Abstract
Accurate polyp segmentation plays a vital role in the early detection and prevention of colorectal cancer. However, the diverse shapes, blurred boundaries, and varying sizes of polyps present significant challenges for automatic segmentation. Existing methods often struggle with effective local feature extraction and modeling long-range dependencies. To overcome these limitations, this paper proposes PolypFormer, which incorporates a local information enhancement module (LIEM) utilizing multi-kernel self-selective attention to better capture texture features, alongside dense channel attention to facilitate more effective feature fusion. Furthermore, a novel cross-shaped windows self-attention mechanism is introduced and integrated into the Transformer architecture to enhance the semantic understanding of polyp regions. Experimental results on five datasets show that the proposed method has good performance in polyp segmentation. On Kvasir-SEG datasets, mDice and mIoU reach 0.920 and 0.886, respectively.
Keywords:
polyp segmentation; local information enhancement; cross-shaped windows self-attention; medical image segmentation; deep learning MSC:
68T45
1. Introduction
Polyps are small, protruding growths originating from the mucosal surface and commonly occur in organs such as the colorectum, stomach, and nasal cavity. Although most polyps are benign lesions, certain types, such as adenomatous polyps, may develop into cancer over time. Therefore, early detection and removal of polyps play a vital role in the prevention of colorectal cancer [1]. Polyp segmentation in medical imaging, which in-volves the automatic identification and extraction of polyp regions, is a crucial step in disease prevention. Accurate polyp segmentation enables clinicians to clearly observe the size, shape, and location of polyps, thereby improving diagnostic accuracy, enhancing colorectal cancer screening efficiency, and reducing the workload of medical professionals [2]. During surgical procedures, segmentation results assist in precise localization of polyps to avoid missing lesions, thereby improving surgical outcomes. Moreover, in post-treatment efficacy evaluation, polyp segmentation can be used to monitor lesion changes and support informed clinical decision making [3]. Consequently, automated polyp segmentation technology is of great significance for reducing cancer risk, improving patient survival rates, and enhancing quality of life.
Traditional polyp segmentation methods primarily rely on simple analyses of surface features such as shape, texture, and clustering to perform basic image segmentation [4]. However, these approaches are limited in accuracy, but with the application of deep learning on medical images, polyp segmentation models have also been greatly advanced. FCN [5] performs image classification through pixel-wise prediction; however, when applied to medical image segmentation, it often results in significant loss of edge information. Since the widespread adoption of the U-Net architecture in biomedical image segmentation in 2015, a multitude of networks based on the encoder-decoder paradigm have emerged. Since the application of Transformer [6] technology to visual tasks, it has effectively overcome the limitations of CNNs in modeling long-range dependencies.
Despite the significant progress made by existing polyp segmentation models in the task of polyp segmentation, several challenges remain:
- Limited ability to capture detailed texture features: Current models struggle to effectively capture the detailed texture features of polyps, particularly when dealing with polyps that have complex shapes or unclear boundaries, which often leads to a decrease in segmentation accuracy;
- Challenges in modeling long-range dependencies: Long-range dependencies in polyp images, such as the overall shape of the polyp and its background context, are often difficult to model effectively. This is crucial for accurate segmentation of polyp regions, particularly in balancing local details with global context.
In response to the above problems, combined with the advantages and disadvantages of each segmentation model, this paper proposes the improved U-Net-based model PolypFormer for polyp segmentation, and the network model innovation and significance are as follows:
- Introduction of a local information enhancement module (LIEM): We design a local information enhancement module that strengthens texture feature extraction through a multi-kernel self-selective attention mechanism. During multi-kernel feature fusion, a dense channel attention mechanism is employed to increase the width of attention computations, enabling better modeling of relationships among different feature channels and enhancing the model’s understanding of polyp morphology and microstructural tissue;
- Replacement of the Transformer module with a cross-shaped windows self-attention mechanism: Considering that polyp regions often exhibit indistinct boundaries, we introduce a Transformer based on cross-shaped windows self-attention to improve the semantic dependency perception of polyp regions, thereby better adapting the Transformer to polyp segmentation tasks.
2. Related Work
2.1. CNN for Polyp Segmentation
With the rapid development of deep learning, convolutional neural networks (CNNs) have been introduced as advanced tools offering more complex feature representations, substantially improving colorectal polyp segmentation performance. Ronneberger et al. [7] proposed the U-Net architecture, which consists of an encoder and decoder connected in a U-shaped framework to facilitate image feature extraction and generate high-resolution segmentation maps. Zhou et al. [8] developed U-Net++, an improved version of U-Net, employing nested skip connections and deep supervision to optimize information flow and enhance the effective fusion of multi-resolution features and demonstrating excellent performance in handling blurred boundaries and complex details. ResUNet, proposed by Diakogiannis et al. [9], integrates residual connections and multi-scale feature extraction to address gradient vanishing issues and improve learning capability. Additionally, architectures such as nn-UNet [10], Attention U-Net [11], and ResUNet++ [12] represent other CNN-based medical image segmentation methods. These networks leverage the advantages of U-shaped architectures for feature extraction in medical images and achieve favorable results. Nevertheless, in polyp segmentation, microstructures and fine tissue details as well as semantic information within polyp regions are important. Previous U-shaped networks only extract relatively simple features and lack the capacity to capture deeper structural information within images.
2.2. Transformer for Polyp Segmentation
Vision Transformer (ViT) [13] and Swin Transformer [14], as commonly used backbone networks in visual tasks, employ different self-attention mechanisms to capture long-distance dependencies. ViT relies on self-attention to extract global information from feature maps, whereas Swin Transformer utilizes window-based self-attention and shifted window self-attention to focus on remote dependencies. Currently, Transformer architectures have also been widely adopted in medical image segmentation. Chen et al. proposed TransUnet [15], which combines the characteristics of Transformer and U-Net by leveraging the Transformer to extract global information while using U-Net’s convolutional feature maps to enhance local detail reconstruction accuracy, ensuring precise localization of targets. Cao et al. developed Swin-Unet [16], in which a hierarchical Swin Transformer encoder captures contextual features through shifted windows and recovers spatial resolution via a symmetric Swin Transformer decoder and patch expanding layers for upsampling. Lin et al. introduced Ds-TransUnet [17], integrating Swin Transformer into U-Net’s encoder and decoder with a dual-scale encoding mechanism and interactive fusion module, and designed a Transformer interaction fusion module to effectively fuse multi-scale information through self-attention, enabling modeling of non-local dependencies and multi-scale context. Other Transformer-based medical image segmentation models include Transfuse [18], UCTransNet [19], MT-UNet [20], and CoTr [21]. However, Transformer models face challenges such as slow convergence during polyp segmentation training. This is mainly because conventional Transformers expend computational resources on global attention, requiring large-scale data for convergence. In polyp segmentation tasks, the critical areas are the edge transition regions between polyps and the background. Conventional Transformers do not incorporate attention priors focused on these edge regions, instead concentrating attention computations globally. Duc et al. [22] proposed the ColonFormer network to address the limitations of existing convolutional network- and Transformer-based methods in polyp segmentation, such as modeling only local appearances or lacking multi-level feature representations with spatial dependencies. However, ColonFormer still employs a self-attention-based Transformer architecture, which results in slow convergence. To tackle this issue, Sanderson et al. [23] introduced a novel full-resolution segmentation framework that combines the strengths of Transformer feature extraction with the compensation ability of convolutional networks in full-resolution prediction by using a main branch and an auxiliary branch for feature extraction. Nevertheless, current research has yet to enhance the extraction of microstructural features in polyp tissues. Additionally, recent algorithms in polyp segmentation, such as SwinE-Net [24], Polypp-PVT [25], and TransResU-Net [26], utilize Vision Transformer or Swin Transformer as backbone networks but still fail to overcome the limitations of using patches as tokens. To enhance the network’s capability in extracting local features such as microstructures and microtextures and to better adapt the Transformer architecture to the morphological characteristics of polyp regions, this study proposes an improved model based on Unet, making it more suitable for polyp segmentation tasks.
