A Review on Nature-Inspired Algorithms for Cancer Disease Prediction and Classification
Abstract
1. Introduction
1.1. Research Issues
- What dimensionality reduction techniques are frequently used to manage high-dimensional gene expression datasets?
- What techniques for feature selection are often used to handle high-dimensional gene expression datasets?
- What datasets have been frequently used to build the model for cancer classification from high-dimensional gene expression data?
- Which models have recently been applied to identify the important set of genes needed to correctly classify cancer disease using DNA high-dimension gene expression data?
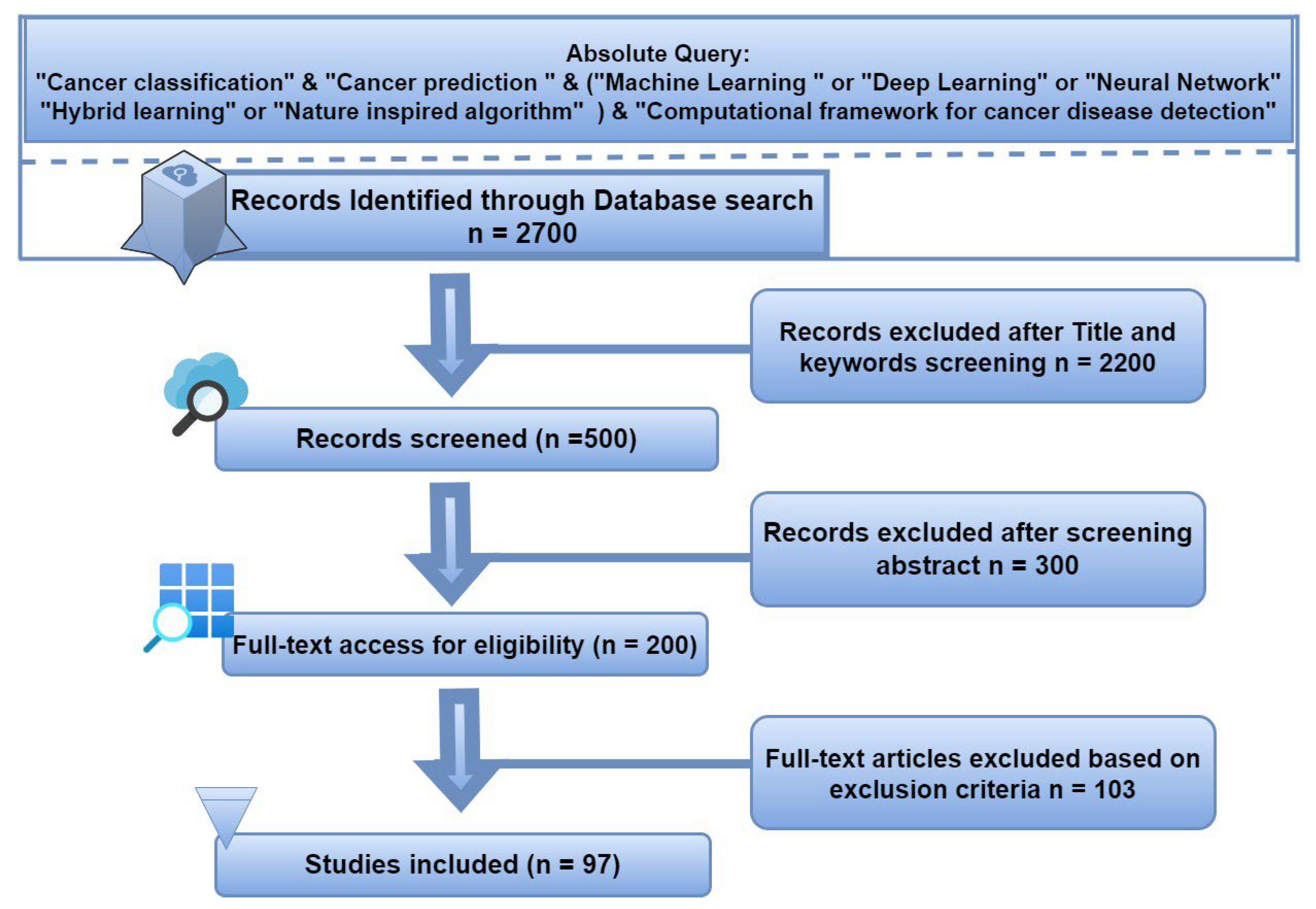
1.2. Methodology for Review and Article Search and Selection Strategy
