Genetics Information with Functional Brain Networks for Dementia Classification
Abstract
1. Introduction
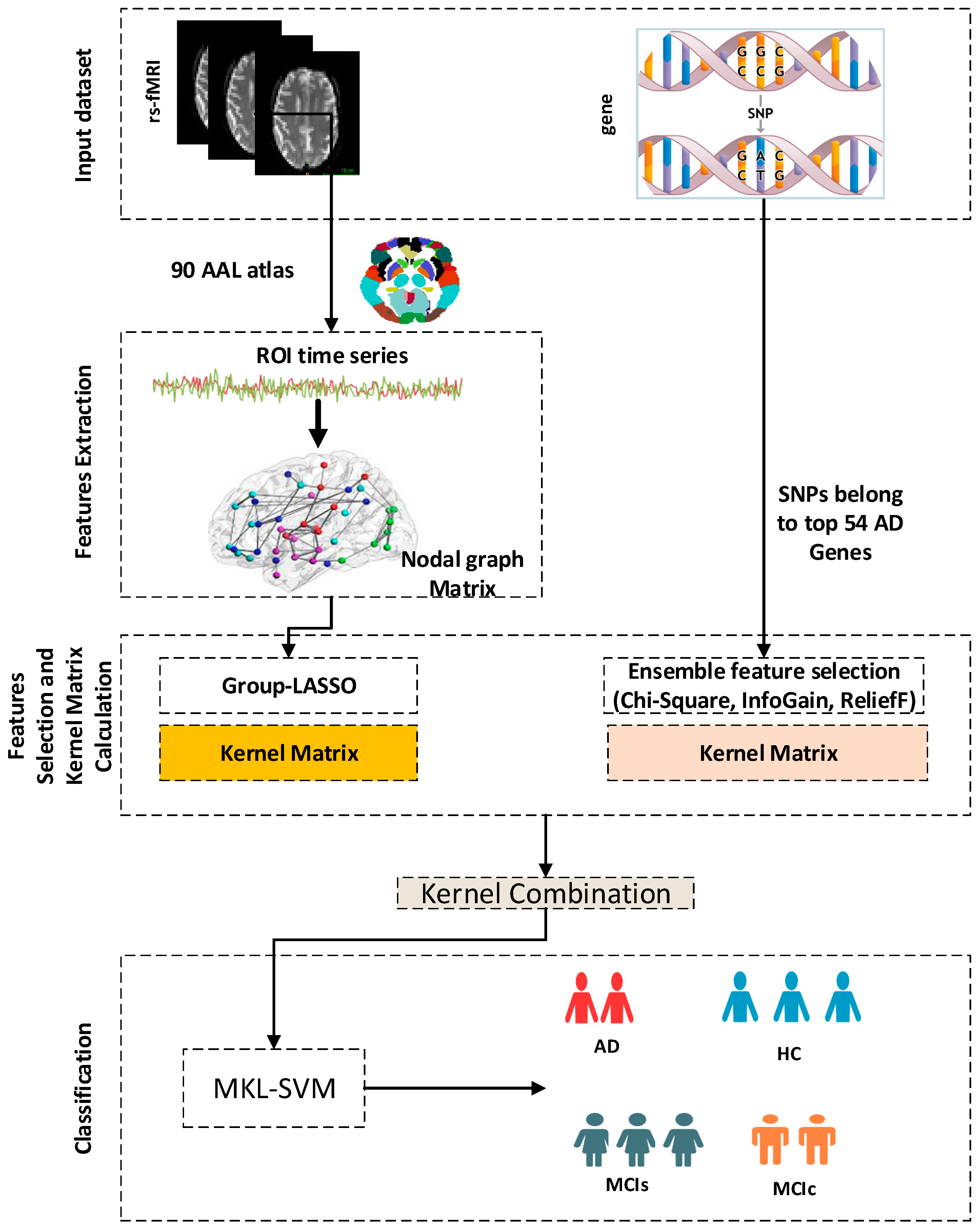
2. Materials and Methods
2.1. Data
2.2. Data Acquisition
2.3. Data Pre-Processing
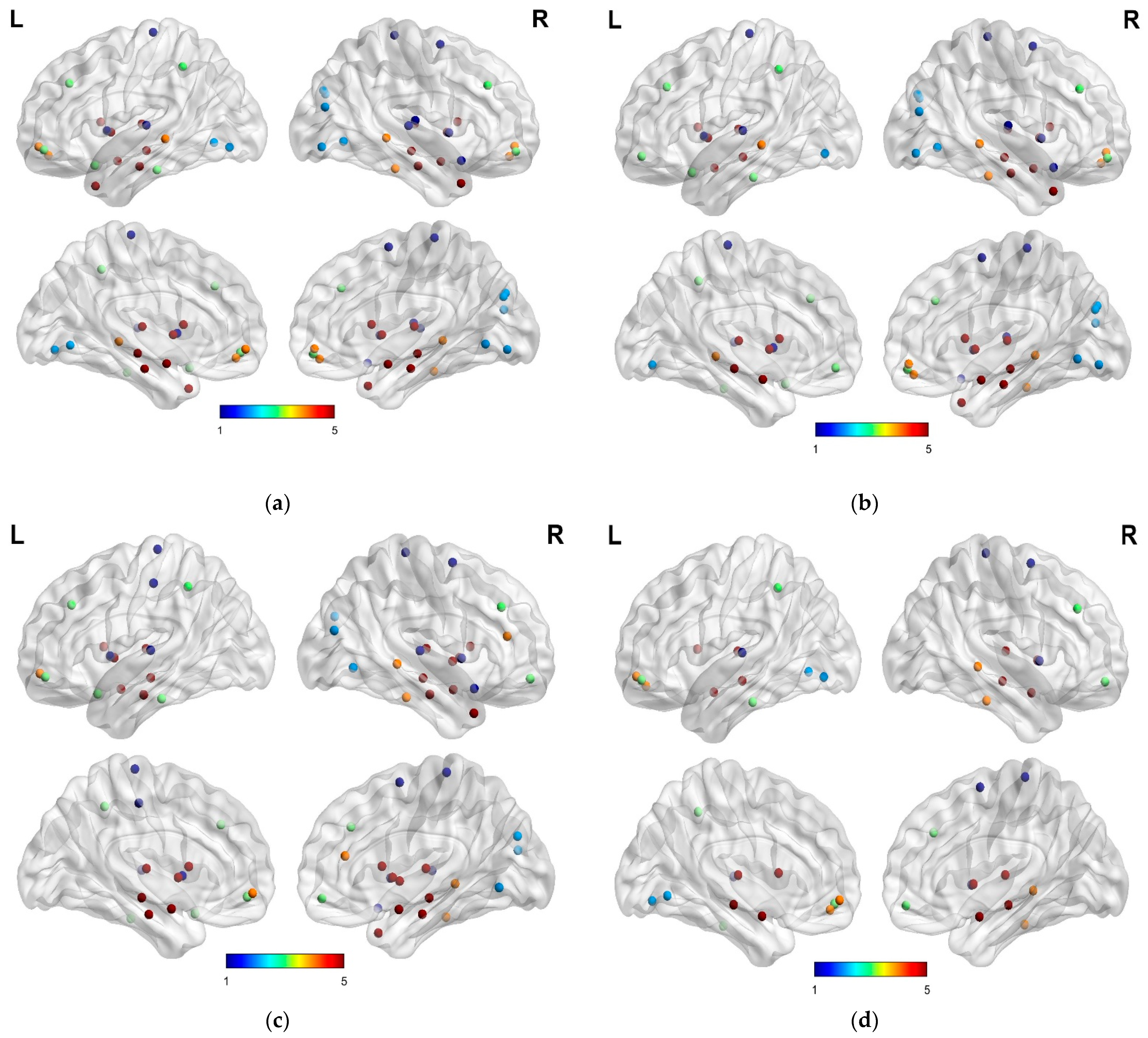
2.4. Construction of Graph Matrix
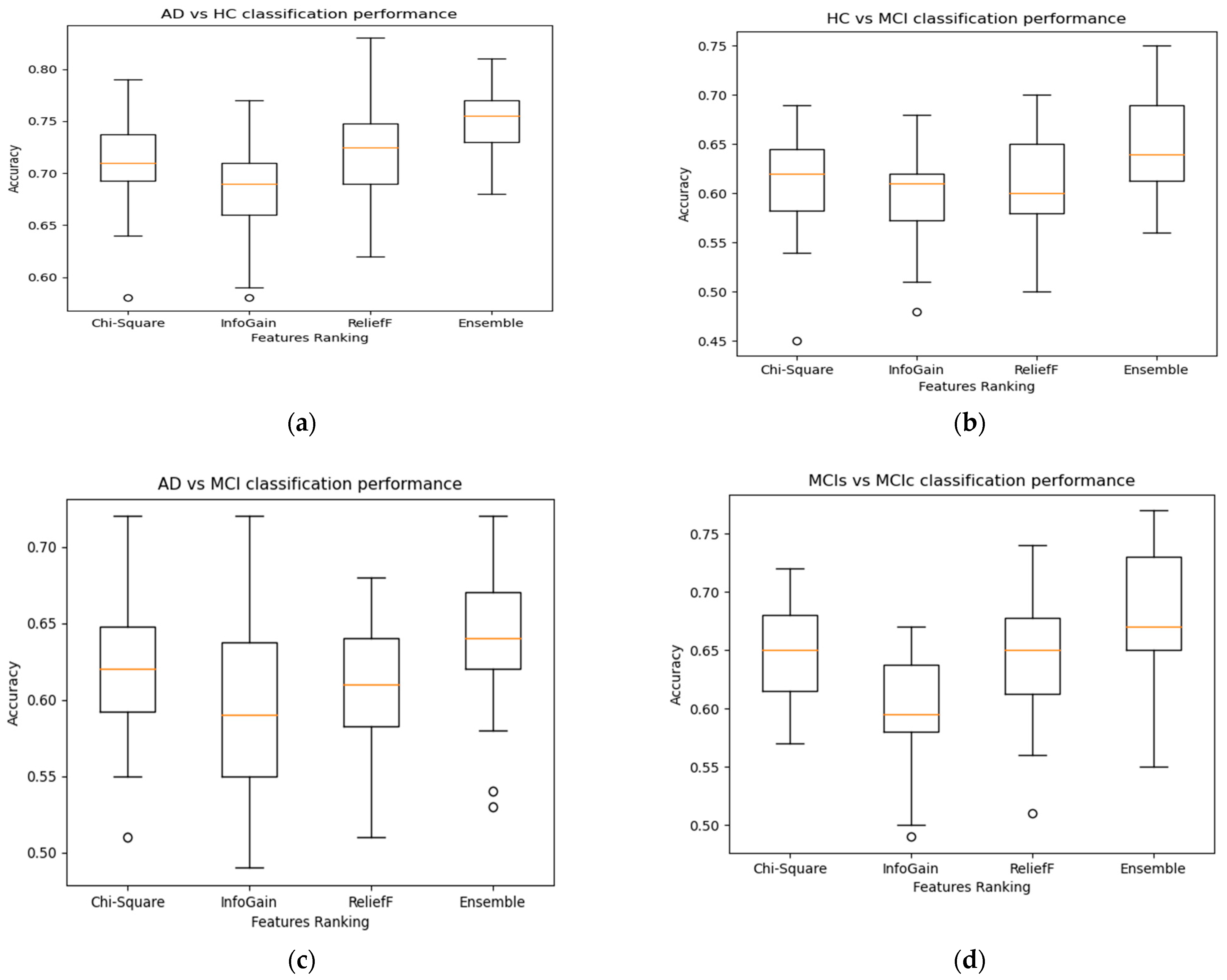
2.5. Features Selection
2.5.1. Group Least Absolute and Shrinkage Selection Operation
2.5.2. Chi-Square
2.5.3. InfoGain
2.5.4. ReliefF
2.6. Random Forest Classifier
2.7. Extreme Gradient Boosting Multi-Classifier (XGB) Classifier
2.8. SVM Classifier
2.9. Evaluation Matrices
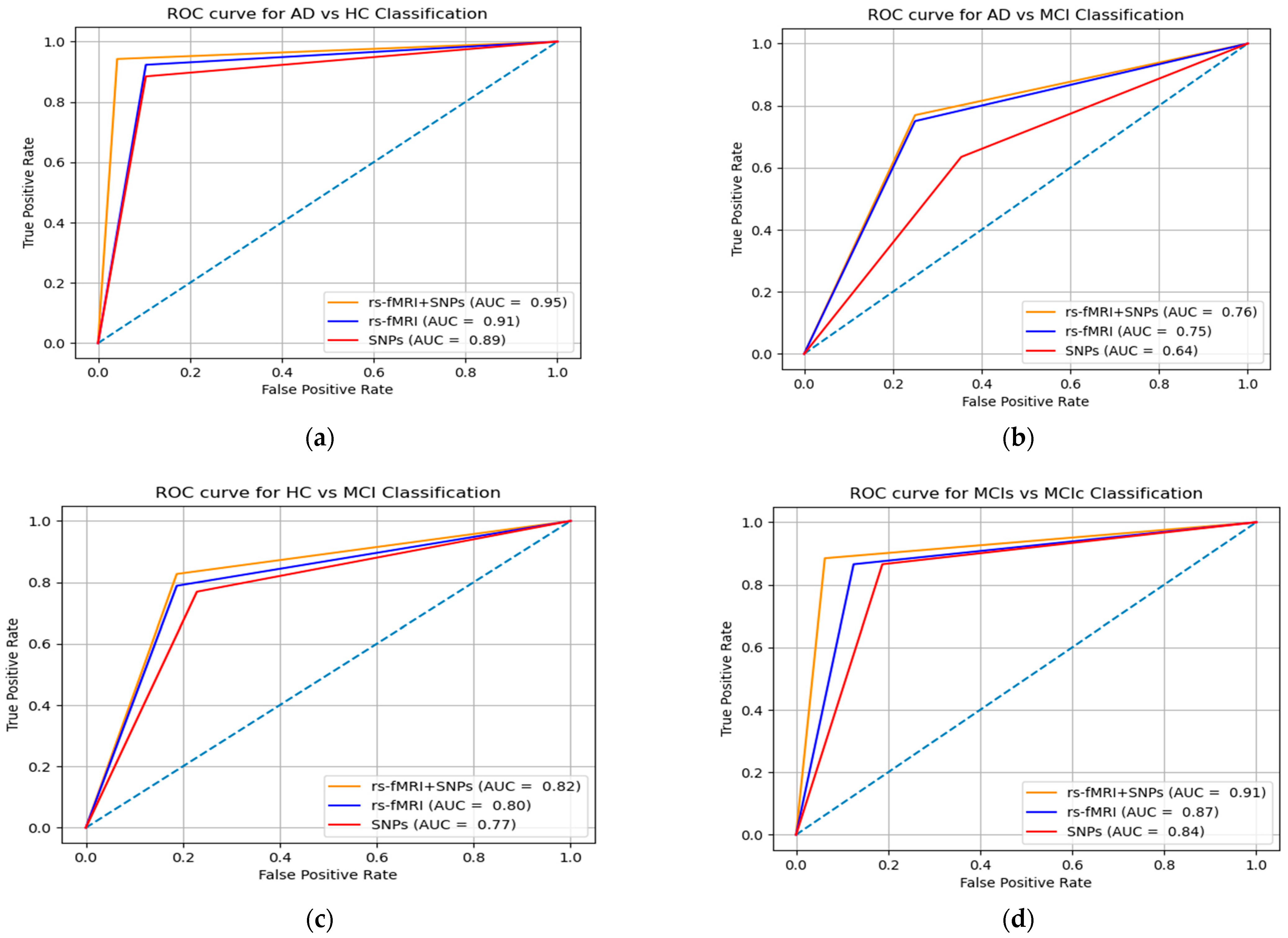
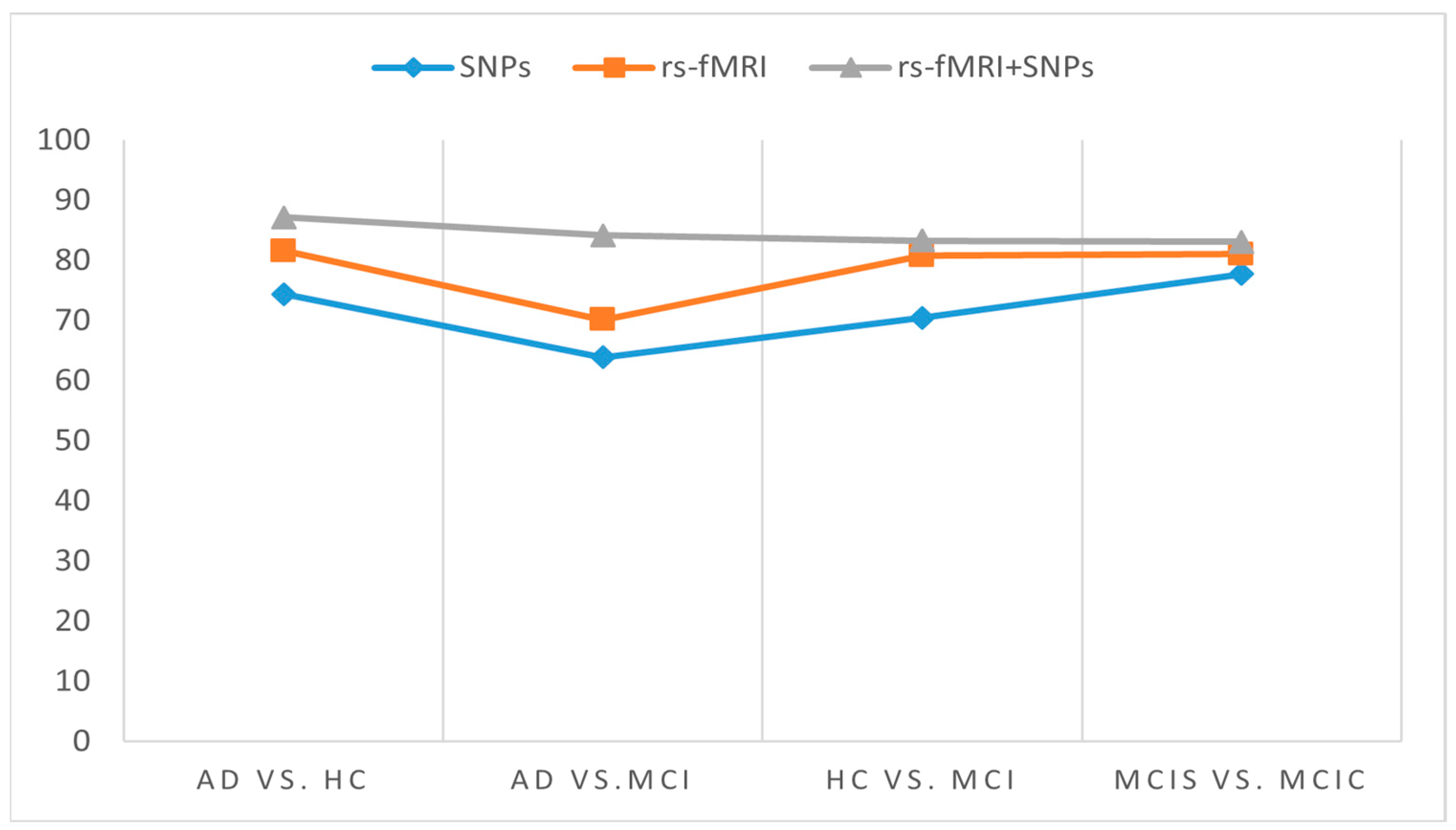
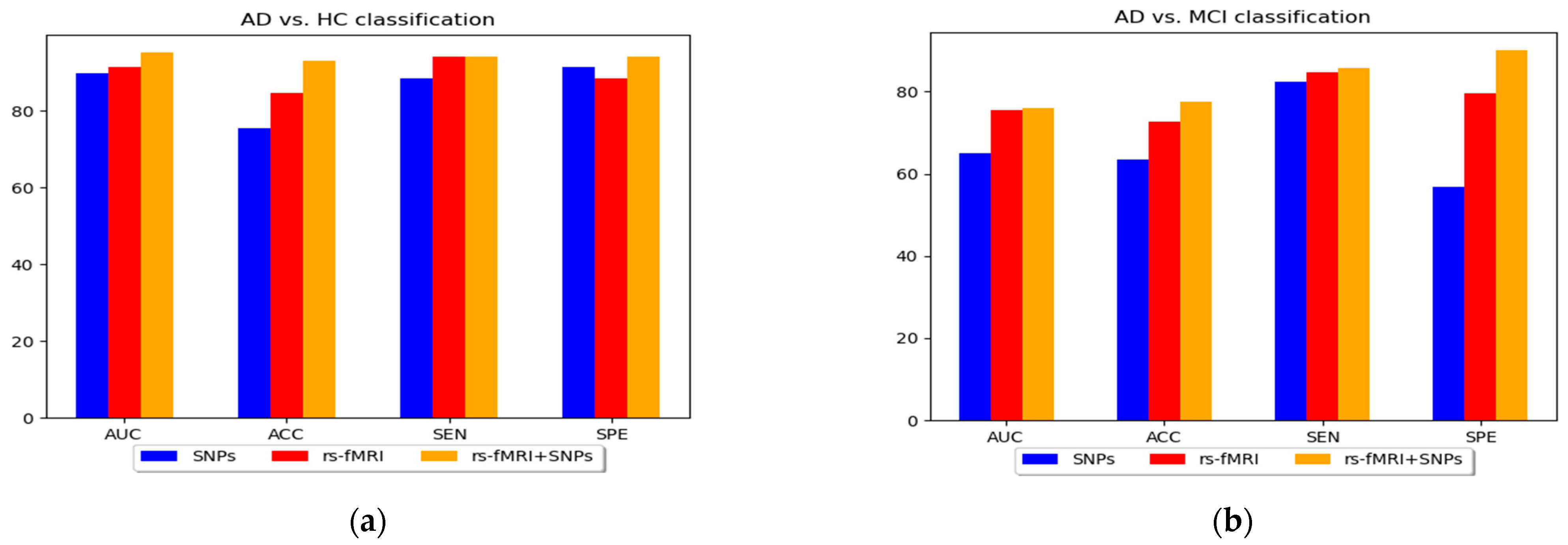
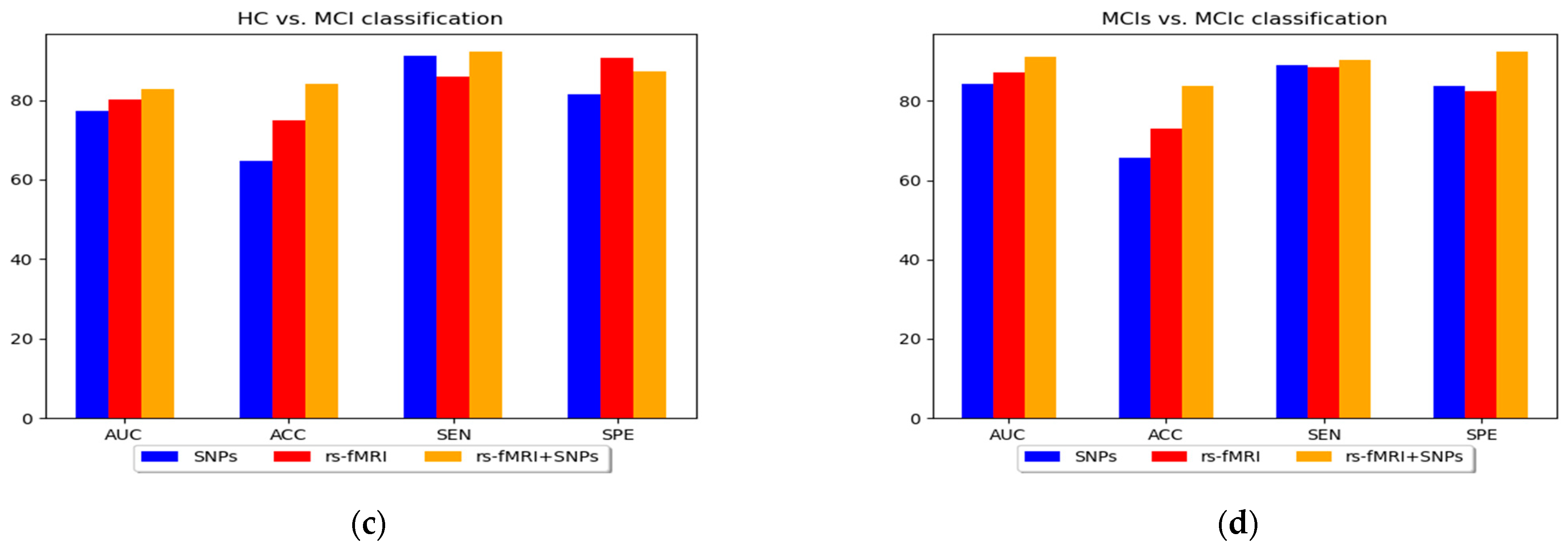
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Association, A. 2019 Alzheimer’s disease facts and figures. Alzheimers Dement. 2019, 15, 321–387. [Google Scholar] [CrossRef]
- Tramutola, A.; Lanzillotta, C.; Perluigi, M.; Butterfield, D.A. Oxidative stress, protein modification and Alzheimer disease. Brain Res. Bull. 2017, 133, 88–96. [Google Scholar] [CrossRef]