3. Methodology
3.1. Overall Architecture
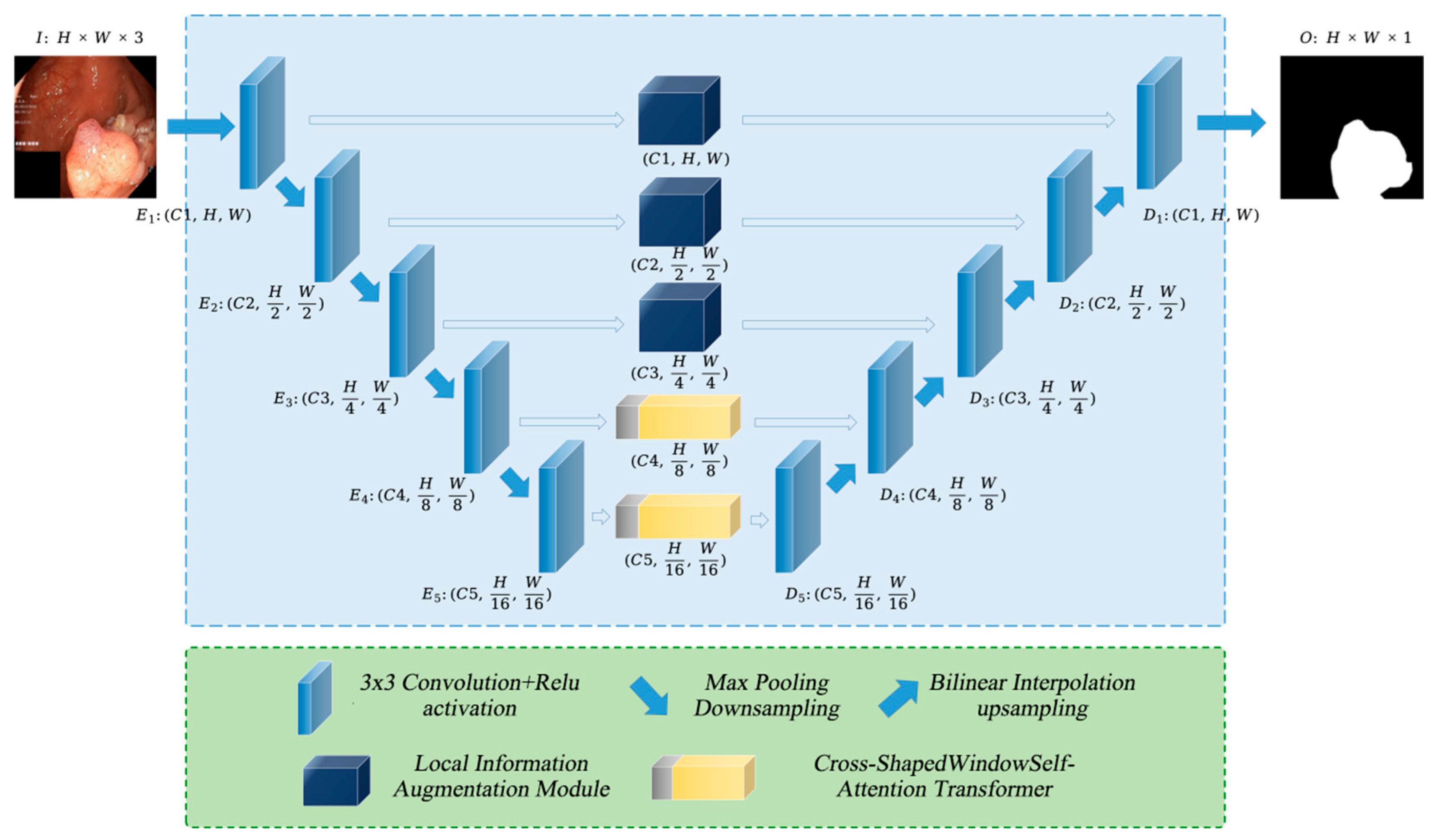
As illustrated in Figure 1, PolypFormer enhances feature extraction from polyp images through a local information enhancement module and a cross-grid self-attention Transformer. In the first three stages of the U-shaped network, the feature maps contain abundant texture detail information; therefore, the local information enhancement module is incorporated into the long skip connections to optimize feature extraction and facilitate transmission to the decoder. In the later stages of the network, the feature maps predominantly consist of high-level semantic information; thus, a cross-shaped windows self-attention Transformer is employed to improve the model’s ability to capture long-range dependencies within polyp regions and their adjacent areas.
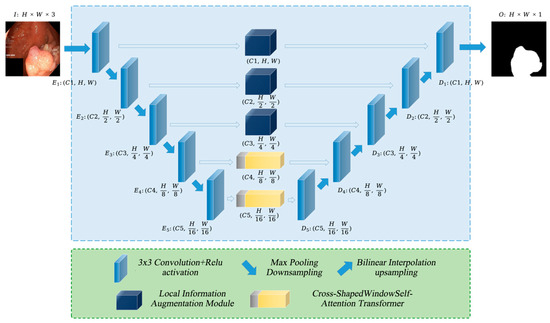
Figure 1.
The overall architecture of PolypFormer.
Assuming the input image is , for the -th stage, the feature map output by each encoder layer has dimensions i, and the output of each decoder layer has dimensions , where , and . After passing through the PolypFormer network, the resulting segmentation map is .
3.2. Local Information Enhancement Module
To strengthen the feature extraction capability of the network architecture, a local information enhancement module is introduced. This module is incorporated into the first three long skip connections of the PolypFormer network to enhance the perception of polyp microstructures and microtissues.
