, fish, insect, etc. | 29,223 | |
| Environmental | Building | Indoor environment, etc. | 17,636 |
| Soil | Grass cover, cropland, soil sediment, etc. | 19,070 | |
| Marine water | Sea water | 8018 | |
| Estuary | Estuary water, estuary sediment, etc. | 475 | |
| Lake | Lake water, lake sediment, etc. | 5022 | |
| Plant | Plant rhizosphere, plant surface, etc. | 6733 | |
| Freshwater | Blank control, tap water, etc. | 5014 | |
| Creek | Creek, small river, etc. | 92 | |
| River | River water, river sediment, etc. | 3631 | |
| Milk | Tanker milk, blended solo milk, etc. | 2883 | |
| Sand | Beach, desert, sand sediment, etc. | 968 | |
| Food | Food surface, etc. | 1981 | |
| Other | Other environment | Mock, hive air, etc. | 7681 |
| Total | 456,702 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Sun, Y.; Luo, W.; Hou, G.; Gao, H.; Su, X. MT-Tracker: A Phylogeny-Aware Algorithm for Quantifying Microbiome Transitions Across Scales and Habitats. Mathematics 2025, 13, 1982. https://doi.org/10.3390/math13121982
Zhu W, Sun Y, Luo W, Hou G, Gao H, Su X. MT-Tracker: A Phylogeny-Aware Algorithm for Quantifying Microbiome Transitions Across Scales and Habitats. Mathematics. 2025; 13(12):1982. https://doi.org/10.3390/math13121982
Chicago/Turabian StyleZhu, Wenjie, Yangyang Sun, Weiwen Luo, Guosen Hou, Hao Gao, and Xiaoquan Su. 2025. "MT-Tracker: A Phylogeny-Aware Algorithm for Quantifying Microbiome Transitions Across Scales and Habitats" Mathematics 13, no. 12: 1982. https://doi.org/10.3390/math13121982
APA StyleZhu, W., Sun, Y., Luo, W., Hou, G., Gao, H., & Su, X. (2025). MT-Tracker: A Phylogeny-Aware Algorithm for Quantifying Microbiome Transitions Across Scales and Habitats. Mathematics, 13(12), 1982. https://doi.org/10.3390/math13121982