- Ju, R.; Hu, C.; Zhou, P.; Li, Q. Early Diagnosis of Alzheimer’s Disease Based on Resting-State Brain Networks and Deep Learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Sims, R.; van der Lee, S.J.; Naj, A.C.; Bellenguez, C.; Badarinarayan, N.; Jakobsdottir, J.; Kunkle, B.W.; Boland, A.; Raybould, R.; Bis, J.C.; et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 2017, 49, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Xin, Y.; Zhang, Q.; Wang, L.; Yang, Z.; Yin, J. Predictive classification of Alzheimer’s disease using brain imaging and genetic data. Sci. Rep. 2022, 12, 2405. [Google Scholar] [CrossRef] [PubMed]
- Huettel, S.A.; Song, A.W.; McCarthy, G. Functional Magnetic Resonance Imaging, 2nd ed.; Oxford University Press: Oxford, UK, 2009. [Google Scholar]
- Ogawa, S.; Lee, T.M.; Kay, A.R.; Tank, D.W. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. USA 1990, 87, 9868–9872. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Meesters, S.; Nicolay, K.; Romeny, B.M.T.H.; Ossenblok, P. The influence of construction methodology on structural brain network measures: A review. J. Neurosci. Methods 2015, 253, 170–182. [Google Scholar] [CrossRef]
- Khazaee, A.; Ebrahimzadeh, A.; Babajani-Feremi, A. Identifying patients with Alzheimer’s disease using resting-state fMRI and graph theory. Clin. Neurophysiol. 2015, 126, 2132–2141. [Google Scholar] [CrossRef]
- Bush, W.S.; Moore, J.H. Chapter 11: Genome-wide association studies. PLoS Comput. Biol. 2012, 8, e1002822. [Google Scholar] [CrossRef]