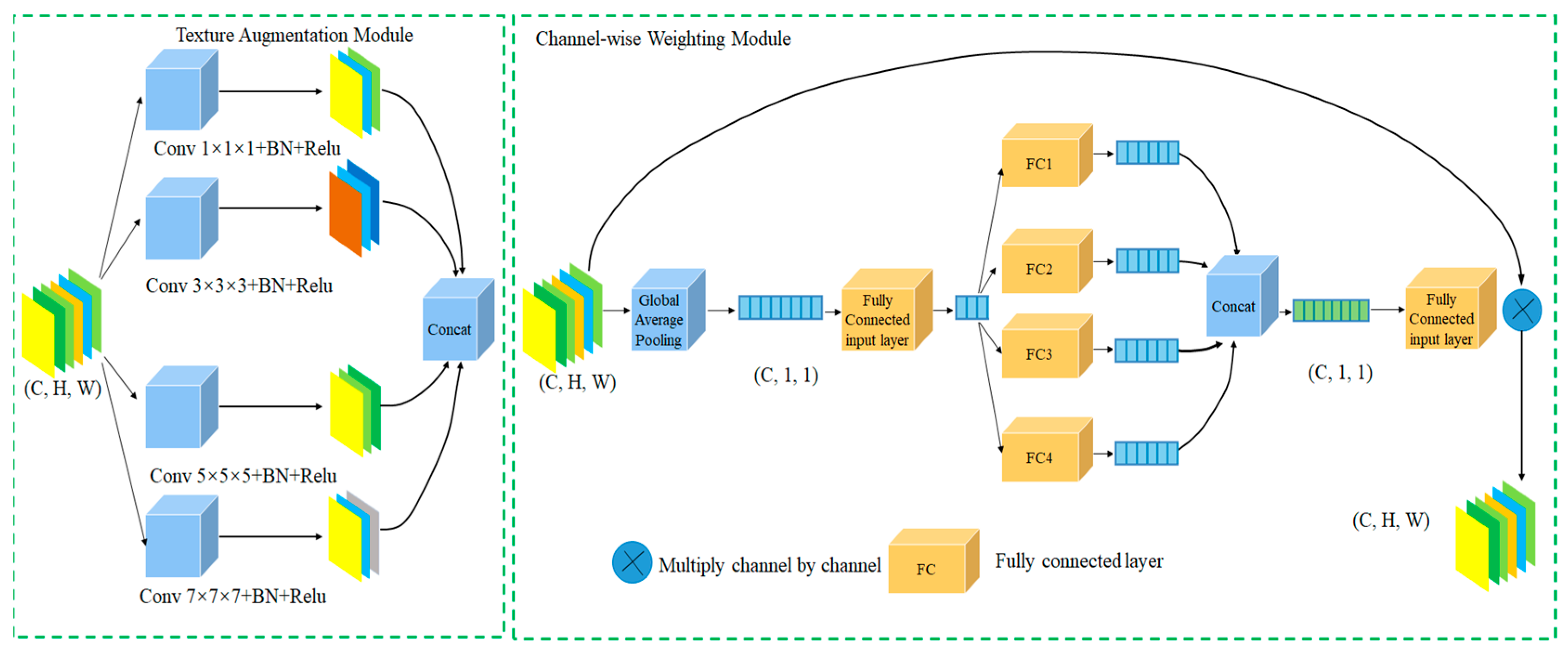
The local information enhancement module is divided into two parts, as illustrated in Figure 2.
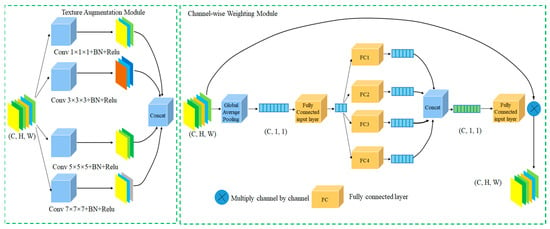
Figure 2.
Local Information Enhancement Module.
The first part is the texture information enhancement module. Given an input feature map , it is processed by four convolutional kernels of different sizes, namely 1 × 1, 3 × 3, 5 × 5, and 7 × 7, to more effectively capture texture features across different receptive fields. Each branch consists of a convolution layer, batch normalization (BN), and ReLU activation function. Features are extracted via these varied kernel sizes, resulting in four feature maps. These four feature maps are then concatenated as follows:
where represent the feature maps from the 1 × 1, 3 × 3, 5 × 5, and 7 × 7 convolutional branches, respectively; Concat denotes the concatenation operation; and is the concatenated feature map. After concatenation, the feature map is fed into the attention calculation layer.
The second part is the channel-wise attention weighting module. The input is the multi-scale feature map , which first undergoes global average pooling to produce . Then, is passed through a fully connected (FC) layer for initial perception without changing its scale. After the initial FC layer, four parallel FC branches are applied; each branch reduces its dimensionality to one quarter of the input channels, producing four intermediate feature maps: . Following these four FC branches, the resulting four feature maps are concatenated and then passed through an output FC layer for dimensionality reduction, yielding the attention score .
The multi-scale feature map is then passed through a 1 × 1 convolution to reduce the channel dimension to one quarter, resulting in . This is multiplied channel-wise by to produce the final locally enhanced feature map :
The global average pooling features are processed from multiple perspectives through four parallel fully connected (FC) branches, where each branch independently computes attention weights. This design enables the model to generate diverse intermediate representations, thereby producing a more expressive and informative attention map. Compared to a single deep FC layer, which may suffer from overfitting or reduced flexibility in modeling heterogeneous channels, our multi-branch structure improves feature channel interaction and enhances adaptivity to complex polyp textures. This approach also encourages a non-redundant activation pattern. It allows multiple semantic information such as edges, texture, and intensity to be expressed together.
The local information enhancement module thus consists of two parts: the first part performs multi-kernel convolutional feature extraction, where four scales of convolutional kernels enrich texture information representation. The second part is the attention calculation layer, inspired by the dense self-selective attention mechanism in SENetV2 [27]. The multi-branch structure facilitates perception of polyp feature information from multiple dimensions.