1.3. Paper Organization
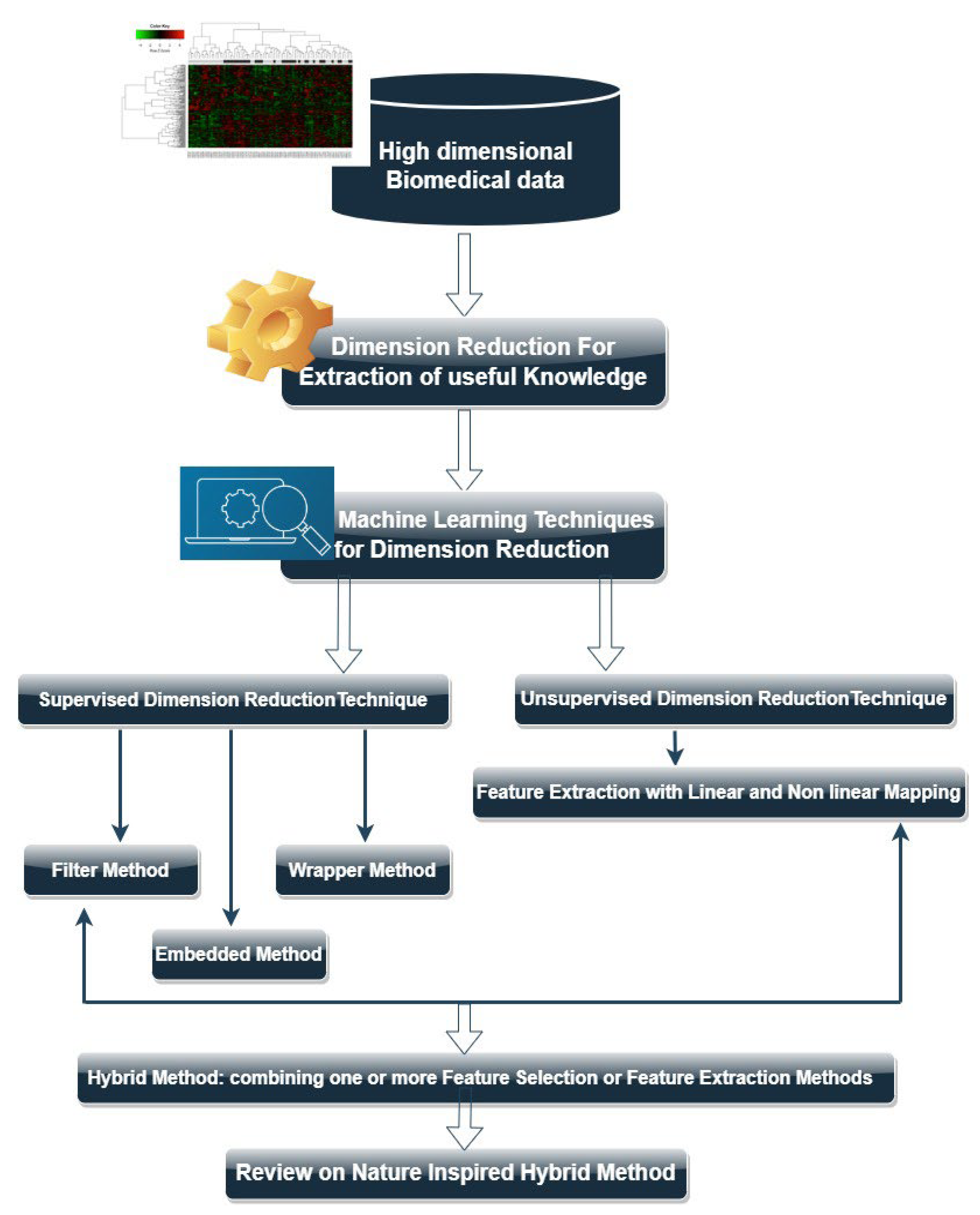
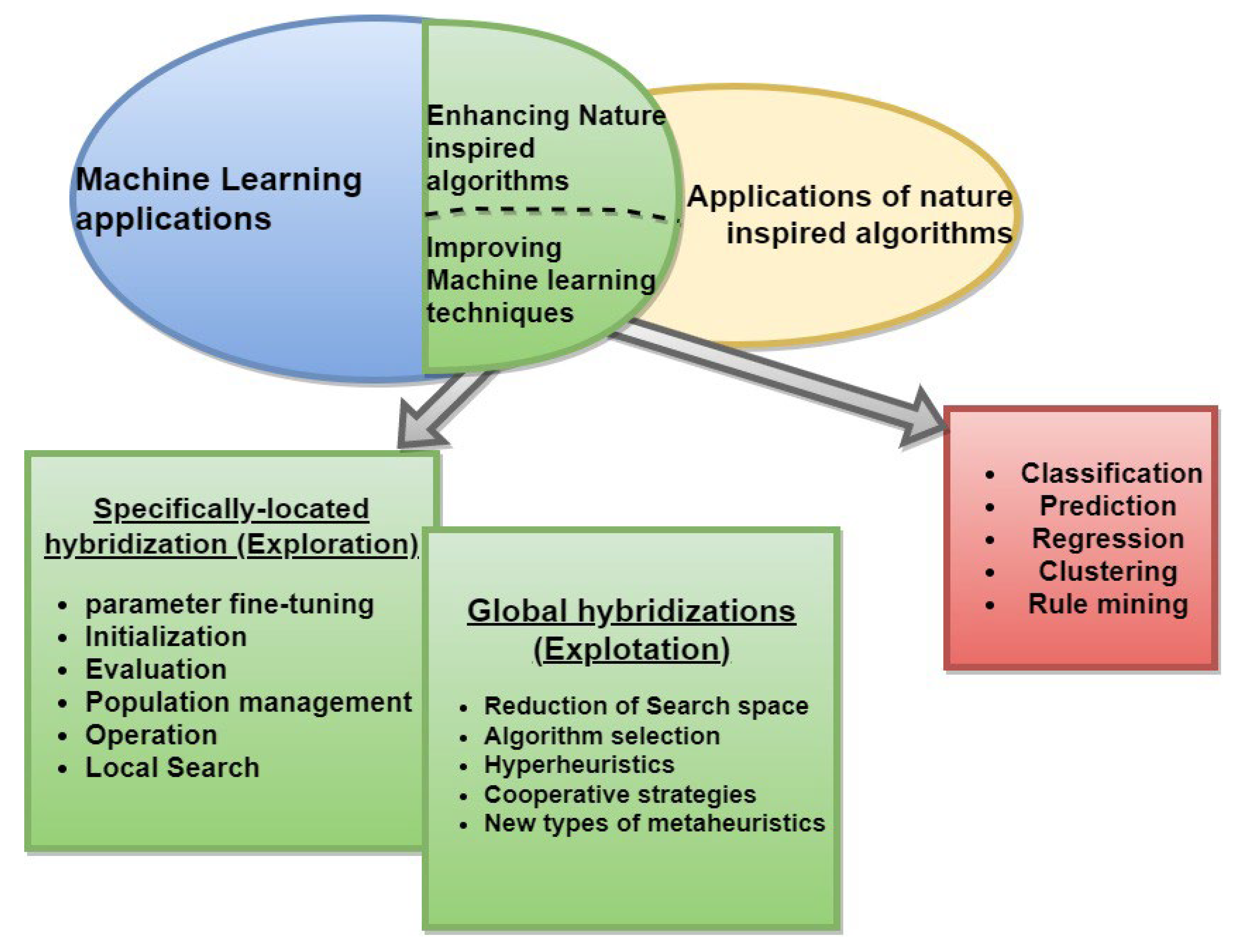
2. Machine Learning Based Dimensionality Reduction Algorithms
3. Types of Dimension Reduction Techniques
3.1. Feature (Gene) Selection Methods
3.1.1. Filter
3.1.2. Wrapper
3.1.3. Embedded
3.2. Feature Extraction
3.3. Hybrid
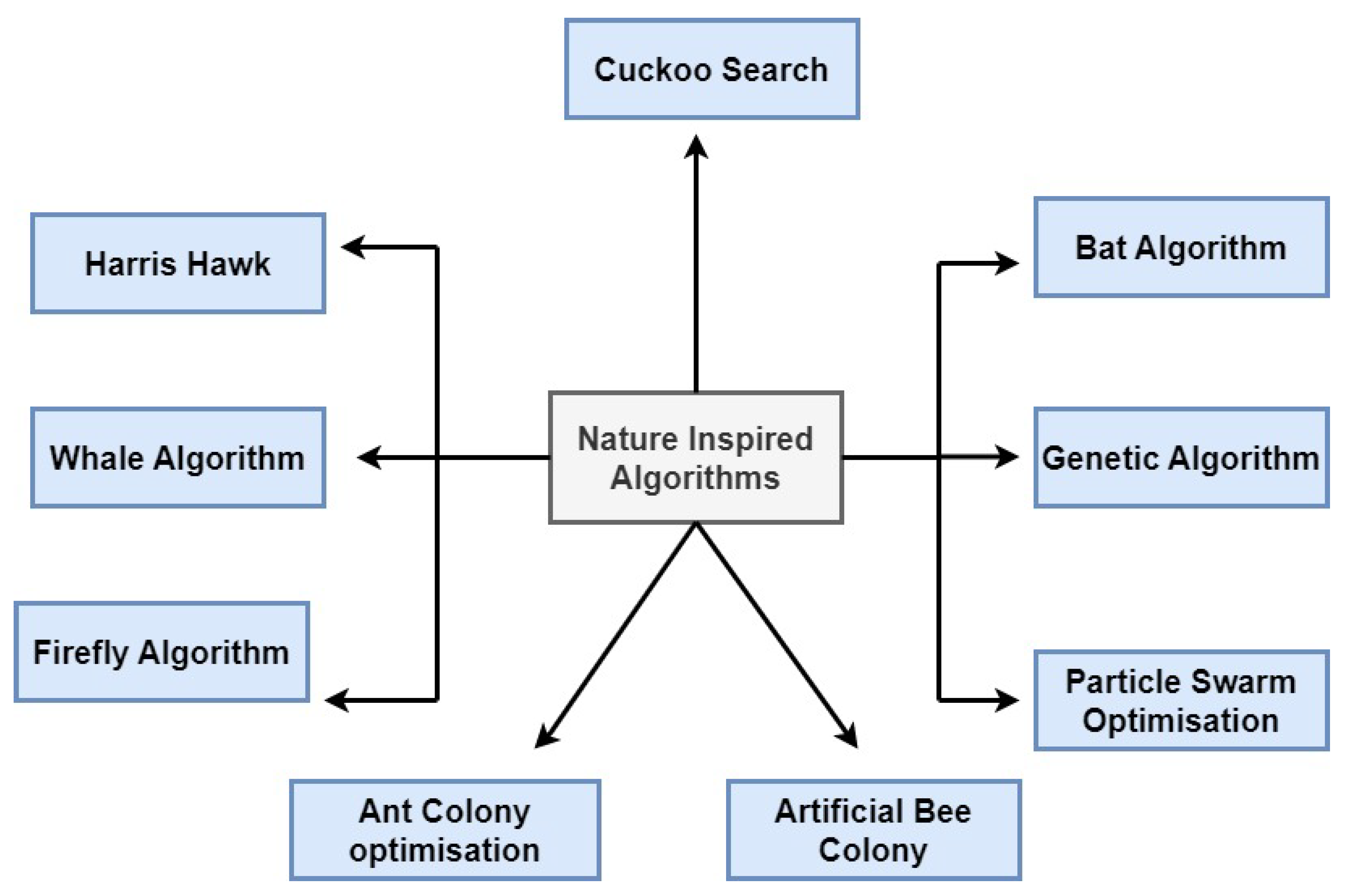
4. Nature-Inspired Algorithms
Characteristics of Nature-Inspired Algorithms (Exploration Phase and Exploitation Phase)
- Each individual in a population of different participants, such as particles, ants, bats, cuckoos, fireflies, bees, etc., signifies an initial solution that has been utilized in every method. In terms of objective fitness, the population usually provides the most accurate information.
