It’s All in the Embedding! Fake News Detection Using Document Embeddings
Abstract
1. Introduction
- (1)
- A simpler neural architecture offers better or at least similar results compared to complex architectures that employ multiple layers, and
- (2)
- The difference in performance lies in the embeddings used to vectorize the textual data and how well these performs in encoding contextual and linguistic features.
- We propose a new document embedding (DocEmb) constructed using word embeddings and transformers. We specifically trained the proposed DocEmb on the five datasets used in the experiments.
- We show empirically that simple Machine Leaning algorithms trained with our proposed DocEmb obtain similar results or better results than deep learning architectures specifically developed for the task of binary and multi-class fake news detection. This contribution is important in the machine learning literature because it changes the focus from the classification architecture to the document encoding architecture.
- We present a new manually filtered dataset. The original dataset is the widely used Fake News Corpus that was annotated with an automatic process.
2. Related Work
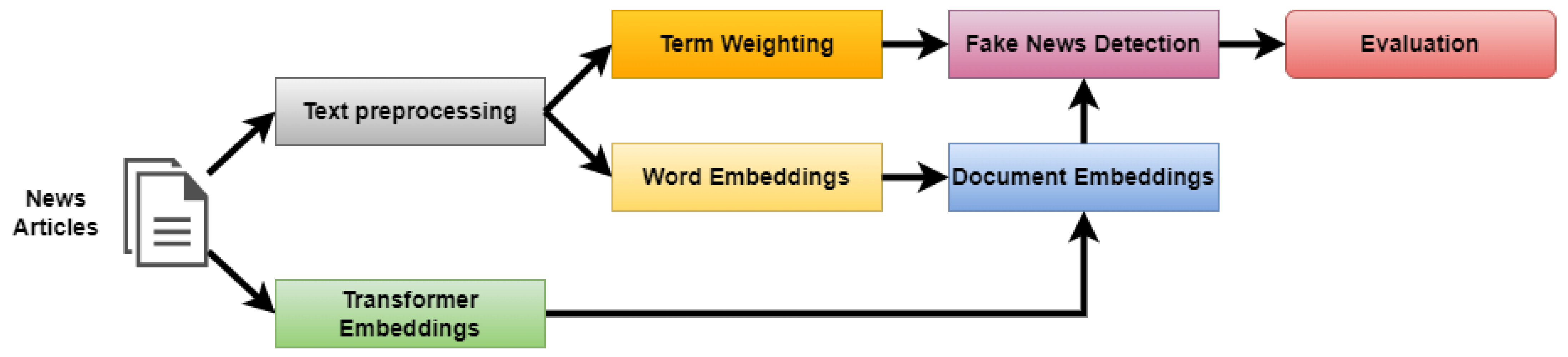
3. Methodology
3.1. Text Preprocessing
3.2. Term Weighting
3.3. Word Embeddings
3.3.1. Word2Vec
CBOW Model
Skip-Gram Model
3.3.2. FastText
3.3.3. GloVe
3.4. Transformers Embeddings
3.4.1. BERT
3.4.2. RoBERTa
3.4.3. BART
3.5. Document Embeddings
3.6. Fake News Detection
3.6.1. Naïve Bayes
3.6.2. Gradient Boosted Trees
3.6.3. Perceptron
3.6.4. Multi-Layer Perceptron
3.6.5. Long Short-Term Memory
- is the input vector of dimension m at step t, with ;
- is the hidden state vector as well as the unit’s output vector of dimension n, where the initial value is ;
- is the input activation vector;
- is the cell state vector, with the initial value ;
- are the weight matrices corresponding to the current input of the input gate, output gate, forget gate, and the cell state;
- are the weight matrices corresponding to the hidden output of the previous state for the current input of the input gate, output gate, forget gate, and the cell state;
- are the bias vectors corresponding to the current input of the input gate, output gate, forget gate, and the cell state;
- is the hyperbolic tangent activation function;
- ⊙ is the Hadamard Product, i.e., element wise product.
3.6.6. Bidirectional LSTM
3.6.7. Gated Recurrent Unit
- is the input vector of dimension m at step t, with ;
- is the input and output of the cell at step t;
- is the candidate hidden state with a cell dimension of n;
- is the current hidden state with a cell dimension of n;
- are the weight matrices corresponding to the current input of the update gate, reset gate, and the hidden state;
- are the weight matrices corresponding to the hidden output of the previous state for the current input of the update gate, reset gate, and the hidden state;
- are the bias vectors corresponding to the current input of the update gate, reset gate, and the hidden state;
- ⊙ is the Hadamard Product.
3.6.8. Bidirectional GRU
3.7. Evaluation Module
- (True Positive) is the number of positive observations that are correctly classified;
- (False Negative) is the number of positive observations that are incorrectly classified as negative;
- (False Positive) is the number of false observations that are incorrectly classified as positive;
- (True Negative) is the number of false observations that are correctly classified.
4. Experimental Results
4.1. Dataset Details
- (1)
- Verify that the title matches the title from the URL;
- (2)
- Verify that the content matches the content from the URL;
- (3)
- Verify that the authors match the authors from the URL;
- (4)
- Verify that the source matches the source from the URL;
- (5)
- Verify if the information is false or reliable;
4.2. Experimental Setup
4.3. Fake News Detection
- (1)
- A simpler neural architecture offers similar or better results compared to complex deep learning architectures that employ multiple layers, i.e., in our comparison, we obtained similar results as the complex MisRoBÆRTa [23] architecture without fine-tuning the transformers;
- (2)
- The embeddings used to vectorize the textual data make all the difference in performance, i.e., the right embedding must be selected to obtain good results with a given model;
- (3)
- We need a data-driven approach to select the best model and the best embedding for our dataset.
4.4. Additional Experiments
- (1)
- A simpler neural architecture offers at least similar or better results as complex architectures that employ multiple layers, and
- (2)
- The difference in performance lies in the embeddings used to vectorize the textual data.