3.3. CSWin Self-Attention Transformer
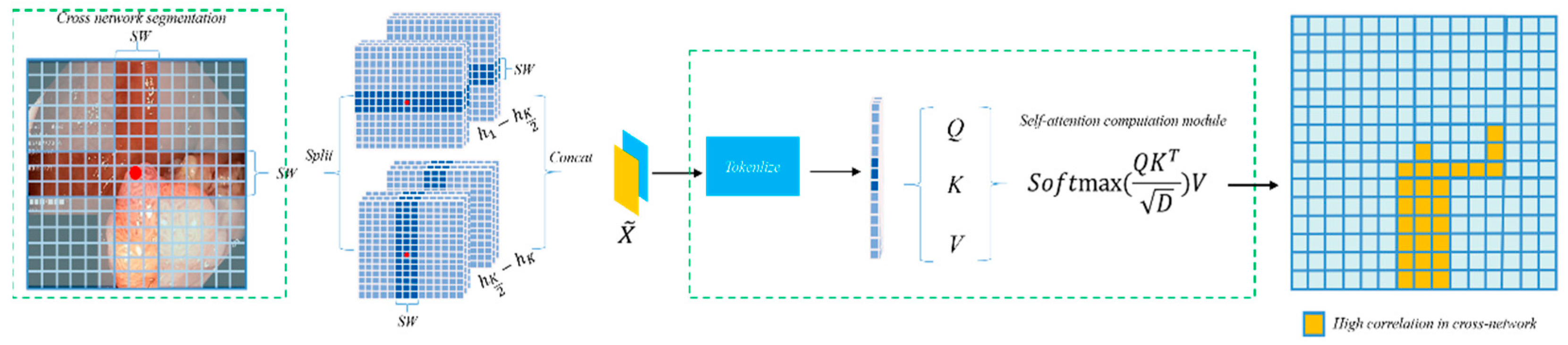
To address the critical issue of long-range feature modeling in polyp segmentation, this paper introduces a CSWin self-attention based Transformer module within PolypFormer to optimize long-distance dependency perception and accelerate convergence [28]. The CSWin self-attention mechanism computes features through horizontal and vertical strip interactions, thereby enhancing the capability of global information modeling.
A wider strip allows each attention head to capture longer spatial dependencies, thereby enhancing global context modeling. However, it also introduces increased computational cost due to the quadratic scaling of attention with respect to the strip length. Conversely, narrower strips reduce computational demand but may limit the receptive field, affecting the model’s ability to perceive long-range semantics. In our implementation, we used a fixed strip width to achieve a good trade-off between modeling power and efficiency. As illustrated in Figure 3, the input feature X undergoes a linear transformation and is divided into several non-overlapping strips of equal width [X1, X2, …, XM], where each strip contains sw × W patches, with sw representing the strip width. Multi-head self-attention computation is performed in parallel within each strip. For the multi-head self-attention calculation on the horizontal strips, the k-th head is represented by Equations (3)–(7):
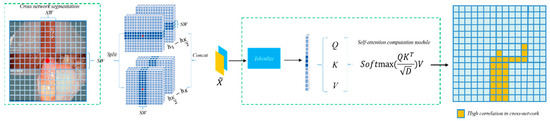
Figure 3.
CSWin Self-Attention.
Here, the matrices , , and denote the projection matrices for the query, key, and value of the k-th head, respectively. The self-attention computation for vertical strips is conducted in the same manner as for horizontal strips. In the multi-head attention calculation with K heads, the K self-attention heads are divided into two distinct groups, each containing K/2 heads. The first group is dedicated to self-attention computation on horizontal strips to capture both local and long-range horizontal features, while the second group handles self-attention on vertical strips to extract vertical correlations within the image. After completing the self-attention calculations on both horizontal and vertical strips, the multi-head self-attention operation is finalized.
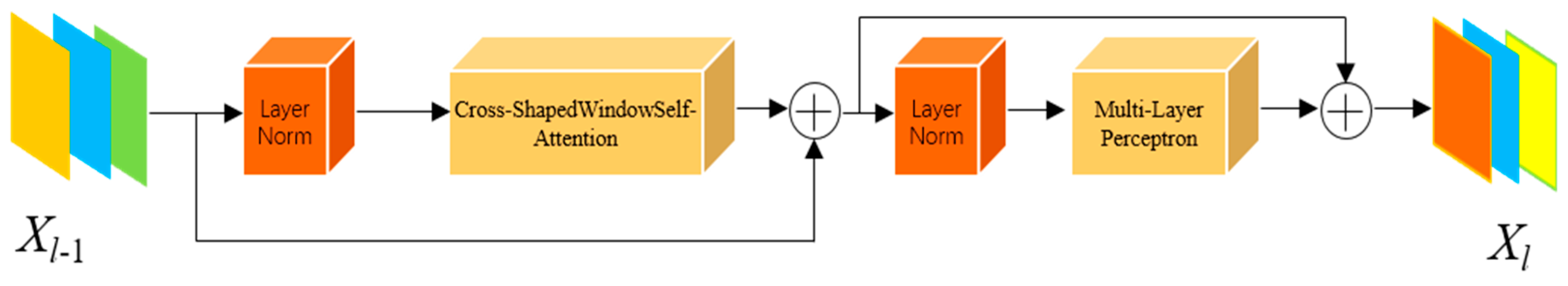
As shown in Figure 4, after completing the cross-grid self-attention computation, it is integrated into the Transformer architecture. This architecture consists of layer normalization (LN), the core component—cross-grid self-attention, and a two-layer Multilayer Perceptron (MLP). The LN layers are equipped with residual connections to mitigate gradient vanishing, while the MLP employs the GELU activation function to enhance nonlinear representation capability. The computation process is as follows:
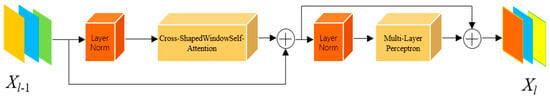
Figure 4.
CSWin Transformer Block.
In Equations (8) and (9), represents the input feature map, while denotes the output of the current layer. The feature maps are fed into the long-range dependency perception module within this structure. By integrating the cross-grid self-attention Transformer into the long skip connections of the last two stages of the U-shaped network, the attention computation within the polyp-adjacent regions is enhanced during feature extraction, thereby improving the semantic perception capability of the polyp regions.
4. Experiments and Results Analysis
In this study, we propose a polyp segmentation network, PolypFormer, and evaluate its effectiveness and robustness across five publicly available datasets. To provide a comprehensive assessment, we compare PolypFormer against various classical and state-of-the-art (SOTA) methods. Additionally, ablation studies and visualization analyses are conducted to further investigate the model’s performance. This section presents a detailed description of the evaluation setup and the corresponding results.
4.1. Experimental Datasets
This study utilizes five publicly available datasets: Kvasir-SEG [29], CVC-ClinicDB [30], CVC-ColonDB [31], EndoScene [32], and ETIS [33].
Kvasir-SEG contains 1000 polyp images with resolutions ranging from 332 × 487 to 1920 × 1072 pixels, exhibiting varying levels of quality and detail. CVC-ClinicDB includes 612 images with a resolution of 384 × 288 pixels, primarily intended for polyp detection in colonoscopy videos. The EndoScene dataset is a composite of CVC-ClinicDB and CVC300, comprising 912 annotated images that span multiple image categories. CVC-ColonDB consists of 380 images extracted from 15 colonoscopy sequences, each with a resolution of 574 × 500 pixels. ETIS contains 192 polyp images of 1225 × 996 pixels, collected from 29 colonoscopy sequences, all with consistent resolution.