- To encourage population development, a variety of operators (such as mutation and crossover) are widely utilized, which are typically stated in relation to computational calculations or algorithms. This kind of growth is usually continuous and produces solutions with a variety of assets. The system is considered to be convergent when all solutions have sufficiently converged.
- The moves of an agent, which are basically preset, form a piecewise zigzag route in the search space. As a result, strategies for randomization are routinely utilized to provide fresh solution vectors or motions. The algorithm can alter its states (or solutions) thanks to this randomization, allowing it to bypass any local optimum.
- Every algorithm tries to do some type of local and/or global search. If the search area is mostly local, there is a higher chance of becoming trapped there. If the search focused too much on global movements, convergence would have been impeded.
5. Review of Nature-Inspired Algorithms for Dimension Reduction of Gene Expression Data
5.1. Cuckoo Search Algorithm (CSA)
- A cuckoo has never laid more than one egg in a random nest.
- The generation that comes after will inherit the best nest and eggs.
- The host bird’s pa is between 0 and 1, which is the chance of finding a cuckoo egg given a certain number of host nests.
5.2. Bat Algorithm
- Bats use echolocation to determine distance. In some amazing ways, they can recognize the distances or intervals between the surrounding obstacles and the prey.
- In search of food, bats fly freely at various velocities and frequencies with varying wavelengths and loudness. They may continuously modify the wavelength and rate of release of loudness of the flashes based on how close their objective.
- Furthermore, loudness could differ in a range of ways. In this case, it is presumed that the volume fluctuates between a high and a low fixed value.
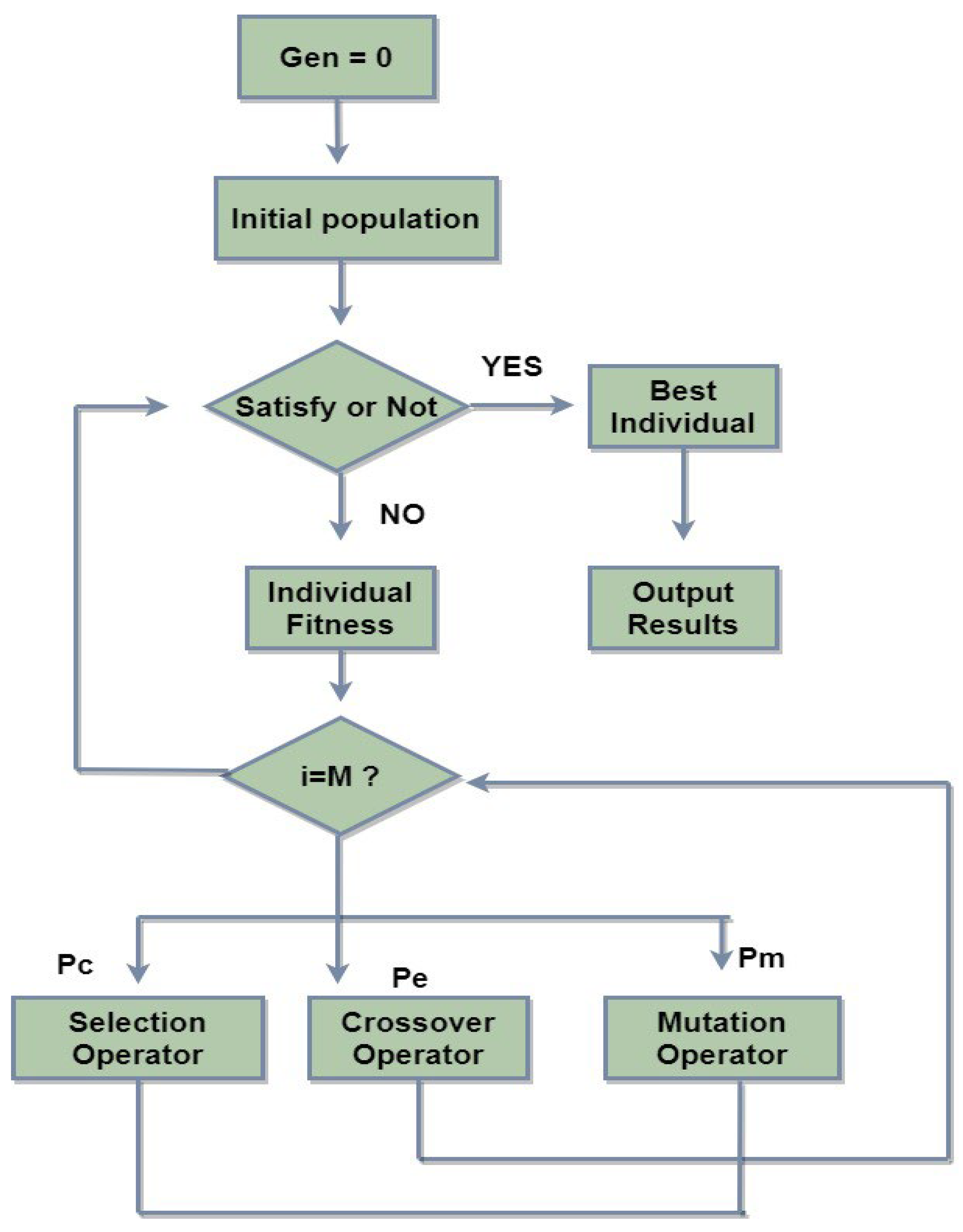
5.3. Genetic Algorithm (GA)
- i.