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.M.; Steinberg, S.; Sealock, J.; Karlsson, I.K.; Hägg, S.; Athanasiu, L.; et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019, 51, 404–413. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Q.; Chen, F.; Meng, X.; Liu, W.; Chen, D.; Yan, J.; Kim, S.; Wang, L.; Feng, W.; et al. Genome-wide association and interaction studies of CSF T-tau/Aβ42 ratio in ADNI cohort. Neurobiol. Aging 2017, 57, 247.e1–247.e8. [Google Scholar] [CrossRef] [PubMed]
- Dukart, J.; Sambataro, F.; Bertolino, A. Accurate Prediction of Conversion to Alzheimer’s Disease using Imaging, Genetic, and Neuropsychological Biomarkers. J. Alzheimers Dis. JAD 2016, 49, 1143–1159. [Google Scholar] [CrossRef] [PubMed]
- Biswal, B.B.; Mennes, M.; Zuo, X.-N.; Gohel, S.; Kelly, C.; Smith, S.M.; Beckmann, C.F.; Adelstein, J.S.; Buckner, R.L.; Colcombe, S.; et al. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. USA 2010, 107, 4734–4739. [Google Scholar] [CrossRef]
- Filippi, M.; Basaia, S.; Canu, E.; Imperiale, F.; Magnani, G.; Falautano, M.; Comi, G.; Falini, A.; Agosta, F. Changes in functional and structural brain connectome along the Alzheimer’s disease continuum. Mol. Psychiatry 2020, 25, 230–239. [Google Scholar] [CrossRef]