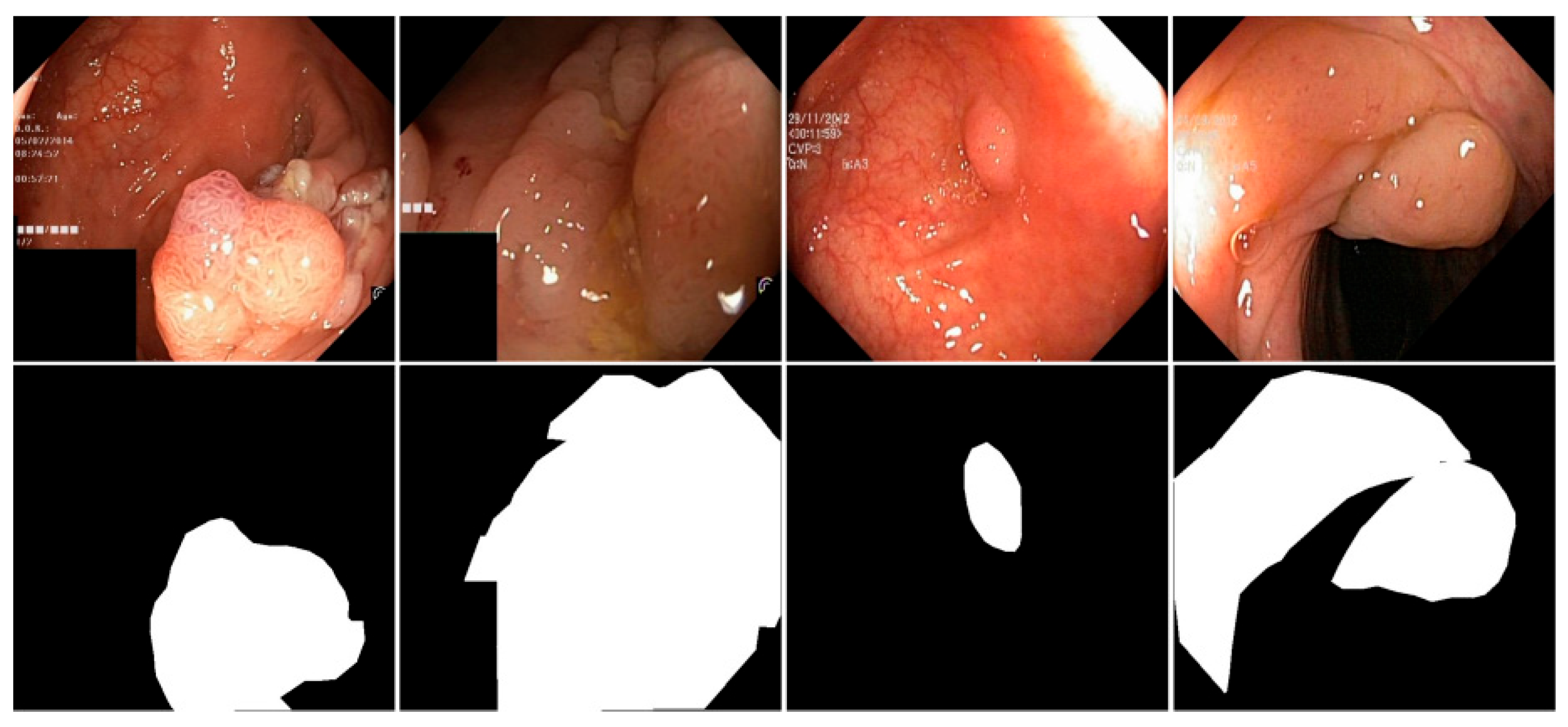
As shown in Figure 5, several representative examples from the Kvasir-SEG and CVC-ClinicDB datasets are presented, including both input images and their corresponding segmentation labels.
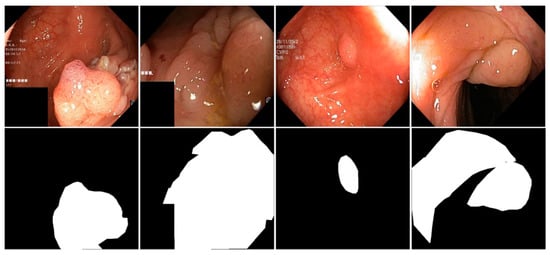
Figure 5.
Images and Segmentation Labels in the Kvasir-SEG and CVC-ClinicDB datasets.
4.2. Experimental Setup and Evaluation Metrics
The proposed PolypFormer was implemented using PyTorch 1.10, along with five baseline models including U-Net, all re-implemented within the MMsegmentation framework to ensure fair comparison. All models were trained with identical input resolutions, preprocessing pipelines, training epochs, loss functions, and evaluation metrics, without using any pretrained weights. Training was conducted on four NVIDIA Quadro RTX 8000 GPUs. The input images were resized to 384 × 384 and normalized with a mean of 0 and a standard deviation of 1. Data augmentation techniques such as random flipping, photometric distortion, padding, and random warping were applied. The Adam optimizer was used with an initial learning rate of 1 × 10−4, and a polynomial decay strategy with a power of 0.9 was adopted. The models performed 5000 iterations with a batch size of 8. Model performance was evaluated using four commonly adopted metrics: mean intersection over union (mIoU), Dice coefficient, precision, and recall, providing a comprehensive assessment of segmentation accuracy, region overlap, and the model’s precision and sensitivity.
For the loss function, considering the binary classification nature of the polyp segmentation task, binary cross-entropy (BCE) was selected as the primary loss function. In addition, Dice loss was incorporated to improve overlap between predicted segmentation and ground truth. BCE effectively addresses binary classification, while Dice loss enhances region consistency. The combination of both contributes to improved segmentation performance and model robustness. The total loss function is formulated as follows:
O denotes the predicted segmentation map by the network, G represents the ground truth label, is the binary cross-entropy loss, and is the Dice loss.
4.3. Ablation Study
PolypFormer integrates a local information enhancement module and a cross-grid self-attention Transformer, with the latter incorporating the cross-grid self-attention mechanism. To analyze the contribution of each component, ablation experiments were conducted on the Kvasir-SEG and CVC-ClinicDB datasets. The results are summarized in Table 1.

Table 1.
The ablation experiment results on the Kvasir-SEG dataset and CVC-ClinicDB datasets.
From the Kvasir-SEG dataset experiments, the baseline model achieved a Dice coefficient of 0.871, mIoU of 0.839, and precision and recall of 0.912 and 0.828, respectively. Incorporating the traditional convolutional, the Dice improved to 0.883, mIoU reached 0.852, and Precision and Recall increased to 0.913 and 0.848, respectively. After incorporating the local information enhancement module, the Dice improved to 0.902, mIoU reached 0.866, and precision and recall increased to 0.916 and 0.868, respectively, indicating the module’s effectiveness in enhancing local feature extraction and proving that the multi-kernel self-selective attention mechanism is superior to traditional convolution. Further, introducing a self-attention Transformer resulted in a Dice of 0.884 and mIoU of 0.845; although precision and recall were slightly improved, the overall gain was limited. This suggests that while global feature extraction offers certain benefits, its impact is not substantial. However, integrating the CSWin self-attention Transformer boosted the Dice to 0.907 and mIoU to 0.871, with precision and recall of 0.902 and 0.884, respectively, verifying the enhancement of segmentation performance through cross-grid attention. Finally, combining both modules in the full PolypFormer architecture yielded the best results, with a Dice coefficient of 0.920, mIoU of 0.886, precision of 0.913, and recall of 0.902, demonstrating a significant improvement in polyp segmentation performance.