- (1)
- (2)
- On the LIAR dataset with 2 labels, Upadhayay and Behzadan [51] obtained an accuracy of using CNN with BERT-base embeddings, while we obtained an accuracy of using LSTM with the document embeddings employing GloVe;
- (3)
- (4)
- On the Buzz Feed News dataset, Horne and Adali [52] obtained an accuracy of using a linear SVM, while we obtained an accuracy of using Perceptron with the document embeddings employing BART;
- (5)
5. Discussion
- (1)
- A simpler neural architecture offers similar if not better results as complex deep learning architectures that employ multiple layers, i.e., in our comparison, we obtained similar results as the complex MisRoBÆRTa [23] architecture, better than state-of-the-art results, i.e., FakeBERT [18], and Poppy [70];
- (2)
- The embeddings used to vectorize the textual data makes all the difference in performance, i.e., the right embedding must be selected to obtain good results with a given model;
- (3)
- We need a data-driven approach to select the best model and the best embedding for our dataset;
- (4)
- The way the word embedding manages to encapsulate the semantic, syntactic, and context features improves the performance of the classification models.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TF | Term Frequency |
| IDF | Inverse Document Frequency |
| TFIDF | Term Frequency-Inverse Document Frequency |
| CLDF | Class Label Frequency Distance |
| SG | Skip-Gram |
| CBOW | Common Bag Of Words |
| GloVe | Global Vectors |
| DocEmb | Document Embedding |
| BERT | Bidirectional Encoder Representations from Transformers |
| RoBERTa | Robustly Optimized BERT pre-training Approach |
| XLM-RoBERTa | Cross-Lingual RoBERTa |
| BART | Bidirectional and Auto-Regressive Transformers |
| NB | Naïve Bayes |
| MNB | Multinomial Naïve Bayes |
| GNB | Gaussian Naïve Bayes |
| SVM | Support Vector Machine |
| LogReg | Logistic Regression |
| UFD | Unsupervised Fake News Detection Framework |
| MLP | Multi-layer Perceptron |
| RNN | Recurrent Neural Network |
| CNN | Convolutional Neural Networks |
| C-CNN | Concatenated CNN |
| LSTM | Long Short-Term Memory |
| BiLSTM | Bidirectional Long Short-Term Memory |
| GRU | Gated Recurrent Unit |
| BiGRU | Bidirectional Gated Recurrent Unit |
References
- Truică, C.O.; Apostol, E.S.; Ștefu, T.; Karras, P. A Deep Learning Architecture for Audience Interest Prediction of News Topic on Social Media. In Proceedings of the International Conference on Extending Database Technology (EDBT2021), Nicosia, Cyprus, 23–26 March 2021; pp. 588–599. [Google Scholar] [CrossRef]
- Mustafaraj, E.; Metaxas, P.T. The Fake News Spreading Plague. In Proceedings of the ACM on Web Science Conference, Troy, NY, USA, 25–28 June 2017; pp. 235–239. [Google Scholar] [CrossRef]
- Ruths, D. The misinformation machine. Science 2019, 363, 348. [Google Scholar] [CrossRef] [PubMed]
- Bastos, M.T.; Mercea, D. The Brexit Botnet and User-Generated Hyperpartisan News. Soc. Sci. Comput. Rev. 2017, 37, 38–54. [Google Scholar] [CrossRef]
- Bovet, A.; Makse, H.A. Influence of fake news in Twitter during the 2016 US presidential election. Nat. Commun. 2019, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Rzymski, P.; Borkowski, L.; Drąg, M.; Flisiak, R.; Jemielity, J.; Krajewski, J.; Mastalerz-Migas, A.; Matyja, A.; Pyrć, K.; Simon, K.; et al. The Strategies to Support the COVID-19 Vaccination with Evidence-Based Communication and Tackling Misinformation. Vaccines 2021, 9, 109. [Google Scholar] [CrossRef]
- Truică, C.O.; Apostol, E.S.; Paschke, A. Awakened at CheckThat! 2022: Fake news detection using BiLSTM and sentence transformer. In Proceedings of the Working Notes of the Conference and Labs of the Evaluation Forum (CLEF2022), Bologna, Italy, 5–8 September 2022; pp. 749–757. [Google Scholar]
- European Commission. Fighting Disinformation; European Commission: Brussels, Belgium, 2020. [Google Scholar]
- Chen, Y.; Conroy, N.K.; Rubin, V.L. News in an online world: The need for an “automatic crap detector”. Proc. Assoc. Inf. Sci. Technol. 2015, 52, 1–4. [Google Scholar] [CrossRef]
- Wang, W.Y. “Liar, Liar Pants on Fire”: A New Benchmark Dataset for Fake News Detection. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Vancouver, BC, Canada, 30 July–4 August 2017; pp. 422–426. [Google Scholar] [CrossRef]
- Conroy, N.K.; Rubin, V.L.; Chen, Y. Automatic deception detection: Methods for finding fake news. Proc. Assoc. Inf. Sci. Technol. 2015, 52, 1–4. [Google Scholar] [CrossRef]
- Kaliyar, R.K.; Goswami, A.; Narang, P.; Sinha, S. FNDNet—A deep convolutional neural network for fake news detection. Cogn. Syst. Res. 2020, 61, 32–44. [Google Scholar] [CrossRef]
- Goldani, M.H.; Safabakhsh, R.; Momtazi, S. Convolutional neural network with margin loss for fake news detection. Inf. Process. Manag. 2021, 58, 102418. [Google Scholar] [CrossRef]
- Saleh, H.; Alharbi, A.; Alsamhi, S.H. OPCNN-FAKE: Optimized convolutional neural network for fake news detection. IEEE Access 2021, 9, 129471–129489. [Google Scholar] [CrossRef]
- Samantaray, S.; Kumar, A. Bi-directional Long Short-Term Memory Network for Fake News Detection from Social Media. In Intelligent and Cloud Computing; Springer: Berlin/Heidelberg, Germany, 2022; pp. 463–470. [Google Scholar] [CrossRef]
- Ilie, V.I.; Truică, C.O.; Apostol, E.S.; Paschke, A. Context-Aware Misinformation Detection: A Benchmark of Deep Learning Architectures Using Word Embeddings. IEEE Access 2021, 9, 162122–162146. [Google Scholar] [CrossRef]
- Jwa, H.; Oh, D.; Park, K.; Kang, J.; Lim, H. exBAKE: Automatic Fake News Detection Model Based on Bidirectional Encoder Representations from Transformers (BERT). Appl. Sci. 2019, 9, 4062. [Google Scholar] [CrossRef]
- Kaliyar, R.K.; Goswami, A.; Narang, P. FakeBERT: Fake news detection in social media with a BERT-based deep learning approach. Multimed. Tools Appl. 2021, 80, 11765–11788. [Google Scholar] [CrossRef] [PubMed]
- Kula, S.; Choraś, M.; Kozik, R. Application of the BERT-Based Architecture in Fake News Detection. In Conference on Complex, Intelligent, and Software Intensive Systems; Springer: Berlin/Heidelberg, Germany, 2020; pp. 239–249. [Google Scholar] [CrossRef]
- Mersinias, M.; Afantenos, S.; Chalkiadakis, G. CLFD: A Novel Vectorization Technique and Its Application in Fake News Detection. In Proceedings of the Language Resources and Evaluation Conference, Marseille, France, 11–16 May 2020; pp. 3475–3483. [Google Scholar]