- Khazaee, A.; Ebrahimzadeh, A.; Babajani-Feremi, A. Alzheimer’s Disease Neuroimaging Initiative Classification of patients with MCI and AD from healthy controls using directed graph measures of resting-state fMRI. Behav. Brain Res. 2017, 322 Pt B, 339–350. [Google Scholar] [CrossRef]
- Liu, X.; Cao, P.; Wang, J.; Kong, J.; Zhao, D. Fused Group Lasso Regularized Multi-Task Feature Learning and Its Application to the Cognitive Performance Prediction of Alzheimer’s Disease. Neuroinformatics 2019, 17, 271–294. [Google Scholar] [CrossRef]
- Zhuo, Z.; Mo, X.; Ma, X.; Han, Y.; Li, H. Identifying aMCI with functional connectivity network characteristics based on subtle AAL atlas. Brain Res. 2018, 1696, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Themistocleous, C.; Eckerström, M.; Kokkinakis, D. Identification of Mild Cognitive Impairment From Speech in Swedish Using Deep Sequential Neural Networks. Front. Neurol. 2018, 9, 975. [Google Scholar] [CrossRef] [PubMed]
- Khazaee, A.; Ebrahimzadeh, A.; Babajani-Feremi, A. Application of advanced machine learning methods on resting-state fMRI network for identification of mild cognitive impairment and Alzheimer’s disease. Brain Imaging Behav. 2016, 10, 799–817. [Google Scholar] [CrossRef]
- Khatri, U.; Kwon, G.-R. Multi-biomarkers-Base Alzheimer’s Disease Classification. J. Multimed. Inf. Syst. 2021, 8, 233–242. [Google Scholar] [CrossRef]