- Initial population: This algorithm starts with the selection of initial sets, which may or may not include the optimal values. These sets of values are called ‘chromosomes’ and the step is called ‘initialize population’.
- ii.
- Fitness function: The value of the objective function for each chromosome has been computed with some fitness value in this step.
- iii.
- Selection: This step is very important and is called ‘selection’ because the fittest chromosomes are selected from the population for subsequent operations.
- iv.
- Crossover: This step is called crossover because, in this step, chromosomes are expressed in terms of genes.
- v.
- Mutation: Mutation is the process of altering the value of a gene to find the best solution.
5.4. Whale Optimization Algorithm (WOA)
5.5. Harris Hawk Optimization (HHO) Algorithm
5.6. Ant Colony Optimization (ACO) Algorithm
5.7. Artificial Bee Colony (ABC)
5.8. Firefly Algorithm (FFA)
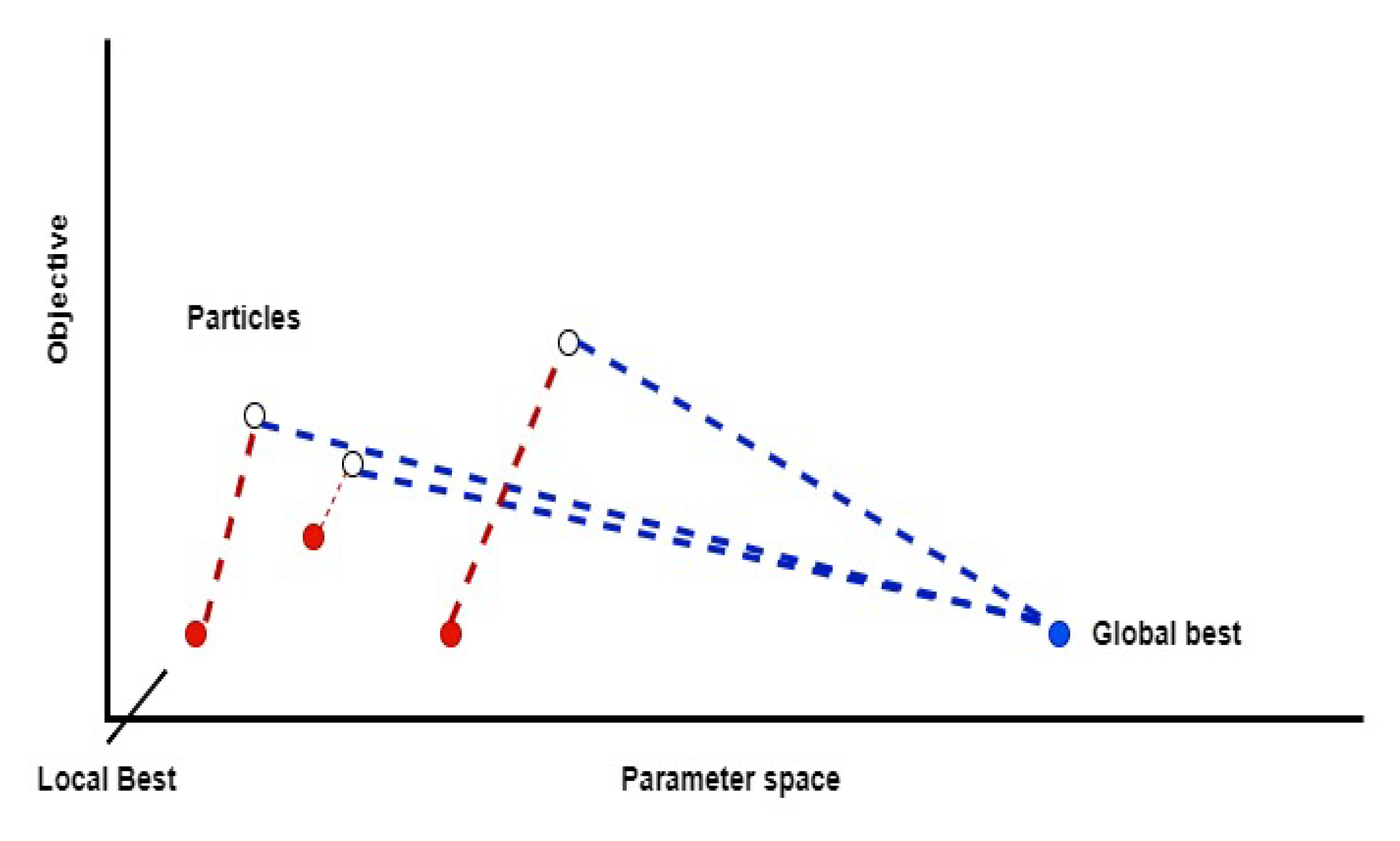
5.9. Particle Swarm Optimization Algorithm
- It uses a number of particles (agents) that constitute a swarm moving around in the search space, looking for the best solution.
- Each particle in the swarm looks for its positional coordinates in the solution space, which are associated with the best solution that has been achieved so far by that particle. It is known as “local best”.
6. Discussion
Implementation Challenges of a Nature-Inspired Algorithm for Gene Expression Data for Cancer Classification and Prediction
7. Advantages and Disadvantages of Nature-Inspired Algorithms
- These algorithms are very effective in locating multi-dimensional and multi-modal issues in high-dimensional gene expression data and finding the optimal solutions.
- Regarding applications of nature-inspired algorithms for gene selection, it was evident that microarray gene expression classification is the most dominant application where nature-inspired algorithms for gene selection are applied and successfully resolve other applications of biomedical data.
- These algorithms have been employed successfully to find different human disorders.
- Compared to the prior option, nature-inspired algorithms make it easier to recognize rational-universal issues in an adequate time frame with greater reliability and precision.
- In the case of high-dimensional biomedical data, the real environments are complicated, and the optimization problems can be high-dimensional, large-scale, multi-modal, and multi-objective; the optimization environments can be dynamic, highly constrained, and uncertain; the fitness evaluations may contain noise and be imprecise and time-consuming.
- The complexity of real environments poses a great challenge to nature-inspired algorithms. Although some researchers have made attempts to solve the aforementioned problems, figuring out how to handle these issues remains a very difficult problem.
- Nature-inspired algorithms currently need a complete mathematical framework for analyzing all methods to fully understand their robustness, consistency, development, and levels of integration.
- Due to the probabilistic character of nature-inspired methods, results are not entirely repeatable; hence, several runs are necessary to acquire useful data.