- Mondal, S.K.; Sahoo, J.P.; Wang, J.; Mondal, K.; Rahman, M.M. Fake News Detection Exploiting TF-IDF Vectorization with Ensemble Learning Models. In Advances in Distributed Computing and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2022; pp. 261–270. [Google Scholar] [CrossRef]
- Aslam, N.; Khan, I.U.; Alotaibi, F.S.; Aldaej, L.A.; Aldubaikil, A.K. Fake Detect: A Deep Learning Ensemble Model for Fake News Detection. Complexity 2021, 2021, 5557784. [Google Scholar] [CrossRef]
- Truică, C.O.; Apostol, E.S. MisRoBÆRTa: Transformers versus Misinformation. Mathematics 2022, 10, 569. [Google Scholar] [CrossRef]
- Sedik, A.; Abohany, A.A.; Sallam, K.M.; Munasinghe, K.; Medhat, T. Deep fake news detection system based on concatenated and recurrent modalities. Expert Syst. Appl. 2022, 208, 117953. [Google Scholar] [CrossRef]
- Verma, P.K.; Agrawal, P.; Amorim, I.; Prodan, R. WELFake: Word Embedding Over Linguistic Features for Fake News Detection. IEEE Trans. Comput. Soc. Syst. 2021, 8, 881–893. [Google Scholar] [CrossRef]
- Shu, K.; Cui, L.; Wang, S.; Lee, D.; Liu, H. dEFEND: Explainable Fake News Detection. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 395–405. [Google Scholar] [CrossRef]
- Khattar, D.; Goud, J.S.; Gupta, M.; Varma, V. MVAE: Multimodal Variational Autoencoder for Fake News Detection. In Proceedings of the World Wide Web Conference, San Francisco, CA, USA, 13–17 May 2019; pp. 2915–2921. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, B.; Yu, P.S. FakeDetector: Effective Fake News Detection with Deep Diffusive Neural Network. In Proceedings of the 2020 IEEE 36th International Conference on Data Engineering (ICDE), Dallas, TX, USA, 20–24 April 2020; pp. 1826–1829. [Google Scholar] [CrossRef]
- Yang, S.; Shu, K.; Wang, S.; Gu, R.; Wu, F.; Liu, H. Unsupervised Fake News Detection on Social Media: A Generative Approach. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 5644–5651. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, S.; Hu, J.; Fang, Q.; Xu, C. Fake News Detection via Knowledge-driven Multimodal Graph Convolutional Networks. In Proceedings of the 2020 International Conference on Multimedia Retrieval, Dublin, Ireland, 8–11 June 2020; pp. 540–547. [Google Scholar] [CrossRef]
- Le, Q.; Mikolov, T. Distributed representations of sentences and documents. In Proceedings of the International Conference on Machine Learning PMLR, Bejing, China, 22–24 June 2014; pp. 1188–1196. [Google Scholar]
- Cui, J.; Kim, K.; Na, S.H.; Shin, S. Meta-Path-based Fake News Detection Leveraging Multi-level Social Context Information. In Proceedings of the Proceedings of the 31st ACM International Conference on Information & Knowledge Management, Atlanta, GA, USA, 17–21 October 2022; pp. 325–334. [Google Scholar]
- Singh, L. Fake news detection: A comparison between available Deep Learning techniques in vector space. In Proceedings of the 2020 IEEE 4th Conference on Information & Communication Technology (CICT), Chennai, India, 3–5 December 2020; pp. 1–4. [Google Scholar]
- Truică, C.O.; Apostol, E.S.; Darmont, J.; Assent, I. TextBenDS: A Generic Textual Data Benchmark for Distributed Systems. Inf. Syst. Front. 2021, 23, 81–100. [Google Scholar] [CrossRef]
- Paltoglou, G.; Thelwall, M. A Study of Information Retrieval Weighting Schemes for Sentiment Analysis. In Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics, Stroudsburg, PA, USA, 11–16 July 2010; pp. 1386–1395. [Google Scholar]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. In Proceedings of the Workshop Proceedings of the International Conference on Learning Representations 2013, Scottsdale, AZ, USA, 2–4 May 2013; pp. 1–12. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed Representations of Words and Phrases and their Compositionality. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 5–10 December 2013; Volume 26, pp. 1–9. [Google Scholar]
- Bojanowski, P.; Grave, E.; Joulin, A.; Mikolov, T. Enriching word vectors with subword information. Trans. Assoc. Comput. Linguist. 2017, 5, 135–146. [Google Scholar] [CrossRef]
- Pennington, J.; Socher, R.; Manning, C.D. GloVe: Global vectors for word representation. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar] [CrossRef]
- Mikolov, T.; Grave, E.; Bojanowski, P.; Puhrsch, C.; Joulin, A. Advances in Pre-Training Distributed Word Representations. In Proceedings of the International Conference on Language Resources and Evaluation, Miyazaki, Japan, 7–12 May 2018; pp. 52–55. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the Conference of the North Association for Computational Linguistics, Minneapolis, MN, USA, 2–7 June 2019; pp. 4171–4186. [Google Scholar] [CrossRef]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Lewis, M.; Liu, Y.; Goyal, N.; Ghazvininejad, M.; Mohamed, A.; Levy, O.; Stoyanov, V.; Zettlemoyer, L. BART: Denoising Sequence-to-Sequence Pre-training for Natural Language Generation, Translation, and Comprehension. In Proceedings of the Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020; pp. 7871–7880. [Google Scholar] [CrossRef]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-Art Natural Language Processing. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Honolulu, HI, USA, 16–20 November 2020; pp. 38–45. [Google Scholar] [CrossRef]
- Rennie, J.D.M.; Shih, L.; Teevan, J.; Karger, D.R. Tackling the Poor Assumptions of Naive Bayes Text Classifiers. In Proceedings of the International Conference on International Conference on Machine Learning, Los Angeles, CA, USA, 23–24 June 2003; pp. 616–623. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Cho, K.; van Merrienboer, B.; Bahdanau, D.; Bengio, Y. On the Properties of Neural Machine Translation: Encoder–Decoder Approaches. In Proceedings of the SSST-8, Eighth Workshop on Syntax, Semantics and Structure in Statistical Translation, Doha, Qatar, 25 October 2014; pp. 103–111. [Google Scholar] [CrossRef]
- Hewamalage, H.; Bergmeir, C.; Bandara, K. Recurrent Neural Networks for Time Series Forecasting: Current status and future directions. Int. J. Forecast. 2021, 37, 388–427. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Szpakowski, M. FakeNewsCorpus. 2020. Available online: https://github.com/several27/FakeNewsCorpus (accessed on 27 December 2022).