- Thompson, P.M.; Ge, T.; Glahn, D.C.; Jahanshad, N.; Nichols, T.E. Genetics of the connectome. NeuroImage 2013, 80, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Vounou, M.; Nichols, T.E.; Montana, G. Alzheimer’s Disease Neuroimaging Initiative Discovering genetic associations with high-dimensional neuroimaging phenotypes: A sparse reduced-rank regression approach. NeuroImage 2010, 53, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.-C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; Jun, G.; DeStefano, A.L.; Bis, J.C.; Beecham, G.W.; et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Chao-Gan, Y.; Yu-Feng, Z. DPARSF: A MATLAB Toolbox for “Pipeline” Data Analysis of Resting-State fMRI. Front. Syst. Neurosci. 2010, 4, 13. [Google Scholar] [CrossRef]
- SPM. Statistical Parametric Mapping. Available online: https://www.fil.ion.ucl.ac.uk/spm/ (accessed on 11 January 2023).
- Wang, J.; Wang, X.; Xia, M.; Liao, X.; Evans, A.; He, Y. GRETNA: A graph theoretical network analysis toolbox for imaging connectomics. Front. Hum. Neurosci. 2015, 9, 386. [Google Scholar] [CrossRef]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 2002, 15, 273–289. [Google Scholar] [CrossRef]