Take-Home Message of the Review
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shah, S.M.; Khan, R.A.; Arif, S.; Sajid, U. Artificial intelligence for breast cancer analysis: Trends & directions. Comput. Biol. Med. 2022, 15, 105221. [Google Scholar] [CrossRef]
- Aziz, R.; Verma, C.K.; Jha, M.; Srivastava, N. Artificial neural network classification of microarray data using new hybrid gene selection method. Int. J. Data Min. Bioinform. 2017, 17, 42–65. [Google Scholar] [CrossRef]
- Tufail, A.B.; Ma, Y.K.; Kaabar, M.K.; Martínez, F.; Junejo, A.R.; Ullah, I.; Khan, R. Deep learning in cancer diagnosis and prognosis prediction: A minireview on challenges, recent trends, and future directions. Comput. Math. Methods Med. 2021, 2021, 9025470. [Google Scholar] [CrossRef] [PubMed]
- Diao, R.; Shen, Q. Nature inspired feature selection meta-heuristics. Artif. Intell. Rev. 2015, 44, 311–340. [Google Scholar] [CrossRef]
- Aziz, R.M. Cuckoo search-based optimization for cancer classification: A new hybrid approach. J. Comput. Biol. 2022, 29, 565–584. [Google Scholar] [CrossRef]
- Elemento, O.; Leslie, C.; Lundin, J.; Tourassi, G. Artificial intelligence in cancer research, diagnosis and therapy. Nat. Rev. Cancer 2021, 21, 747–752. [Google Scholar] [CrossRef]
- Aziz, R.; Verma, C.K.; Srivastava, N. Dimension reduction methods for microarray data: A review. AIMS Bioeng. 2017, 4, 179–197. [Google Scholar] [CrossRef]
- Alomari, O.A.; Khader, A.T.; Al-Betar, M.A.; Abualigah, L.M. Gene selection for cancer classification by combining minimum redundancy maximum relevancy and bat-inspired algorithm. Int. J. Data Min. Bioinform. 2017, 19, 32–51. [Google Scholar] [CrossRef]
- Aziz, R.M.; Mahto, R.; Goel, K.; Das, A.; Kumar, P.; Saxena, A. Modified Genetic Algorithm with Deep Learning for Fraud Transactions of Ethereum Smart Contract. Appl. Sci. 2023, 13, 697. [Google Scholar] [CrossRef]
- Musheer, R.A.; Verma, C.K.; Srivastava, N. Novel machine learning approach for classification of high-dimensional microarray data. Soft Comput. 2019, 23, 13409–13421. [Google Scholar] [CrossRef]
- Ramachandran, A.; Rustum, R.; Adeloye, A.J. Review of anaerobic digestion modeling and optimization using nature-inspired techniques. Processes 2019, 12, 953. [Google Scholar] [CrossRef]
- Mohamad, A.B.; Zain, A.M.; Nazira Bazin, N.E. Cuckoo search algorithm for optimization problems a literature review and its applications. Appl. Artif. Intell. 2014, 28, 419–448. [Google Scholar] [CrossRef]
- Aziz, R.; Verma, C.K.; Srivastava, N. Artificial neural network classification of high dimensional data with novel optimization approach of dimension reduction. Ann. Data Sci. 2018, 5, 615–635. [Google Scholar] [CrossRef]
- Naik, M.; Nath, M.R.; Wunnava, A.; Sahany, S.; Panda, R. A new adaptive Cuckoo search algorithm. In Proceedings of the 2015 IEEE 2nd International Conference on Recent Trends in Information Systems (ReTIS), Kolkata, India, 9–11 July 2015; IEEE: New York, NY, USA; pp. 1–5. [Google Scholar] [CrossRef]
- Heyn, P.C.; Meeks, S.; Pruchno, R. Methodological guidance for a quality review article. Gerontologist 2019, 59, 197–201. [Google Scholar] [CrossRef]
- Yildiz, A.R. Cuckoo search algorithm for the selection of optimal machining parameters in milling operations. Int. J. Adv. Manuf. Technol. 2013, 64, 55–61. [Google Scholar] [CrossRef]
- Fister, I.; Yang, X.S.; Fister, D.; Fister, I. Cuckoo search: A brief literature review. In Cuckoo Search and Firefly Algorithm: Theory and Applications; Springer: Cham, Switzerland, 2014; pp. 49–62. [Google Scholar] [CrossRef]
- Civicioglu, P.; Besdok, E. Comparative analysis of the cuckoo search algorithm. In Cuckoo Search and Firefly Algorithm: Theory and Applications; Springer: Cham, Switzerland, 2014; pp. 85–113. [Google Scholar] [CrossRef]
- Aziz, R.M.; Baluch, M.F.; Patel, S.; Ganie, A.H. LGBM: A approach for Ethereum fraud detection. Int. J. Inf. Technol. 2022, 14, 3321–3331. [Google Scholar] [CrossRef]
- Aziz, R.; Verma, C.K.; Srivastava, N. A novel approach for dimension reduction of microarray. Comput. Biol. Chem. 2017, 71, 161–169. [Google Scholar] [CrossRef]
- Aziz, R.; Srivastava, N.; Verma, C.K. T-independent component analysis for svm classification of DNA-microarray data. Int. J. Bioinform. Res. 2015, 6, 305–312. [Google Scholar] [CrossRef]
- Desai, N.P.; Baluch, M.F.; Makrariya, A.; Musheer Aziz, R. Image processing model with deep learning approach for fish species classification. Turk. J. Comput. Math. Educ. 2022, 13, 85–99. [Google Scholar]
- Aziz, R.M.; Hussain, A.; Sharma, P.; Kumar, P. Machine learning based soft computing regression analysis approach for crime data prediction. Karbala Int. J. Mod. Sci. 2022, 8, 1–19. [Google Scholar] [CrossRef]
- Aziz, R.M.; Baluch, M.F.; Patel, S.; Kumar, P. A based approach to detect the Ethereum fraud transactions with limited attributes. Karbala Int. J. Mod. Sci. 2022, 8, 139–151. [Google Scholar] [CrossRef]
- Luan, J.; Yao, Z.; Zhao, F.; Song, X. A novel method to solve supplier selection problem: Hybrid algorithm of genetic algorithm and ant colony optimization. Math. Comput. Simul. 2019, 156, 294–309. [Google Scholar] [CrossRef]
- Aziz, R.M.; Sharma, P.; Hussain, A. Algorithms for Crime Prediction under Indian Penal Code. Ann. Data Sci. 2022, 1–32. [Google Scholar] [CrossRef]
- Deepak, M.; Rustum, R. Review of Latest Advances in Nature-Inspired Algorithms for Optimization of Activated Sludge Processes. Processes 2022, 11, 77. [Google Scholar] [CrossRef]
- Sagu, A.; Gill, N.S.; Gulia, P.; Singh, P.K.; Hong, W.C. Design of Metaheuristic Optimization Algorithms for Deep Learning Model for Secure IoT Environment. Sustainability 2023, 15, 2204. [Google Scholar] [CrossRef]
- Aziz, R.; Verma, C.; Srivastava, N. A weighted-SNR feature selection from independent component subspace for nb classification of microarray data. Int. J. Biotechnol. Res. 2015, 6, 245–255. [Google Scholar] [CrossRef]
- Prabukumar, M.; Agilandeeswari, L.; Ganesan, K. An intelligent lung cancer diagnosis system using cuckoo search optimization and support vector machine classifier. J. Ambient Intell. Humaniz. Comput. 2019, 10, 267–293. [Google Scholar] [CrossRef]
- Alzaqebah, M.; Briki, K.; Alrefai, N.; Brini, S.; Jawarneh, S.; Alsmadi, M.K.; Mohammad, R.M.A.; ALmarashdeh, I.; Alghamdi, F.A.; Aldhafferi, N.; et al. Memory based cuckoo search algorithm for feature selection of gene expression dataset. Inform. Med. Unlocked 2021, 24, 100572. [Google Scholar] [CrossRef]