- Upadhayay, B.; Behzadan, V. Sentimental LIAR: Extended Corpus and Deep Learning Models for Fake Claim Classification. In Proceedings of the 2020 IEEE International Conference on Intelligence and Security Informatics (ISI), Virtual Event, 9–10 November 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Horne, B.; Adali, S. This Just In: Fake News Packs A Lot In Title, Uses Simpler, Repetitive Content in Text Body, More Similar to Satire Than Real News. In Proceedings of the International AAAI Conference on Web and Social Media, Montreal, QC, Canada, 15–18 May 2017; pp. 759–766. [Google Scholar]
- Rashkin, H.; Choi, E.; Jang, J.Y.; Volkova, S.; Choi, Y. Truth of Varying Shades: Analyzing Language in Fake News and Political Fact-Checking. In Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, Copenhagen, Denmark, 7–11 September 2017; pp. 2931–2937. [Google Scholar] [CrossRef]
- Barrón-Cedeño, A.; Jaradat, I.; Da San Martino, G.; Nakov, P. Proppy: Organizing the news based on their propagandistic content. Inf. Process. Manag. 2019, 56, 1849–1864. [Google Scholar] [CrossRef]
- Kurasinski, L.; Mihailescu, R.C. Towards Machine Learning Explainability in Text Classification for Fake News Detection. In Proceedings of the 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 14–17 December 2020; pp. 775–781. [Google Scholar] [CrossRef]
- Nørregaard, J.; Horne, B.D.; Adalı, S. NELA-GT-2018: A Large Multi-Labelled News Dataset for the Study of Misinformation in News Articles. In Proceedings of the International AAAI Conference on Web and Social Media, Münich, Germany, 11–14 June 2019; pp. 630–638. [Google Scholar]
- Kwak, H.; An, J.; Ahn, Y.Y. A Systematic Media Frame Analysis of 1. In 5 Million New York Times Articles from 2000 to 2017. In Proceedings of the ACM Conference on Web Science, Southampton, UK, 6–10 July 2020; pp. 305–314. [Google Scholar] [CrossRef]
- Reed, R.D.; Marks II, R.J. Neural Smithing: Supervised Learning in Feedforward Artificial Neural Networks; MIT Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Arora, S.; Ge, R.; Moitra, A. Learning Topic Models – Going beyond SVD. In Proceedings of the Annual Symposium on Foundations of Computer Science, Washington, DC, USA, 20–23 October 2012; pp. 1–10. [Google Scholar] [CrossRef]
- Bird, S.; Loper, E.; Klein, E. Natural Language Processing with Python; O’Reilly: Sebastopol, CA, USA, 2009. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Grootendorst, M. PolyFuzz. 2020. Available online: https://maartengr.github.io/PolyFuzz/ (accessed on 27 December 2022).
- Řehůřek, R.; Sojka, P. Software Framework for Topic Modelling with Large Corpora. In Proceedings of the Workshop on New Challenges for NLP Frameworks, Valletta, Malta, 22 May 2010; pp. 45–50. [Google Scholar]
- Kula, M. Python-Glove. 2020. Available online: https://github.com/maciejkula/glove-python (accessed on 27 December 2022).
- Rajapakse, T. SimpleTransformers. 2021. Available online: https://simpletransformers.ai/ (accessed on 27 December 2022).
- Reimers, N.; Gurevych, I. Sentence-BERT: Sentence Embeddings using Siamese BERT-Networks. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing, Hong Kong, China, 3–7 November 2019; pp. 3982–3992. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- D’Ulizia, A.; Caschera, M.C.; Ferri, F.; Grifoni, P. Fake news detection: A survey of evaluation datasets. PeerJ Comput. Sci. 2021, 7, e518. [Google Scholar] [CrossRef]
- Alhindi, T.; Petridis, S.; Muresan, S. Where is Your Evidence: Improving Fact-checking by Justification Modeling. In Proceedings of the First Workshop on Fact Extraction and VERification (FEVER), Brussels, Belgium, 24 July 2018; pp. 85–90. [Google Scholar] [CrossRef]