- Dai, Z.; He, Y. Disrupted structural and functional brain connectomes in mild cognitive impairment and Alzheimer’s disease. Neurosci. Bull. 2014, 30, 217–232. [Google Scholar] [CrossRef]
- Newman, M.E.J. Finding community structure in networks using the eigenvectors of matrices. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2006, 74 Pt 2, 036104. [Google Scholar] [CrossRef]
- Bechtel, W. Modules, Brain Parts, and Evolutionary Psychology. In Evolutionary Psychology: Alternative Approaches; Scher, S.J., Rauscher, F., Eds.; Springer: Boston, MA, USA, 2003; pp. 211–227. [Google Scholar] [CrossRef]
- Newman, M.E.J. Fast algorithm for detecting community structure in networks. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2004, 69 Pt 2, 066133. [Google Scholar] [CrossRef]
- Seijo-Pardo, B.; Bolón-Canedo, V.; Porto-Díaz, I.; Alonso-Betanzos, A. Ensemble Feature Selection for Rankings of Features. In Advances in Computational Intelligence, Proceedings of the 13th International Work-Conference on Artificial Neural Networks, IWANN 2015, Palma de Mallorca, Spain, 10–12 June 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 29–42. [Google Scholar]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- SLEP: Sparse Learning with Efficient Projections. Available online: http://www.yelabs.net/software/SLEP/ (accessed on 11 January 2023).