- Gunavathi, C.; Premalatha, K. Cuckoo search optimisation for feature selection in cancer classification: A new approach. Int. J. Data Min. Bioinform. 2015, 13, 248–265. [Google Scholar] [CrossRef]
- Arjmand, A.; Meshgini, S.; Afrouzian, R.; Farzamnia, A. Breast tumor segmentation using K-means clustering and cuckoo search optimization. In Proceedings of the 2019 9th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 24–25 October 2019; IEEE: New York, NY, USA; pp. 305–308. [Google Scholar] [CrossRef]
- Sampathkumar, A.; Rastogi, R.; Arukonda, S.; Shankar, A.; Kautish, S.; Sivaram, M. An efficient hybrid methodology for detection of cancer-causing gene using CSC for micro array data. J. Ambient Intell. Humaniz. Comput. 2020, 11, 4743–4751. [Google Scholar] [CrossRef]
- Akila, S.; Christe, S.A. A wrapper based binary bat algorithm with greedy crossover for attribute selection. Expert Syst. Appl. 2022, 187, 115828. [Google Scholar] [CrossRef]
- Al-Betar, M.A.; Alomari, O.A.; Abu-Romman, S.M. A TRIZ-inspired bat algorithm for gene selection in cancer classification. Genomics 2020, 112, 114–126. [Google Scholar] [CrossRef]
- Jeyasingh, S.; Veluchamy, M. Modified bat algorithm for feature selection with the wisconsin diagnosis breast cancer (WDBC) dataset. Asian Pac. J. Cancer Prev. 2017, 18, 1257. [Google Scholar] [CrossRef]
- Alhassan, A.M.; Zainon, W.M.N.W. BAT algorithm with fuzzy C-ordered means (BAFCOM) clustering segmentation and enhanced capsule networks (ECN) for brain cancer MRI images classification. IEEE Access 2020, 8, 201741–201751. [Google Scholar] [CrossRef]
- Hambali, M.A.; Oladele, T.O.; Adewole, K.S.; Sangaiah, A.K.; Gao, W. Feature selection and computational optimization in high-dimensional microarray cancer datasets via InfoGain-modified bat algorithm. Multimed. Tools Appl. 2022, 81, 36505–36549. [Google Scholar] [CrossRef]
- Chatra, K.; Kuppili, V.; Edla, D.R.; Verma, A.K. Cancer data classification using binary bat optimization and extreme learning machine with a novel fitness function. Med. Biol. Eng. Comput. 2019, 57, 2673–2682. [Google Scholar] [CrossRef]
- Ahmad, F.; Mat Isa, N.A.; Hussain, Z.; Osman, M.K.; Sulaiman, S.N. A GA-based feature selection and parameter optimization of an ANN in diagnosing breast cancer. Pattern Anal. Appl. 2015, 18, 861–870. [Google Scholar] [CrossRef]
- Mohammadi, S.; Hejazi, S.R. Using particle swarm optimization and genetic algorithms for optimal control of non-linear fractional-order chaotic system of cancer cells. Math. Comput. Simul. 2023, 206, 538–560. [Google Scholar] [CrossRef]
- Rojas, M.G.; Olivera, A.C.; Carballido, J.A.; Vidal, P.J. Memetic micro-genetic algorithms for cancer data classification. Intell. Syst. Appl. 2023, 17, 200173. [Google Scholar] [CrossRef]
- Zexuan, Z.; Ong, Y.S.; Dash, M. Markov blanket-embedded genetic algorithm for gene selection. Pattern Recognit. 2007, 40, 3236–3248. [Google Scholar] [CrossRef]
- Motieghader, H.; Najafi, A.; Sadeghi, B.; Masoudi-Nejad, A. A hybrid gene selection algorithm for microarray cancer classification using genetic algorithm and learning automata. Inform. Med. Unlocked 2017, 9, 246–254. [Google Scholar] [CrossRef]
- Jansi Rani, M.; Devaraj, D. Two-stage hybrid gene selection using mutual information and genetic algorithm for cancer data classification. J. Med. Syst. 2019, 43, 235. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Su, R.; Romain, M.; Sébastien, V.; Pierre, V.; Isabelle, G. Feature selection for outcome prediction in oesophageal cancer using genetic algorithm and random forest classifier. Comput. Med. Imaging Graph. 2017, 60, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Zhu, Z.; Gu, X. An intelligent system for lung cancer diagnosis using a new genetic algorithm based feature selection method. J. Med. Syst. 2018, 38, 97. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.; Jalil, Z.; Javed, A.R.; Batool, I.; Khan, M.Z.; Noorwali, A.; Gadekallu, T.R.; Akbar, A. BCD-WERT: A novel approach for breast cancer detection using whale optimization based efficient features and extremely randomized tree algorithm. PeerJ Comput. Sci. 2021, 7, e390. [Google Scholar] [CrossRef]
- Fang, H.; Fan, H.; Lin, S.; Qing, Z.; Sheykhahmad, F.R. Automatic breast cancer detection based on optimized neural network using whale optimization algorithm. Int. J. Imaging Syst. Technol. 2021, 31, 425–438. [Google Scholar] [CrossRef]
- Stephan, P.; Stephan, T.; Kannan, R.; Abraham, A. A hybrid artificial bee colony with whale optimization algorithm for improved breast cancer diagnosis. Neural Comput. Appl. 2021, 33, 13667–13691. [Google Scholar] [CrossRef]
- Sayed, G.I.; Darwish, A.; Hassanien, A.E. Binary whale optimization algorithm and binary moth flame optimization with clustering algorithms for clinical breast cancer diagnoses. J. Classif. 2020, 37, 66–96. [Google Scholar] [CrossRef]
- Sayed, G.I.; Darwish, A.; Hassanien, A.E.; Pan, J.S. Breast cancer diagnosis approach based on meta-heuristic optimization algorithm inspired by the bubble-net hunting strategy of whales. In Genetic and Evolutionary Computing, Proceedings of the Tenth International Conference on Genetic and Evolutionary Computing, Fuzhou, China, 7–9 November 2016; Springer International Publishing: Cham, Switzerland, 2017; pp. 306–313. [Google Scholar] [CrossRef]
- Kaur, N.; Kaur, L.; Cheema, S.S. An enhanced version of Harris Hawks optimization by dimension learning-based hunting for breast cancer detection. Sci. Rep. 2021, 11, 21933. [Google Scholar] [CrossRef]
- Rammurthy, D.; Mahesh, P.K. Whale Harris hawks optimization based deep learning classifier for brain tumor detection using MRI images. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 3259–3272. [Google Scholar] [CrossRef]
- Ibrahim, H.T.; Mazher, W.J.; Jassim, E.M. Modified Harris Hawks optimizer for feature selection and support vector machine kernels. Indones. J. Electr. Eng. Comput. Sci. 2023, 29, 942–953. [Google Scholar]
- Hussien, A.G.; Abualigah, L.; Abu Zitar, R.; Hashim, F.A.; Amin, M.; Saber, A.; Almotairi, K.H.; Gandomi, A.H. Recent advances in harris hawks optimization: A comparative study and applications. Electronics 2022, 11, 1919. [Google Scholar] [CrossRef]