- Barrón-Cedeño, A.; Martino, G.D.S.; Jaradat, I.; Nakov, P. Proppy: A System to Unmask Propaganda in Online News. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; pp. 9847–9848. [Google Scholar] [CrossRef]
- Truică, C.O.; Apostol, E.S.; Șerban, M.L.; Paschke, A. Topic-Based Document-Level Sentiment Analysis Using Contextual Cues. Mathematics 2021, 9, 2722. [Google Scholar] [CrossRef]
| Statistics before Preprocessing | #Tokens per Document | #Tokens per Class | ||||||
|---|---|---|---|---|---|---|---|---|
| Class | Encoding | Description | Mean | Min | Max | StdDev | Unique | All |
| Fake News | 1 | Fabricated or distorted information | 517.83 | 6 | 8 812 | 883.35 | 119 283 | 5 178 300 |
| Reliable | 0 | Reliable information | 575.66 | 7 | 10 541 | 602.16 | 82 203 | 5 756 643 |
| Entire Dataset Statistics | 546.75 | 6 | 10 541 | 618.63 | 159 113 | 10 934 943 | ||
| Textual Information after Preprocessing | Sim (FT) | Sim (BERT) | ||||||
| Top-10 Unigrams | Fake News | people time government world year story market American God day | 0.83 | 0.93 | ||||
| Reliable | people God Christian government American time world war America political | |||||||
| Top-1 Topic | Fake News | people Trump year day government time state world market war | 0.84 | 0.94 | ||||
| Reliable | church Trump people God president war state year Bush government | |||||||
| Naïve Bayes | Gradient Boosted Trees | |||||
|---|---|---|---|---|---|---|
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 92.69 ± 0.25 | 91.72 ± 0.39 | 93.86 ± 0.41 | 98.76 ± 0.21 | 99.79 ± 0.08 | 97.72 ± 0.43 |
| DocEmbWord2VecCBOW | 66.29 ± 0.53 | 60.78 ± 0.42 | 91.85 ± 0.27 | 95.67 ± 0.18 | 96.17 ± 0.37 | 95.13 ± 0.34 |
| DocEmbWord2VecSG | 53.10 ± 0.37 | 51.74 ± 0.20 | 92.46 ± 0.77 | 97.10 ± 0.21 | 97.61 ± 0.31 | 96.56 ± 0.16 |
| DocEmbFastTextCBOW | 56.13 ± 0.53 | 53.59 ± 0.33 | 91.45 ± 0.58 | 94.90 ± 0.24 | 95.49 ± 0.39 | 94.24 ± 0.46 |
| DocEmbFastTextSG | 54.00 ± 0.69 | 52.23 ± 0.38 | 93.66 ± 0.98 | 97.05 ± 0.26 | 97.45 ± 0.32 | 96.64 ± 0.49 |
| DocEmbGloVe | 53.43 ± 0.42 | 51.98 ± 0.24 | 89.94 ± 0.78 | 96.02 ± 0.30 | 96.73 ± 0.35 | 95.26 ± 0.43 |
| DocEmbBERT | 80.90 ± 0.64 | 74.94 ± 0.67 | 92.87 ± 0.58 | 97.43 ± 0.21 | 97.75 ± 0.29 | 97.10 ± 0.30 |
| DocEmbRoBERTa | 91.98 ± 0.31 | 94.05 ± 0.44 | 89.63 ± 0.60 | 97.38 ± 0.22 | 98.72 ± 0.31 | 96.02 ± 0.42 |
| DocEmbBART | 89.13 ± 0.32 | 83.71 ± 0.46 | 97.19 ± 0.37 | 98.26 ± 0.19 | 98.18 ± 0.31 | 98.35 ± 0.21 |
| Perceptron | Multi-Layer Perceptron | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 95.79 ± 0.33 | 96.55 ± 0.46 | 94.98 ± 0.42 | 98.04 ± 0.19 | 98.37 ± 0.28 | 97.70 ± 0.33 |
| DocEmbWord2VecCBOW | 93.61 ± 0.27 | 94.22 ± 0.53 | 92.93 ± 0.49 | 94.96 ± 0.31 | 95.26 ± 0.36 | 94.63 ± 0.64 |
| DocEmbWord2VecSG | 92.04 ± 0.34 | 94.65 ± 0.49 | 89.12 ± 0.91 | 95.88 ± 0.30 | 96.34 ± 0.65 | 95.40 ± 1.09 |
| DocEmbFastTextCBOW | 93.46 ± 0.30 | 94.48 ± 0.64 | 92.33 ± 0.67 | 94.92 ± 0.23 | 95.12 ± 0.72 | 94.71 ± 0.67 |
| DocEmbFastTextSG | 91.60 ± 0.49 | 94.06 ± 0.54 | 88.81 ± 0.76 | 96.00 ± 0.30 | 96.48 ± 0.35 | 95.48 ± 0.74 |
| DocEmbGloVe | 89.57 ± 0.50 | 92.57 ± 0.60 | 86.04 ± 1.18 | 94.05 ± 0.38 | 94.29 ± 0.89 | 93.79 ± 0.60 |
| DocEmbBERT | 97.09 ± 0.21 | 97.50 ± 0.62 | 96.66 ± 0.62 | 98.34 ± 0.19 | 98.56 ± 0.62 | 98.11 ± 0.56 |
| DocEmbRoBERTa | 96.19 ± 0.50 | 96.89 ± 1.62 | 95.49 ± 0.99 | 97.28 ± 0.55 | 98.51 ± 1.32 | 96.04 ± 0.47 |
| DocEmbBART | 98.57 ± 0.15 | 98.71 ± 0.29 | 98.43 ± 0.33 | 98.93 ± 0.16 | 99.07 ± 0.55 | 98.80 ± 0.39 |
| LSTM | Bidirectional LSTM | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 97.88 ± 0.23 | 98.20 ± 0.19 | 97.55 ± 0.40 | 97.84 ± 0.24 | 98.21 ± 0.30 | 97.46 ± 0.49 |
| DocEmbWord2VecCBOW | 96.59 ± 0.36 | 96.38 ± 1.05 | 96.84 ± 1.17 | 96.89 ± 0.26 | 96.89 ± 0.53 | 96.89 ± 0.38 |