- Liu, H.; Setiono, R. Chi2: Feature selection and discretization of numeric attributes. In Proceedings of the 7th IEEE International Conference on Tools with Artificial Intelligence, Herndon, VA, USA, 5–8 November 1995; pp. 388–391. [Google Scholar] [CrossRef]
- Witten, I.H.; Frank, E. Data mining: Practical machine learning tools and techniques with Java implementations. ACM SIGMOD Rec. 2002, 31, 76–77. [Google Scholar] [CrossRef]
- Robnik-Šikonja, M.; Kononenko, I. Theoretical and Empirical Analysis of ReliefF and RReliefF. Mach. Learn. 2003, 53, 23–69. [Google Scholar] [CrossRef]
- Schonlau, M.; Zou, R.Y. The random forest algorithm for statistical learning. Stata J. 2020, 20, 3–29. [Google Scholar] [CrossRef]
- Zhang, L.; Zhan, C. Machine Learning in Rock Facies Classification: An Application of XGBoost. In Proceedings of the International Geophysical Conference, Qingdao, China, 17–20 April 2017; SEG Global Meeting Abstracts. Society of Exploration Geophysicists and Chinese Petroleum Society: Beijing, China, 2017; pp. 1371–1374. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Gonen, M.; Alpaydın, E. Multiple Kernel Learning Algorithms. J. Mach. Learn. Res. 2011, 12, 2211–2268. [Google Scholar]
- Peng, Z.; Hu, Q.; Dang, J. Multi-kernel SVM based depression recognition using social media data. Int. J. Mach. Learn. Cybern. 2019, 10, 43–57. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Xu, X.; Li, W.; Mei, J.; Tao, M.; Wang, X.; Zhao, Q.; Liang, X.; Wu, W.; Ding, D.; Wang, P. Feature Selection and Combination of Information in the Functional Brain Connectome for Discrimination of Mild Cognitive Impairment and Analyses of Altered Brain Patterns. Front. Aging Neurosci. 2020, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Brand, L.; O’Callaghan, B.; Sun, A.; Wang, H. Task Balanced Multimodal Feature Selection to Predict the Progression of Alzheimer’s Disease. In Proceedings of the 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), Cincinnati, OH, USA, 26–28 October 2020; pp. 196–203. [Google Scholar] [CrossRef]
- Bi, X.-A.; Hu, X.; Wu, H.; Wang, Y. Multimodal Data Analysis of Alzheimer’s Disease Based on Clustering Evolutionary Random Forest. IEEE J. Biomed. Health Inform. 2020, 24, 2973–2983. [Google Scholar] [CrossRef] [PubMed]

| Group | HC | MCIs | MCIc | AD |
|---|---|---|---|---|
| Nos. of Subjects | 32 | 33 | 35 | 45 |
| Male/Female | 17/15 | 18/17 | 20/15 | 23/22 |
| Age | 74.82 ± 7.13 | 73.50 ± 7.4 | 73.35 ± 12 | 75.67 ± 8.71 |
| CDR | 0 | 0.5 | 0.5 ± 0.4 | 0.7 ± (0.3) |
| Education | 17.1 | 16.03 | 16.50 | 15.83 |
| MMSE | 18.30 ± 5.1 | 29 ± 1.0 | 24.8 ± 3.31 | 29.1 ± 1.7 |
| Gene Symbol | rs-ID | Position | Gene Symbol | rs-ID | Position |
|---|---|---|---|---|---|
| ADAMTS4 | rs4575098 | 161155392 | ECHDC3 | rs7920721 | 11720308 |
| CR1 | rs6656401 | 207692049 | SPI1 | rs3740688 | 47380340 |
| CR1 | rs2093760 | 207786828 | CELF1 | rs10838725 | 47557871 |
| CR1 | rs4844610 | 207802552 | MS4A6A | rs983392 | 59923508 |
| BIN1 | rs4663105 | 127891427 | MS4A2 | rs7933202 | 59936926 |
| BIN1 | rs6733839 | 127892810 | MS4A6A | rs2081545 | 59958380 |
| INPP5D | rs10933431 | 233981912 | PICALM | rs867611 | 85776544 |
| INPP5D | rs35349669 | 234068476 | PICALM | rs10792832 | 85867875 |
| CLNK | rs6448453 | 11026028 | PICALM | rs3851179 | 85868640 |
| MEF2C-AS1 | rs190982 | 88223420 | FERMT2 | rs17125924 | 53391680 |
| HLA-DRB1 | rs9271058 | 32575406 | FERMT2 | rs17125944 | 53400629 |
| CD2AP | rs9473117 | 47431284 | SLC24A4 | rs10498633 | 92926952 |
| CD2AP | rs9381563 | 47432637 | SLC24A4 | rs12881735 | 92932828 |
| CD2AP | rs10948363 | 47487762 | SLC24A4 | rs12590654 | 92938855 |
| GPR141 | rs2718058 | 37841534 | ADAM10 | rs442495 | 59022615 |
| GPR141 | rs4723711 | 37844263 | KAT8 | rs59735493 | 31133100 |
| PILRA | rs1859788 | 99971834 | SCIMP | rs113260531 | 5138980 |
| ZCWPW1 | rs1476679 | 100004446 | ABI3 | rs28394864 | 47450775 |
| NYAP1 | rs12539172 | 100091795 | ABCA7 | rs111278892 | 1039323 |
| EPHA1 | rs10808026 | 143099133 | ABCA7 | rs3752246 | 1056492 |
| EPHA1-AS1 | rs7810606 | 143108158 | ABCA7 | rs4147929 | 1063443 |
| EPHA1-AS1 | rs11771145 | 143110762 | PVRL2 | rs41289512 | 45351516 |
| PTK2B | rs28834970 | 27195121 | CD33 | rs3865444 | 51727962 |
| PTK2B | rs73223431 | 27219987 | CASS4 | rs6024870 | 54997568 |