- Jiang, F.; Zhu, Q.; Tian, T. Breast Cancer Detection Based on Modified Harris Hawks Optimization and Extreme Learning Machine Embedded with Feature Weighting. Neural Process. Lett. 2022, 23, 1–24. [Google Scholar] [CrossRef]
- Sun, L.; Kong, X.; Xu, J.; Xue, Z.A.; Zhai, R.; Zhang, S. A hybrid gene selection method based on ReliefF and ant colony optimization algorithm for tumor classification. Sci. Rep. 2019, 9, 8978. [Google Scholar] [CrossRef]
- Sumeyya, İ.; Gençtürk, T.H.; Gülağız, F.K.; Özcan, H.; Altuncu, M.A.; Şahin, S. hybSVM: Bacterial colony optimization algorithm based SVM for malignant melanoma detection. Eng. Sci. Technol. Int. J. 2021, 24, 1059–1071. [Google Scholar] [CrossRef]
- Fallahzadeh, O.; Dehghani-Bidgoli, Z.; Assarian, M. Raman spectral feature selection using ant colony optimization for breast cancer diagnosis. Lasers Med. Sci. 2018, 33, 1799–1806. [Google Scholar] [CrossRef]
- Zainuddin, S.; Nhita, F.; Wisesty, U.N. Classification of gene expressions of lung cancer and colon tumor using Adaptive-Network-Based Fuzzy Inference System (ANFIS) with Ant Colony Optimization (ACO) as the feature selection. J. Phys. Conf. Ser. 2019, 1192, 012019. [Google Scholar] [CrossRef]
- Rajagopal, R. Glioma brain tumor detection and segmentation using weighting random forest classifier with optimized ant colony features. Int. J. Imaging Syst. Technol. 2019, 29, 353–359. [Google Scholar] [CrossRef]
- Coleto-Alcudia, V.; Vega-Rodríguez, M.A. Artificial bee colony algorithm based on dominance (ABCD) for a hybrid gene selection method. Knowl. Based Syst. 2020, 205, 106323. [Google Scholar] [CrossRef]
- Aziz, R.M. Application of nature inspired soft computing techniques for gene selection: A novel frame work for classification of cancer. Soft Comput. 2022, 26, 12179–12196. [Google Scholar] [CrossRef]
- Punitha, S.; Al-Turjman, F.; Stephan, T. An automated breast cancer diagnosis using feature selection and parameter optimization in ANN. Comput. Electr. Eng. 2021, 90, 106958. [Google Scholar] [CrossRef]
- Karthiga, B.; Rekha, M. Feature extraction and I-NB classification of CT images for early lung cancer detection. Mater. Today Proc. 2020, 33, 3334–3341. [Google Scholar] [CrossRef]
- Aljanabi, M.; Jumaa, F.A.; Abed, J.K.; Al-Hamadani, H. Analysis of Automatic Detection of Tumour Lesions Images using Bee Colony Technique. J. Phys. Conf. Ser. 2020, 1530, 012012. [Google Scholar] [CrossRef]
- Al-Thanoon, N.A.; Qasim, O.S.; Algamal, Z.Y. Tuning parameter estimation in SCAD-support vector machine using firefly algorithm with application in gene selection and cancer classification. Comput. Biol. Med. 2018, 103, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Sawhney, R.; Mathur, P.; Shankar, R. A firefly algorithm based wrapper-penalty feature selection method for cancer diagnosis. In Computational Science and Its Applications–ICCSA 2018, Proceedings of the 18th International Conference, Melbourne, VIC, Australia, 2–5 July 2018; Proceedings, Part I 18; Springer International Publishing: Cham, Switzerland, 2018; pp. 438–449. [Google Scholar] [CrossRef]
- Farouk, R.M.; Mustafa, H.I.; Ali, A.E. Hybrid Firefly and Swarm Algorithms for Breast Cancer Mammograms Classification Based on Rough Set Theory Features Selection. In Proceedings of the Future Technologies Conference (FTC), Vancouver, BC, Canada, 28–29 October 2021; Springer International Publishing: Cham, Switzerland; Volume 3, pp. 849–867. [Google Scholar] [CrossRef]
- Alshamlan, H.M.; Badr, G.H.; Alohali, Y.A. Genetic Bee Colony (GBC) algorithm: A new gene selection method for microarray cancer classification. Comput. Biol. Chem. 2015, 56, 49–60. [Google Scholar] [CrossRef]
- Nguyun, L.C.; Meesad, P.; Unger, H. A highly accurate firefly based algorithm for heart disease prediction. Expert Syst. Appl. 2015, 42, 8221–8231. [Google Scholar] [CrossRef]
- Banu, P.N.; Azar, A.T.; Inbarani, H.H. Fuzzy firefly clustering for tumour and cancer analysis. Int. J. Model. Identif. Control. 2017, 27, 92–103. [Google Scholar] [CrossRef]
- Almugren, N.; Alshamlan, H.M. New bio-marker gene discovery algorithms for cancer gene expression profile. IEEE Access 2019, 7, 136907–136913. [Google Scholar] [CrossRef]
- Osama, S.; Shaban, H.; Ali, A.A. Gene reduction and algorithms for cancer classification based on microarray gene expression data: A comprehensive review. Expert Syst. Appl. 2022, 213, 118946. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Y.; Gao, X.; Gong, D.; Song, X.; Guo, Y.; Wang, J. A federated feature selection algorithm based on particle swarm optimization under privacy protection. Knowl. Based Syst. 2023, 260, 110122. [Google Scholar] [CrossRef]
- Al-Thanoon, N.A.; Qasim, O.S.; Algamal, Z.Y. A new hybrid firefly algorithm and particle swarm optimization for tuning parameter estimation in penalized support vector machine with application in chemometrics. Chemom. Intell. Lab. Syst. 2019, 184, 142–152. [Google Scholar] [CrossRef]
- Kar, S.; Sharma, K.D.; Maitra, M. Gene selection from microarray gene expression data for classification of cancer subgroups employing PSO and adaptive K-nearest neighborhood technique. Expert Syst. Appl. 2015, 42, 612–627. [Google Scholar] [CrossRef]
- Ansari, A.S.; Zamani, A.S.; Mohammadi, M.S.; Ritonga, M.; Ahmed, S.S.; Pounraj, D.; Kaliyaperumal, K. Detection of Pancreatic Cancer in CT Scan Images Using PSO SVM and Image Processing. BioMed Res. Int. 2022, 2022, 8544337. [Google Scholar] [CrossRef]
- Houssein, E.H.; Abdelminaam, D.S.; Hassan, H.N.; Al-Sayed, M.M.; Nabil, E. A hybrid barnacles mating optimizer algorithm with support vector machines for gene selection of microarray cancer classification. IEEE Access 2021, 9, 64895–64905. [Google Scholar] [CrossRef]
- Kaul, S.; Kumar, Y.; Ghosh, U.; Alnumay, W. Nature-inspired optimization algorithms for different computing systems: Novel perspective and systematic review. Multimed. Tools Appl. 2022, 81, 26779–26801. [Google Scholar] [CrossRef]
- Lazebnik, T. Cell-Level Spatio-Temporal Model for a Bacillus Calmette–Guérin-Based Immunotherapy Treatment Protocol of Superficial Bladder Cancer. Cells 2022, 11, 2372. [Google Scholar] [CrossRef]
- Vahmiyan, M.; Kheirabadi, M.; Akbari, E. Feature selection methods in microarray gene expression data: A systematic mapping study. Neural Comput. Appl. 2022, 34, 19675–19702. [Google Scholar] [CrossRef]