| DocEmbWord2VecSG | 96.23 ± 0.31 | 96.70 ± 0.97 | 95.76 ± 1.32 | 96.39 ± 0.37 | 96.81 ± 1.30 | 95.98 ± 1.32 |
| DocEmbFastTextCBOW | 96.16 ± 0.26 | 96.22 ± 0.55 | 96.11 ± 0.85 | 96.30 ± 0.32 | 96.63 ± 1.19 | 95.98 ± 1.14 |
| DocEmbFastTextSG | 96.61 ± 0.27 | 96.52 ± 0.91 | 96.72 ± 0.90 | 96.79 ± 0.22 | 96.78 ± 0.91 | 96.82 ± 0.88 |
| DocEmbGloVe | 94.66 ± 0.48 | 94.92 ± 2.02 | 94.45 ± 1.72 | 94.86 ± 0.37 | 95.24 ± 1.67 | 94.51 ± 1.63 |
| DocEmbBERT | 98.57 ± 0.34 | 98.59 ± 0.76 | 98.57 ± 1.10 | 98.72 ± 0.40 | 98.90 ± 0.81 | 98.55 ± 0.87 |
| DocEmbRoBERTa | 96.88 ± 1.57 | 98.02 ± 2.95 | 95.80 ± 1.56 | 96.97 ± 1.33 | 97.78 ± 2.85 | 96.22 ± 0.74 |
| DocEmbBART | 99.29 ± 0.10 | 99.46 ± 0.13 | 99.13 ± 0.14 | 99.34 ± 0.08 | 99.48 ± 0.12 | 99.20 ± 0.11 |
| GRU | Bidirectional GRU | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 97.88 ± 0.24 | 98.28 ± 0.27 | 97.47 ± 0.45 | 97.84 ± 0.30 | 98.04 ± 0.60 | 97.64 ± 0.58 |
| DocEmbWord2VecCBOW | 96.56 ± 0.29 | 96.43 ± 0.75 | 96.71 ± 1.01 | 96.57 ± 0.23 | 96.37 ± 0.90 | 96.81 ± 0.69 |
| DocEmbWord2VecSG | 96.21 ± 0.44 | 95.91 ± 1.44 | 96.58 ± 0.97 | 96.35 ± 0.38 | 96.45 ± 1.09 | 96.26 ± 1.47 |
| DocEmbFastTextCBOW | 96.12 ± 0.16 | 96.54 ± 1.04 | 95.70 ± 1.00 | 96.20 ± 0.33 | 96.47 ± 0.88 | 95.91 ± 1.00 |
| DocEmbFastTextSG | 96.40 ± 0.35 | 96.11 ± 1.22 | 96.74 ± 1.32 | 96.76 ± 0.22 | 96.76 ± 0.59 | 96.77 ± 0.50 |
| DocEmbGloVe | 94.62 ± 0.64 | 95.60 ± 2.30 | 93.66 ± 1.81 | 94.84 ± 0.60 | 95.80 ± 1.74 | 93.86 ± 2.01 |
| DocEmbBERT | 98.82 ± 0.11 | 98.71 ± 0.52 | 98.92 ± 0.48 | 98.61 ± 0.41 | 99.17 ± 0.47 | 98.05 ± 1.14 |
| DocEmbRoBERTa | 97.37 ± 0.60 | 99.17 ± 0.43 | 95.56 ± 1.47 | 97.31 ± 0.44 | 98.34 ± 1.24 | 96.26 ± 0.72 |
| DocEmbBART | 99.31 ± 0.10 | 99.39 ± 0.23 | 99.22 ± 0.14 | 99.36 ± 0.08 | 99.50 ± 0.09 | 99.22 ± 0.09 |
| Accuracy | Precision | Recall | ||||
| MisRoBÆRTa [23] | 99.34 ± 0.03 | 99.34 ± 0.03 | 99.34 ± 0.02 | |||
| Naïve Bayes | Gradient Boosted Trees | |||||
|---|---|---|---|---|---|---|
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 23.07 ± 0.70 | 24.16 ± 2.03 | 23.07 ± 0.70 | 23.00 ± 0.93 | 22.86 ± 0.89 | 23.00 ± 0.93 |
| DocEmbWord2VecCBOW | 18.32 ± 1.01 | 21.62 ± 1.42 | 18.32 ± 1.01 | 22.40 ± 0.59 | 22.37 ± 0.68 | 22.40 ± 0.59 |
| DocEmbWord2VecSG | 20.42 ± 0.96 | 21.74 ± 0.84 | 20.42 ± 0.96 | 23.12 ± 0.69 | 23.29 ± 0.69 | 23.12 ± 0.69 |
| DocEmbFastTextCBOW | 17.19 ± 0.63 | 21.89 ± 1.45 | 17.19 ± 0.63 | 22.68 ± 0.55 | 22.63 ± 0.71 | 22.68 ± 0.55 |
| DocEmbFastTextSG | 19.85 ± 1.10 | 21.57 ± 1.22 | 19.85 ± 1.10 | 22.93 ± 0.83 | 23.00 ± 0.80 | 22.93 ± 0.83 |
| DocEmbGloVe | 17.60 ± 0.77 | 21.31 ± 1.18 | 17.60 ± 0.77 | 21.99 ± 0.63 | 21.72 ± 0.71 | 21.99 ± 0.63 |
| DocEmbBERT | 20.58 ± 0.71 | 22.40 ± 1.17 | 20.58 ± 0.71 | 23.78 ± 0.82 | 24.03 ± 0.97 | 23.78 ± 0.82 |
| DocEmbRoBERTa | 15.91 ± 1.02 | 20.31 ± 1.38 | 15.91 ± 1.02 | 21.09 ± 1.08 | 20.51 ± 0.85 | 21.09 ± 1.08 |
| DocEmbBART | 21.79 ± 0.90 | 24.07 ± 1.09 | 21.79 ± 0.90 | 24.93 ± 0.74 | 25.26 ± 0.83 | 24.93 ± 0.74 |
| Perceptron | Multi-Layer Perceptron | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 23.71 ± 0.68 | 24.79 ± 1.26 | 23.71 ± 0.68 | 23.04 ± 0.66 | 23.08 ± 0.77 | 23.04 ± 0.66 |
| DocEmbWord2VecCBOW | 22.83 ± 0.65 | 22.37 ± 0.66 | 22.83 ± 0.65 | 22.70 ± 0.91 | 22.56 ± 0.87 | 22.70 ± 0.91 |
| DocEmbWord2VecSG | 23.46 ± 0.73 | 23.19 ± 0.68 | 23.46 ± 0.73 | 23.26 ± 0.65 | 23.15 ± 1.09 | 23.26 ± 0.65 |
| DocEmbFastTextCBOW | 22.34 ± 0.49 | 21.63 ± 0.81 | 22.34 ± 0.49 | 22.72 ± 0.89 | 22.16 ± 1.19 | 22.72 ± 0.89 |
| DocEmbFastTextSG | 23.62 ± 0.82 | 23.29 ± 1.08 | 23.62 ± 0.82 | 23.48 ± 0.92 | 23.47 ± 1.13 | 23.48 ± 0.92 |
| DocEmbGloVe | 23.64 ± 0.55 | 22.54 ± 0.97 | 23.64 ± 0.55 | 23.24 ± 0.71 | 21.94 ± 1.31 | 23.24 ± 0.71 |