In the CVC-ClinicDB dataset ablation study, the baseline model achieved a Dice of 0.910, mIoU of 0.879, precision of 0.898, and recall of 0.892. After incorporating the traditional convolutional, the Dice improved to 0.912, mIoU reached 0.881, and precision and recall increased to 0.899 and 0.894, respectively. Incorporating the local information enhancement module increased the Dice to 0.915 and mIoU to 0.884, with precision and recall improving to 0.902and 0.896, highlighting enhanced capability in capturing fine details and complex boundaries. When the self-attention Transformer was added, the model achieved a Dice of 0.913 and mIoU of 0.882, with precision and recall of 0.900 and 0.894, showing marginal improvement but proving less effective than the local enhancement. Introducing the CSWin self-attention Transformer further elevated the Dice to 0.919, mIoU to 0.889, and precision and recall to 0.909 and 0.905, respectively, significantly boosting segmentation capability. Ultimately, the proposed PolypFormer achieved the best performance, with a Dice of 0.928, mIoU of 0.899, precision of 0.918, and recall of 0.935, showcasing the combined advantages of both local enhancement and cross-grid self-attention mechanisms.
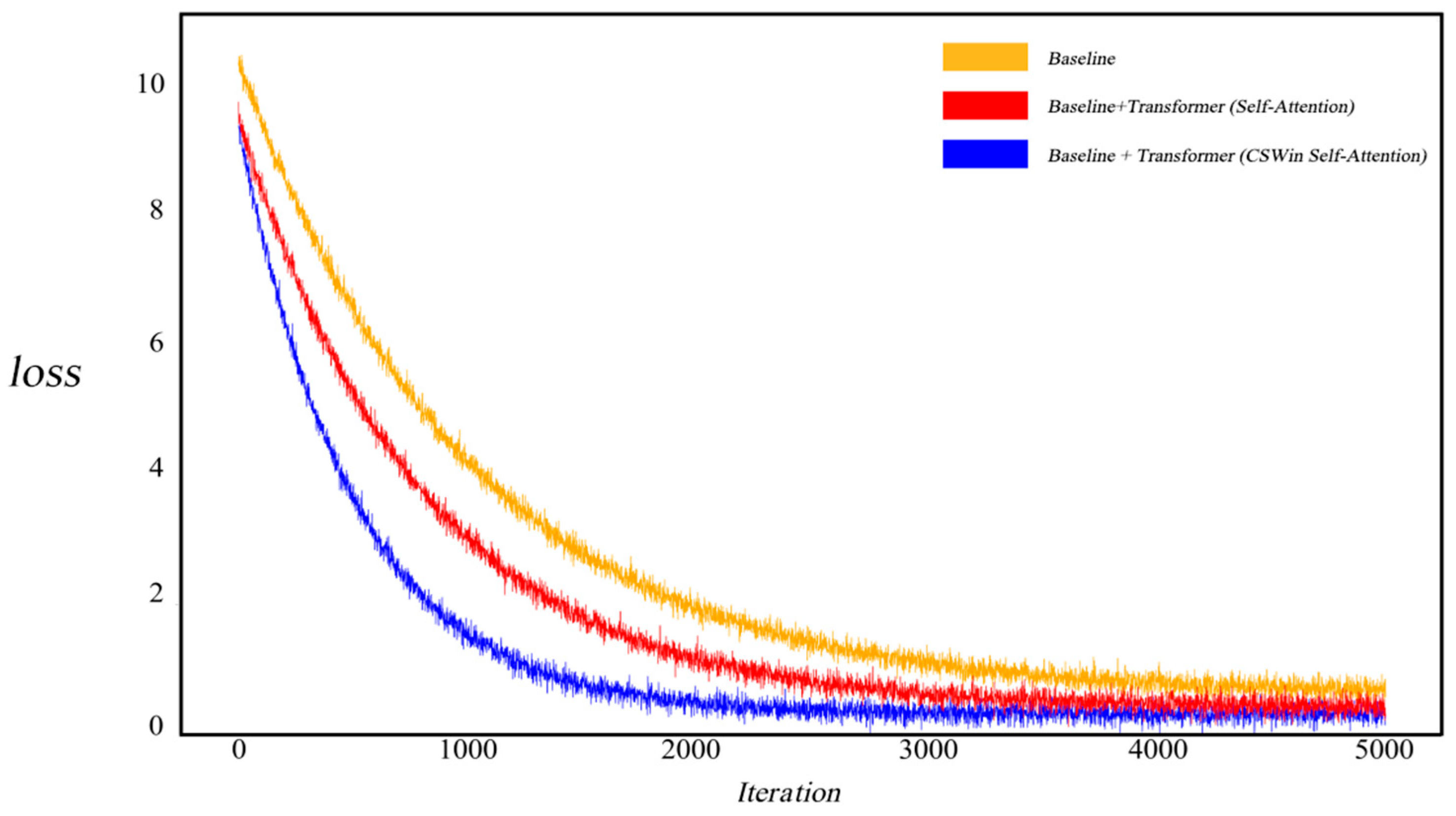
As illustrated in Figure 6, to investigate the impact of the CSWin self-attention mechanism on model convergence, we visualized the training loss curves. When employing a standard self-attention-based Transformer, the model exhibited relatively slower convergence. In contrast, after integrating the cross-grid self-attention Transformer, the model demonstrated accelerated convergence and a notably reduced loss value, indicating that this mechanism can effectively enhance training efficiency and improve segmentation accuracy.
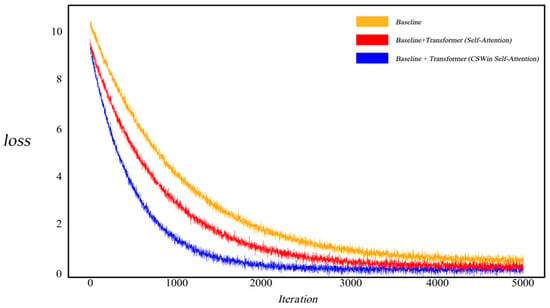
Figure 6.
Images and Segmentation Labels in the Kvasir-SEG and CVC-ClinicDB Datasets. Comparison of Loss Variation between Self-Attention-based Transformer and CSWin Self-Attention-based Transformer.