- Abu Khurma, R.; Aljarah, I.; Sharieh, A.; Abd Elaziz, M.; Damaševičius, R.; Krilavičius, T. A review of the modification strategies of the nature inspired algorithms for feature selection problem. Mathematics 2022, 10, 464. [Google Scholar] [CrossRef]
- Nave, O.; Elbaz, M. Artificial immune system features added to breast cancer clinical data for machine learning (ML) applications. Biosystems 2021, 202, 104341. [Google Scholar] [CrossRef]
- Almazrua, H.; Alshamlan, H. A comprehensive survey of recent hybrid feature selection methods in cancer microarray gene expression data. IEEE Access 2022, 10, 71427–71449. [Google Scholar] [CrossRef]
- Alrefai, N.; Ibrahim, O. Optimized feature selection method using particle swarm intelligence with ensemble learning for cancer classification based on microarray datasets. Neural Comput. Appl. 2022, 34, 13513–13528. [Google Scholar] [CrossRef]
- Nassif, A.B.; Talib, M.A.; Nasir, Q.; Afadar, Y.; Elgendy, O. Breast cancer detection using artificial intelligence techniques: A systematic literature review. Artif. Intell. Med. 2022, 127, 102276. [Google Scholar] [CrossRef] [PubMed]
- Lazebnik, T.; Bunimovich-Mendrazitsky, S. Improved Geometric Configuration for the Bladder Cancer BCG-Based Immunotherapy Treatment Model. In Mathematical and Computational Oncology, Proceedings of the Third International Symposium, ISMCO 2021, Virtual Event, 11–13 October 2021; Springer International Publishing: Cham, Switzerland, 2021; pp. 63–67. [Google Scholar] [CrossRef]
- Aziz, R.M.; Desai, N.P.; Baluch, M.F. Computer vision model with novel cuckoo search based deep learning approach for classification of fish image. Multimed. Tools Appl. 2022, 82, 3677–3696. [Google Scholar] [CrossRef]
- Adhikari, M.; Srirama, S.N.; Amgoth, T. A comprehensive survey on nature-inspired algorithms and their applications in edge computing: Challenges and future directions. Softw. Pract. Exp. 2022, 52, 1004–1034. [Google Scholar] [CrossRef]
- Guzev, E.; Halachmi, S.; Bunimovich-Mendrazitsky, S. Additional extension of the mathematical model for BCG immunotherapy of bladder cancer and its validation by auxiliary tool. Int. J. Nonlinear Sci. Numer. Simul. 2019, 20, 675–689. [Google Scholar] [CrossRef]
- Aziz, R.M. Nature-inspired metaheuristics model for gene selection and classification of biomedical microarray data. Med. Biol. Eng. Comput. 2022, 60, 1627–1646. [Google Scholar] [CrossRef]
- Hameed, S.S.; Hassan, W.H.; Latiff, L.A.; Muhammadsharif, F.F. A comparative study of nature-inspired metaheuristic algorithms using a three-phase hybrid approach for gene selection and classification in high-dimensional cancer datasets. Soft Comput. 2021, 25, 8683–8701. [Google Scholar] [CrossRef]
- Ghosh, M.; Sen, S.; Sarkar, R.; Maulik, U. Quantum squirrel inspired algorithm for gene selection in methylation and expression data of prostate cancer. Appl. Soft Comput. 2021, 105, 107221. [Google Scholar] [CrossRef]
- Jain, I.; Jain, V.K.; Jain, R. Correlation feature selection based improved-binary particle swarm optimization for gene selection and cancer classification. Appl. Soft Comput. 2018, 62, 203–215. [Google Scholar] [CrossRef]




| Study Types | Advantages | Drawbacks | Regular Search | Effectiveness |
|---|---|---|---|---|
| Filter Method | Autonomous Neglects classification Quick calculation times | No communication with the classification model Disregards important features | Univariate Multivariate | Quicker than other feature selection techniques lowers the feature’s significance |
| Wrapper Method | Dependent on feature Computerized and time consuming Interacts with the classifier and feature selection | Overfitting risk Dependent on the classifier Lengthy calculation time Time that is complex exponential overfitting risk Dependent on the classifiers | Deterministic Stochastic | Superior to the filter method Extraordinary efficiency |
| Embedded Method | Having a low chance of overfitting Interacting with the classifier Using the best FS method with the classification model | Classifier depends on the technique of selection Propensity for overfitting | Integrated model Simplified model | Computations are less expensive than wrapping |
| Feature Extraction | Higher discriminating power Control over fitting problem | Loss of data interpretability The transformation may be expensive | PCA, Linear discriminant analysis, ICA | Used in an effective way in the hybrid algorithm |
| Hybrid Method | Combines multiple feature selection and extraction techniques. | Time-consuming and challenging | Searching in depth Ideal FS | Complexity Reduced mistake |
| Algorithm | Introduced By | Year | Inspired By |
|---|---|---|---|
| Genetic Algorithm | John Holland | 1960 | Process of natural selection |
| Ant Colony Optimization | Marco Dorigo | 1992 | Foraging behavior of natural ants |
| Particle Swarm Algorithm | Kennedy and Eberhart | 1995 | Social behavior of birds |
| Artificial Bee Colony | Karaboga | 2005 | Intelligent foraging behavior of bees |
| Fire fly Algorithm | Xin-She Yang | 2008 | Flashing behavior of fireflies |
| Cuckoo Search | Yang and Suash deb | 2009 | Obligate brood parasitism |
| Bat Algorithm | Xin She yang | 2010 | Echolocation behavior of microbats |
| Whale Algorithm | Mirjalli and Lewis | 2016 | Hunting mechanism of humpback whales in nature |
| Harris Hawk | Heidar | 2019 | Harris hawks hunting as a group |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yaqoob, A.; Aziz, R.M.; Verma, N.K.; Lalwani, P.; Makrariya, A.; Kumar, P. A Review on Nature-Inspired Algorithms for Cancer Disease Prediction and Classification. Mathematics 2023, 11, 1081. https://doi.org/10.3390/math11051081
Yaqoob A, Aziz RM, Verma NK, Lalwani P, Makrariya A, Kumar P. A Review on Nature-Inspired Algorithms for Cancer Disease Prediction and Classification. Mathematics. 2023; 11(5):1081. https://doi.org/10.3390/math11051081
Chicago/Turabian StyleYaqoob, Abrar, Rabia Musheer Aziz, Navneet Kumar Verma, Praveen Lalwani, Akshara Makrariya, and Pavan Kumar. 2023. "A Review on Nature-Inspired Algorithms for Cancer Disease Prediction and Classification" Mathematics 11, no. 5: 1081. https://doi.org/10.3390/math11051081
APA StyleYaqoob, A., Aziz, R. M., Verma, N. K., Lalwani, P., Makrariya, A., & Kumar, P. (2023). A Review on Nature-Inspired Algorithms for Cancer Disease Prediction and Classification. Mathematics, 11(5), 1081. https://doi.org/10.3390/math11051081