| CLU | rs4236673 | 27464929 | CASS4 | rs6014724 | 54998544 |
| CLU | rs9331896 | 27467686 | CASS4 | rs7274581 | 55018260 |
| ECHDC3 | rs11257238 | 11717397 | APOE | rs429358 | 45411941 |
| Group | Features | Classifiers | AUC | ACC | SEN | SPEC | Cohen’s Kappa |
|---|---|---|---|---|---|---|---|
| AD vs. HC | SNPs | MKL-SVM | 89.73 | 75.5 | 88.33 | 91.28 | 74.41 |
| RF | 84.55 | 72.31 | 86.82 | 89.11 | 70.12 | ||
| XGB | 87.13 | 74.17 | 87.31 | 90.32 | 73.13 | ||
| rs-fMRI | MKL-SVM | 91.51 | 84.51 | 94.17 | 88.54 | 81.57 | |
| RF | 86.04 | 83.12 | 90.19 | 92.03 | 82.48 | ||
| XGB | 92.31 | 73.95 | 91.57 | 92.48 | 85.01 | ||
| rs-fMRI + SNPs | MKL-SVM | 95.13 | 93.03 | 94.16 | 94.17 | 87.17 | |
| RF | 87.08 | 88.53 | 86.93 | 89.05 | 83.07 | ||
| XGB | 90.45 | 92.71 | 90.91 | 93.75 | 85.01 | ||
| AD vs.MCI | SNPs | MKL-SVM | 65.12 | 63.35 | 82.37 | 56.78 | 63.87 |
| RF | 60.14 | 61.05 | 65.41 | 60.14 | 65.78 | ||
| XGB | 62.05 | 63.03 | 67.14 | 63.71 | 65.01 | ||
| rs-fMRI | MKL-SVM | 75.37 | 72.71 | 84.7 | 79.55 | 70.13 | |
| RF | 69.45 | 72.41 | 75.12 | 80.01 | 68.01 | ||
| XGB | 73.14 | 72.03 | 78.45 | 78.47 | 69.71 | ||
| rs-fMRI + SNPs | MKL-SVM | 76 | 77.43 | 85.75 | 90.01 | 84.15 | |
| RF | 74.01 | 75.45 | 90.04 | 88.56 | 83.89 | ||
| XGB | 74.92 | 76.03 | 88.01 | 91.88 | 85.03 | ||
| HC vs. MCI | SNPs | MKL-SVM | 77.21 | 64.71 | 91.02 | 81.45 | 70.44 |
| RF | 75.71 | 60.14 | 80.78 | 80.09 | 77.31 | ||
| XGB | 77.02 | 63.72 | 88.67 | 80.78 | 78.85 | ||
| rs-fMRI | MKL-SVM | 80.11 | 75.03 | 85.78 | 90.55 | 80.77 | |
| RF | 75.53 | 71.23 | 83.78 | 85.47 | 78.23 | ||
| XGB | 78.18 | 74.11 | 81.91 | 87.98 | 80.07 | ||
| rs-fMRI + SNPs | MKL-SVM | 82.79 | 84.01 | 92.13 | 87.17 | 83.31 | |
| RF | 76.17 | 80.28 | 90.03 | 90.78 | 80.47 | ||
| XGB | 78.5 | 81.55 | 92.07 | 94.01 | 81.85 | ||
| MCIs vs. MCIc | SNPs | MKL-SVM | 84.37 | 65.73 | 89.05 | 83.7 | 77.75 |
| RF | 74.23 | 63.62 | 87.78 | 75.45 | 75.48 | ||
| XGB | 83.37 | 63.78 | 89.03 | 84.77 | 78.97 | ||
| rs-fMRI | MKL-SVM | 87.1 | 73.08 | 88.51 | 82.33 | 81.05 | |
| RF | 81.54 | 70.98 | 85.33 | 82.78 | 82.77 | ||
| XGB | 85.09 | 72.54 | 87.07 | 87.41 | 80.32 | ||
| rs-fMRI + SNPs | MKL-SVM | 91.07 | 83.73 | 90.31 | 92.37 | 83.09 | |
| RF | 88.45 | 82.47 | 91.04 | 93.01 | 78.75 | ||
| XGB | 88.98 | 82.97 | 88.97 | 92.88 | 82.79 |
| Reference | Methods | Modality | No of Subjects | Group | ACC | SEN | SPE |
|---|---|---|---|---|---|---|---|
| Dukarat et al. [13] | Bayesian-Markov-Blanket+Navie Bayes | FDG-PAET, AV45-PET, SMRI, APOE | 122 HC/265 sMCI/177 cMCI/144 AD | AD vs. NC | 86.8 | 87.5 | 86.1 |
| Brand et al. [49] | Task-balanced multi-modal feature selection | sMRI, SNPs | 201 HC/170 AD/352 MCI | AD vs. HC/MCI | 72.8 | - | - |
| Sheng et al. [5] | Fisher score+Multitask feature seletion+SVM | sMRI, SNPs | 25 AD/25 EMCI/25 EMCI/25 HC | AD vs. HC | 98 | 100 | 96 |
| LMCI vs. EMCI | 80 | 88 | 72 | ||||
| LMCI vs. HC | 86 | 88 | 84 | ||||
| EMCI vs. HC | 82 | 80 | 84 | ||||
| Bi et al. [50] | Cluster evolutionary random forest (CERF)+SVM | fMRI, SNPs | 37 AD/37 EMCI/35 HC | AD vs. HC | 81 | - | - |
| EMCI vs. HC | 80 | - | - | ||||
| EMCI vs. NC | 0.803 | 0.794 | 0.856 | ||||
| our method | Ensemble, group-LASSO, MKL-SVM | fMRI, SNPs | 32 HC/33 MCIs/35 MCIc/45 AD | AD vs. HC | 93.03 | 95.15 | 94.17 |
| AD vs. MCI | 77.43 | 85.75 | 90.01 | ||||
| HC vs. MCI | 84.01 | 92.13 | 87.17 | ||||
| MCIs vs. MCIc | 83.73 | 90.31 | 92.37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khatri, U.; Kim, J.-I.; Kwon, G.-R. Genetics Information with Functional Brain Networks for Dementia Classification. Mathematics 2023, 11, 1529. https://doi.org/10.3390/math11061529
Khatri U, Kim J-I, Kwon G-R. Genetics Information with Functional Brain Networks for Dementia Classification. Mathematics. 2023; 11(6):1529. https://doi.org/10.3390/math11061529
Chicago/Turabian StyleKhatri, Uttam, Ji-In Kim, and Goo-Rak Kwon. 2023. "Genetics Information with Functional Brain Networks for Dementia Classification" Mathematics 11, no. 6: 1529. https://doi.org/10.3390/math11061529
APA StyleKhatri, U., Kim, J.-I., & Kwon, G.-R. (2023). Genetics Information with Functional Brain Networks for Dementia Classification. Mathematics, 11(6), 1529. https://doi.org/10.3390/math11061529