| DocEmbBERT | 24.06 ± 1.03 | 24.33 ± 1.00 | 24.06 ± 1.03 | 23.58 ± 0.66 | 23.88 ± 0.84 | 23.58 ± 0.66 |
| DocEmbRoBERTa | 21.66 ± 1.59 | 21.01 ± 1.60 | 21.66 ± 1.59 | 22.82 ± 0.61 | 20.83 ± 1.22 | 22.82 ± 0.61 |
| DocEmbBART | 25.60 ± 0.41 | 25.84 ± 0.18 | 25.60 ± 0.41 | 25.89 ± 0.75 | 26.15 ± 0.72 | 25.89 ± 0.75 |
| LSTM | Bidirectional LSTM | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 21.77 ± 0.50 | 21.77 ± 0.48 | 21.77 ± 0.50 | 21.62 ± 0.49 | 21.60 ± 0.48 | 21.62 ± 0.49 |
| DocEmbWord2VecCBOW | 22.70 ± 0.95 | 22.53 ± 0.94 | 22.70 ± 0.95 | 22.65 ± 0.60 | 22.57 ± 0.62 | 22.65 ± 0.60 |
| DocEmbWord2VecSG | 23.66 ± 0.99 | 23.46 ± 1.02 | 23.66 ± 0.99 | 23.51 ± 0.69 | 23.14 ± 0.84 | 23.51 ± 0.69 |
| DocEmbFastTextCBOW | 22.40 ± 1.06 | 22.23 ± 1.07 | 22.40 ± 1.06 | 22.53 ± 0.85 | 22.49 ± 0.86 | 22.53 ± 0.85 |
| DocEmbFastTextSG | 23.50 ± 0.76 | 23.24 ± 0.90 | 23.50 ± 0.76 | 23.45 ± 0.61 | 23.41 ± 0.89 | 23.45 ± 0.61 |
| DocEmbGloVe | 23.59 ± 0.46 | 22.90 ± 1.40 | 23.59 ± 0.46 | 23.08 ± 0.42 | 22.77 ± 0.88 | 23.08 ± 0.42 |
| DocEmbBERT | 23.21 ± 0.55 | 23.37 ± 0.52 | 23.21 ± 0.55 | 23.31 ± 0.70 | 23.25 ± 0.76 | 23.31 ± 0.70 |
| DocEmbRoBERTa | 22.94 ± 1.00 | 21.65 ± 1.08 | 22.94 ± 1.00 | 22.96 ± 0.61 | 19.77 ± 1.66 | 22.96 ± 0.61 |
| DocEmbBART | 25.02 ± 0.57 | 25.08 ± 0.59 | 25.02 ± 0.57 | 25.75 ± 0.61 | 25.78 ± 0.62 | 25.75 ± 0.61 |
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 21.62 ± 0.50 | 21.63 ± 0.47 | 21.62 ± 0.50 | 21.43 ± 0.44 | 21.44 ± 0.46 | 21.43 ± 0.44 |
| DocEmbWord2VecCBOW | 22.36 ± 1.00 | 22.14 ± 0.94 | 22.36 ± 1.00 | 22.51 ± 0.76 | 22.46 ± 0.79 | 22.51 ± 0.76 |
| DocEmbWord2VecSG | 23.47 ± 0.53 | 23.02 ± 0.72 | 23.47 ± 0.53 | 23.47 ± 0.71 | 23.16 ± 0.66 | 23.47 ± 0.71 |
| DocEmbFastTextCBOW | 22.62 ± 0.77 | 22.48 ± 0.72 | 22.62 ± 0.77 | 22.51 ± 0.53 | 22.42 ± 0.44 | 22.51 ± 0.53 |
| DocEmbFastTextSG | 23.34 ± 0.55 | 23.08 ± 0.76 | 23.34 ± 0.55 | 23.54 ± 0.71 | 23.28 ± 0.58 | 23.54 ± 0.71 |
| DocEmbGloVe | 23.47 ± 0.64 | 22.84 ± 1.76 | 23.47 ± 0.64 | 23.21 ± 0.56 | 22.93 ± 1.27 | 23.21 ± 0.56 |
| DocEmbBERT | 23.64 ± 0.36 | 23.90 ± 0.53 | 23.64 ± 0.36 | 23.00 ± 0.72 | 23.21 ± 0.88 | 23.00 ± 0.72 |
| DocEmbRoBERTa | 22.69 ± 0.68 | 19.84 ± 1.95 | 22.69 ± 0.68 | 22.73 ± 0.72 | 21.55 ± 2.26 | 22.73 ± 0.72 |
| DocEmbBART | 24.99 ± 0.66 | 25.00 ± 0.69 | 24.99 ± 0.66 | 25.20 ± 0.88 | 25.26 ± 0.86 | 25.20 ± 0.88 |
| Accuracy | Precision | Recall | ||||
| MisRoBÆRTa [23] | 24.62 ± 0.39 | 25.87 ± 0.67 | 24.61 ± 0.39 | |||
| F1-Score | ||||||
| Hybrid CNNs [10] | 27.70 | |||||
| BiLSTM [69] | 26.00 | |||||
| Naïve Bayes | Gradient Boosted Trees | |||||
|---|---|---|---|---|---|---|
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 83.92 ± 0.04 | 83.96 ± 0.01 | 99.94 ± 0.04 | 83.64 ± 0.20 | 84.09 ± 0.08 | 99.31 ± 0.19 |
| DocEmbWord2VecCBOW | 67.02 ± 1.21 | 86.09 ± 0.43 | 72.43 ± 1.76 | 83.28 ± 0.19 | 84.15 ± 0.07 | 98.68 ± 0.25 |
| DocEmbWord2VecSG | 64.57 ± 3.54 | 86.07 ± 0.31 | 68.96 ± 5.01 | 83.30 ± 0.33 | 84.15 ± 0.11 | 98.70 ± 0.35 |
| DocEmbFastTextCBOW | 67.89 ± 2.19 | 85.60 ± 0.32 | 74.26 ± 3.20 | 83.25 ± 0.28 | 84.16 ± 0.12 | 98.61 ± 0.29 |
| DocEmbFastTextSG | 65.19 ± 4.32 | 86.17 ± 0.43 | 69.75 ± 6.17 | 83.28 ± 0.23 | 84.16 ± 0.10 | 98.65 ± 0.32 |
| DocEmbGloVe | 59.71 ± 1.80 | 85.89 ± 0.52 | 62.24 ± 2.54 | 83.05 ± 0.26 | 84.10 ± 0.08 | 98.42 ± 0.29 |
| DocEmbBERT | 61.21 ± 0.91 | 86.76 ± 0.58 | 63.49 ± 1.14 | 83.38 ± 0.19 | 84.16 ± 0.09 | 98.80 ± 0.19 |
| DocEmbRoBERTa | 60.04 ± 3.98 | 85.30 ± 0.53 | 63.35 ± 6.07 | 83.36 ± 0.20 | 84.02 ± 0.08 | 99.01 ± 0.25 |
| DocEmbBART | 62.11 ± 1.03 | 87.65 ± 0.52 | 63.87 ± 1.25 | 83.46 ± 0.19 | 84.36 ± 0.12 | 98.58 ± 0.32 |
| Perceptron | Multi-Layer Perceptron | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 83.97 ± 0.01 | 83.97 ± 0.01 | 99.99 ± 0.01 | 80.87 ± 0.69 | 84.50 ± 0.15 | 94.58 ± 1.13 |