As shown in Figure 7, after training, we converted the output features of the CSWin Transformer into heatmaps to intuitively visualize the network’s attention to different regions during feature extraction. The results indicate that, with the incorporation of the CSWin self-attention mechanism, high-response regions are primarily concentrated around the polyp areas, suggesting that the model effectively focuses on the target regions. Compared with conventional methods, this structure demonstrates superior capability in capturing the shape, location, and boundary information of polyps, thereby significantly enhancing the model’s semantic perception and performance in polyp segmentation tasks.

Figure 7.
Visualization of the feature maps generated by the CSWin Transformer as heatmaps.
4.4. Comparative Experiments
In the comparative experiments, we implemented PolypFormer and reproduced five baseline algorithms, including the convolutional neural network-based U-Net, the Transformer-based TransUnet and SwinUnet, as well as two recent state-of-the-art (SOTA) methods in the field of polyp segmentation: PraNet and Polyp-PVT. These models were employed to objectively evaluate the performance of PolypFormer. The results are summarized in Table 2.

Table 2.
The comparative Experiment results on the Kvasir-SEG dataset and CVC-ClinicDB datasets.
Table 2 presents the comparative experimental results on the Kvasir-SEG and CVC-ClinicDB datasets.
On the Kvasir-SEG dataset, PolypFormer achieved the highest Dice coefficient of 0.920, outperforming both Polyp-PVT and PraNet, thus demonstrating superior segmentation accuracy. In contrast, conventional methods such as U-Net and U-Net++ obtained lower Dice scores of 0.895 and 0.878, respectively. PolypFormer also led in the mIoU metric, reaching 0.886, surpassing Polyp-PVT (0.860) and PraNet (0.858). In terms of precision, PolypFormer and Polyp-PVT achieved comparable and high scores of 0.913 and 0.912, respectively. PolypFormer also attained the highest recall of 0.938, indicating strong sensitivity. Both PraNet and Polyp-PVT fell slightly behind in recall. TransUnet and U-Net++ performed poorly in recall, which may contribute to missed detections of small or low-contrast polyps.
On the CVC-ClinicDB dataset, PolypFormer again achieved the highest Dice coefficient of 0.928, indicating outstanding performance. Polyp-PVT and PraNet followed closely, while U-Net (0.839) and U-Net++ (0.878) showed relatively inferior performance, suggesting that advancements in network architecture and attention mechanisms are more effective in handling complex boundaries and morphological variations. In terms of mIoU, PolypFormer remained in the lead with a score of 0.899, clearly outperforming Polyp-PVT (0.887) and PraNet (0.882). SwinUnet, with an mIoU of only 0.795, showed a limited ability to capture spatial information. Regarding precision, PolypFormer (0.918), Polyp-PVT (0.910), and ResUnet (0.908) all demonstrated excellent performance, while SwinUnet’s precision was relatively low (0.833), suggesting limitations in fine-detail representation. PolypFormer also achieved the highest recall of 0.935, reflecting its exceptional sensitivity. Overall, PolypFormer demonstrated robust and superior performance on the CVC-ClinicDB dataset, validating the effectiveness of the local information enhancement module and CSWin self-attention Transformer in improving polyp segmentation.
In Table 3, we compare the performance of PolypFormer against existing methods on three commonly used test datasets: CVC-ColonDB, EndoScene, and ETIS. The experimental results demonstrate that PolypFormer achieves superior performance across all evaluation metrics. Notably, in terms of Dice coefficient and mIoU, PolypFormer attained scores of 0.917, 0.852, and 0.693 and 0.617, 0.911, and 0.868 on the CVC-ColonDB, EndoScene, and ETIS datasets respectively, significantly outperforming current state-of-the-art methods such as Polypp-PVT and PraNet. These results indicate that by integrating the local information enhancement module and cross-grid self-attention mechanism, PolypFormer effectively improves the handling of complex boundaries and microstructures in polyp images, leading to more precise segmentation outcomes. Compared to traditional algorithms like U-Net, PolypFormer demonstrates substantial performance gains across diverse datasets, validating its potential and advantage in polyp segmentation tasks.

Table 3.
The comparative experiment results on the CVC-ColonDB, EndoScene, and ETIS datasets.
4.5. Model Complexity
Table 4 provides a comparative overview of the parameter count and floating-point operations per second for PolypFormer and several representative segmentation models.

Table 4.
Comparison of parameter count and computational cost across different segmentation models.
U-Net contains approximately 31.54 M parameters with a computational cost of 74.77 G, while ResUNet requires 34.23 M parameters and 84.57 G. These conventional convolution-based models are computationally lightweight but typically lack the capacity to capture long-range dependencies, which limits their segmentation performance in complex scenes. FCN, based on a ResNet-50 backbone, has 33.30 M parameters and 80.05 G FLOPs, also remaining within the lightweight category. In contrast, transformer-based models significantly increase both parameter count and computational burden. TransUNet reaches 105.93 M parameters with 154.26 G FLOPs, while Swin-Unet comprises 62.38 M parameters and 124.10 G FLOPs. Polyp-PVT, utilizing the PVTv2-B2 backbone, provides a more balanced design with 52.91 M parameters and 99.01 G FLOPs. The proposed PolypFormer integrates a cross-shaped window attention mechanism based on CSWin along with a local information enhancement module (LIEM), resulting in a total complexity of 85.62 M parameters and 105.32 G FLOPs. Although not the most lightweight among the compared models, PolypFormer achieves a favorable trade-off between accuracy and computational cost. It consistently delivers superior segmentation performance compared to both lighter convolutional networks and heavier transformer-based architectures.
4.6. Visualization Analysis
To provide a qualitative assessment of PolypFormer, segmentation results of multiple algorithms on the Kvasir-SEG and CVC-ClinicDB datasets are presented in Figure 8 and Figure 9, respectively.

Figure 8.
Partial visualization of segmentation results on the Kvasir-SEG dataset.

Figure 9.
Partial visualization of segmentation results on the CVC-ClinicDB dataset.
In Figure 8 and Figure 9, the convolutional neural network-based method U-Net commonly exhibits missed detections and false positives, resulting in noticeable holes or artifacts in the segmentation outputs and overall poor segmentation quality. Such issues not only affect the accuracy of the results but also reduce the model’s reliability in practical applications. In contrast, Transformer-based methods demonstrate relatively better performance in polyp segmentation tasks. These methods more effectively capture complex structures and fine details within the images, significantly reducing missed detections and false positives, thereby ensuring the integrity and accuracy of the segmentation results. The proposed PolypFormer achieves segmentation results closest to the ground truth, highlighting its superiority in handling complex images and extracting effective features. In summary, through comparative analysis, it is evident that PolypFormer delivers outstanding performance in polyp segmentation tasks.
5. Conclusions
In this paper, we propose a novel polyp segmentation network, PolypFormer, which significantly enhances segmentation performance by integrating a local information enhancement module and a CSWin self-attention Transformer. The local information enhancement module effectively captures detailed texture features within images, while the CSwin self-attention Transformer strengthens the model’s semantic understanding. Compared with conventional self-attention Transformers, the CSWin self-attention Transformer demonstrates faster convergence and stronger feature perception. The combination of these two components results in a powerful model capable of comprehensive analysis of polyp images. Extensive experiments on the Kvasir-SEG and CVC-ClinicDB datasets show that PolypFormer achieves outstanding results across multiple evaluation metrics, highlighting its potential in polyp segmentation tasks.
PolypFormer not only overcomes the limitations of traditional U-shaped networks but also offers new insights and methodologies for future medical image segmentation research. With ongoing model refinement and clinical validation, PolypFormer is expected to play a significant role in practical applications, advancing the field of medical image analysis.
Author Contributions
Conceptualization, L.F.; methodology, L.F.; software, L.F.; validation, L.F.; formal analysis, L.F.; investigation, L.F.; resources, L.F.; data curation, L.F.; writing—original draft preparation, L.F. and Y.J.; writing—review and editing, L.F. and Y.J.; visualization, L.F.; supervision, L.F. and Y.J.; project administration, L.F. and Y.J.; funding acquisition, L.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
This study utilizes publicly accessible anonymized datasets, including Kvasir-SEG (https://datasets.simula.no/kvasir-seg/), CVC-ClinicDB (https://polyp.grand-challenge.org/CVCClinicDB/), CVC-ColonDB (http://vi.cvc.uab.es/colon-qa/cvccolondb/), EndoScene (https://pages.cvc.uab.es/CVC-Colon/), and ETIS-Larib Polyp DB (https://www.kaggle.com/datasets/nguyenvoquocduong/etis-laribpolypdb). These datasets are publicly available for academic use and were utilized in full compliance with their respective licenses.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gupta, M.; Mishra, A. A systematic review of deep learning based image segmentation to detect Polypp. Artif. Intell. Rev. 2024, 57, 7. [Google Scholar] [CrossRef]
- Sánchez-Peralta, L.F.; Bote-Curiel, L.; Picón, A.; Sánchez-Margallo, F.M.; Pagador, J.B. Deep learning to find colorectal Polypps in colonoscopy: A systematic literature review. Artif. Intell. Med. 2020, 108, 101923. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lv, F.; Chen, C.; Hao, A.; Li, S. Colorectal Polypp Segmentation in the Deep Learning Era: A Comprehensive Survey. arXiv preprint 2024, arXiv:2401.11734. [Google Scholar]
- Sasmal, P.; Bhuyan, M.K.; Dutta, S.; Iwahori, Y. An unsupervised approach of colonic Polypp segmentation using adaptive markov random fields. Pattern Recognit. Lett. 2022, 154, 7–15. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Vaswani, A. Attention is all you need. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Proceedings 4. Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C. ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm. Remote Sens. 2020, 162, 94–114. [Google Scholar] [CrossRef]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; De Lange, T.; Halvorsen, P.; Johansen, H.D. Resunet++: An advanced architecture for medical image segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; IEEE: New York, NU, USA, 2019; pp. 225–2255. [Google Scholar]
- Sharir, G.; Noy, A.; Zelnik-Manor, L. An image is worth 16 × 16 words, what is a video worth? arXiv 2021, arXiv:2103.13915. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF international conference on computer vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer Nature: Cham, Switzerland, 2023; pp. 205–218. [Google Scholar]
- Lin, A.; Chen, B.; Xu, J.; Zhang, Z.; Lu, G.; Zhang, D. Ds-transunet: Dual swin transformer u-net for medical image segmentation. IEEE Trans. Instrum. Meas. 2022, 71, 1–15. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, H.; Hu, Q. Transfuse: Fusing transformers and CNNs for medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24. Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 14–24. [Google Scholar]
- Wang, H.; Cao, P.; Wang, J.; Zaiane, O.R. Uctransnet: Rethinking the skip connections in u-net from a channel-wise perspective with transformer. In Proceedings of the AAAI Conference on Artificial Intelligence, Online, 22 February–1 March 2022; Volume 36, pp. 2441–2449. [Google Scholar]
- Jha, A.; Kumar, A.; Pande, S.; Banerjee, B.; Chaudhuri, S. Mt-unet: A novel u-net based multi-task architecture for visual scene understanding. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Abu Dhabi, United Arab Emirates, 25–28 October 2020; IEEE: New York, NU, USA, 2020; pp. 2191–2195. [Google Scholar]
- Xie, Y.; Zhang, J.; Shen, C.; Xia, Y. Cotr: Efficiently bridging CNN and transformer for 3d medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part III 24. Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 171–180. [Google Scholar]
- Duc, N.T.; Oanh, N.T.; Thuy, N.T.; Triet, T.M.; Dinh, V.S. Colonformer: An efficient transformer based method for colon Polypp segmentation. IEEE Access 2022, 10, 80575–80586. [Google Scholar] [CrossRef]
- Sanderson, E.; Matuszewski, B.J. FCN-transformer feature fusion for Polypp segmentation. In Annual Conference on Medical Image Understanding and Analysis; Springer International Publishing: Cham, Switzerland, 2022; pp. 892–907. [Google Scholar]
- Park, K.B.; Lee, J.Y. SwinE-Net: Hybrid deep learning approach to novel Polypp segmentation using convolutional neural network and Swin Transformer. J. Comput. Des. Eng. 2022, 9, 616–632. [Google Scholar] [CrossRef]
- Dong, B.; Wang, W.; Fan, D.P.; Li, J.; Fu, H.; Shao, L. Polypp-pvt: Polypp segmentation with pyramid vision transformers. arXiv 2021, arXiv:2108.06932. [Google Scholar]
- Tomar, N.K.; Shergill, A.; Rieders, B.; Bagci, U.; Jha, D. TransResU-Net: Transformer based ResU-Net for real-time colonoscopy Polypp segmentation. arXiv 2022, arXiv:2206.08985. [Google Scholar]
- Narayanan, M. SENetV2: Aggregated dense layer for channelwise and global representations. arXiv 2023, arXiv:2311.10807. [Google Scholar]
- Dong, X.; Bao, J.; Chen, D.; Zhang, W.; Yu, N.; Yuan, L.; Chen, D.; Guo, B. CSWin transformer: A general vision transformer backbone withcross-shaped windows. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 12124–12134. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; De Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented Polypp dataset. In Proceedings of the MultiMedia modeling: 26th international conference, MMM 2020, Daejeon, Republic of Korea, 5–8 January 2020; Proceedings, Part II 26. Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 451–462. [Google Scholar]
- Silva, J.; Histace, A.; Romain, O.; Dray, X. Granado, Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Gurudu, S.R.; Liang, J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging 2015, 35, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, D.; Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; López, A.M.; Romero, A.; Drozdzal, M.; Courville, A. A benchmark for endoluminal scene segmentation of colonoscopy images. J. Healthc. Eng. 2017, 2017, 4037190. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Pranet: Parallel reverse attention network for Polypp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer International Publishing: Cham, Switzerland, 2020; pp. 263–273. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).