| DocEmbWord2VecCBOW | 83.88 ± 0.06 | 83.96 ± 0.02 | 99.88 ± 0.06 | 83.96 ± 0.04 | 83.98 ± 0.02 | 99.97 ± 0.03 |
| DocEmbWord2VecSG | 83.94 ± 0.04 | 83.97 ± 0.01 | 99.95 ± 0.06 | 83.90 ± 0.10 | 83.99 ± 0.04 | 99.86 ± 0.10 |
| DocEmbFastTextCBOW | 83.87 ± 0.06 | 83.98 ± 0.03 | 99.82 ± 0.08 | 83.95 ± 0.06 | 83.99 ± 0.03 | 99.94 ± 0.07 |
| DocEmbFastTextSG | 83.97 ± 0.02 | 83.97 ± 0.01 | 99.99 ± 0.01 | 83.93 ± 0.08 | 83.99 ± 0.02 | 99.91 ± 0.10 |
| DocEmbGloVe | 83.97 ± 0.01 | 83.97 ± 0.01 | 99.99 ± 0.01 | 83.96 ± 0.01 | 83.97 ± 0.01 | 99.99 ± 0.01 |
| DocEmbBERT | 83.81 ± 0.11 | 83.98 ± 0.05 | 99.74 ± 0.14 | 83.18 ± 0.52 | 84.25 ± 0.25 | 98.37 ± 1.11 |
| DocEmbRoBERTa | 83.96 ± 0.03 | 83.97 ± 0.01 | 99.99 ± 0.03 | 83.97 ± 0.01 | 83.97 ± 0.01 | 99.99 ± 0.01 |
| DocEmbBART | 83.44 ± 0.55 | 84.37 ± 0.28 | 98.53 ± 1.17 | 81.63 ± 1.01 | 84.97 ± 0.33 | 94.91 ± 1.52 |
| LSTM | Bidirectional LSTM | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 77.05 ± 0.89 | 84.75 ± 0.20 | 88.62 ± 1.13 | 76.96 ± 0.86 | 84.75 ± 0.17 | 88.49 ± 1.08 |
| DocEmbWord2VecCBOW | 81.43 ± 0.57 | 84.44 ± 0.13 | 95.47 ± 0.88 | 80.51 ± 1.02 | 84.63 ± 0.18 | 93.83 ± 1.65 |
| DocEmbWord2VecSG | 83.86 ± 0.13 | 84.04 ± 0.05 | 99.72 ± 0.13 | 83.79 ± 0.16 | 84.07 ± 0.05 | 99.57 ± 0.17 |
| DocEmbFastTextCBOW | 81.20 ± 0.74 | 84.45 ± 0.23 | 95.13 ± 1.03 | 80.77 ± 1.12 | 84.49 ± 0.34 | 94.44 ± 1.63 |
| DocEmbFastTextSG | 83.88 ± 0.14 | 84.01 ± 0.04 | 99.79 ± 0.16 | 83.78 ± 0.19 | 84.00 ± 0.06 | 99.66 ± 0.19 |
| DocEmbGloVe | 83.99 ± 0.02 | 83.98 ± 0.02 | 99.99 ± 0.01 | 83.98 ± 0.04 | 83.98 ± 0.02 | 99.99 ± 0.01 |
| DocEmbBERT | 80.37 ± 1.53 | 84.84 ± 0.49 | 93.31 ± 2.74 | 79.69 ± 1.76 | 85.05 ± 0.35 | 92.00 ± 2.95 |
| DocEmbRoBERTa | 83.97 ± 0.01 | 83.97 ± 0.01 | 99.99 ± 0.01 | 83.97 ± 0.01 | 83.97 ± 0.01 | 99.99 ± 0.01 |
| DocEmbBART | 81.41 ± 0.85 | 84.93 ± 0.22 | 94.66 ± 1.21 | 81.43 ± 0.94 | 85.14 ± 0.22 | 94.34 ± 1.35 |
| GRU | Bidirectional GRU | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 76.99 ± 0.59 | 84.71 ± 0.19 | 88.58 ± 0.65 | 76.87 ± 0.66 | 84.72 ± 0.22 | 88.39 ± 0.94 |
| DocEmbWord2VecCBOW | 81.52 ± 0.67 | 84.49 ± 0.24 | 95.54 ± 0.95 | 80.52 ± 1.01 | 84.58 ± 0.15 | 93.92 ± 1.53 |
| DocEmbWord2VecSG | 83.91 ± 0.13 | 84.04 ± 0.04 | 99.80 ± 0.14 | 83.81 ± 0.16 | 84.06 ± 0.05 | 99.60 ± 0.18 |
| DocEmbFastTextCBOW | 80.93 ± 1.14 | 84.44 ± 0.24 | 94.74 ± 1.93 | 80.20 ± 0.96 | 84.67 ± 0.27 | 93.31 ± 1.55 |
| DocEmbFastTextSG | 83.89 ± 0.12 | 84.01 ± 0.04 | 99.80 ± 0.13 | 83.82 ± 0.16 | 84.02 ± 0.05 | 99.70 ± 0.16 |
| DocEmbGloVe | 83.98 ± 0.03 | 83.98 ± 0.02 | 99.99 ± 0.01 | 83.98 ± 0.03 | 83.99 ± 0.02 | 99.99 ± 0.01 |
| DocEmbBERT | 79.86 ± 2.30 | 84.73 ± 0.40 | 92.75 ± 3.83 | 79.94 ± 1.25 | 84.93 ± 0.36 | 92.54 ± 2.08 |
| DocEmbRoBERTa | 83.97 ± 0.01 | 83.97 ± 0.01 | 99.99 ± 0.01 | 83.97 ± 0.01 | 83.97 ± 0.01 | 99.99 ± 0.01 |
| DocEmbBART | 81.22 ± 0.75 | 85.13 ± 0.27 | 94.06 ± 1.19 | 80.60 ± 0.90 | 85.24 ± 0.22 | 93.01 ± 1.34 |
| Accuracy | Precision | Recall | ||||
| MisRoBÆRTa [23] | 81.15 ± 0.07 | 81.15 ± 0.07 | 81.16 ± 0.07 | |||
| Accuracy | ||||||
| CNN with BERT-base embeddings [51] | 70.00 | |||||
| UFD [29] | 75.90 | |||||
| Naïve Bayes | Gradient Boosted Trees | |||||
|---|---|---|---|---|---|---|
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 89.54 ± 0.20 | 92.94 ± 0.33 | 84.93 ± 0.67 | 96.74 ± 0.15 | 96.22 ± 0.36 | 97.10 ± 0.29 |
| DocEmbWord2VecCBOW | 71.14 ± 0.41 | 78.89 ± 0.63 | 55.41 ± 0.62 | 91.90 ± 0.23 | 92.01 ± 0.46 | 91.24 ± 0.40 |
| DocEmbWord2VecSG | 60.91 ± 0.53 | 85.19 ± 1.69 | 23.66 ± 1.16 | 93.31 ± 0.37 | 93.81 ± 0.32 | 92.33 ± 0.55 |
| DocEmbFastTextCBOW | 67.45 ± 0.67 | 77.26 ± 1.05 | 46.75 ± 1.06 | 91.45 ± 0.29 | 91.32 ± 0.68 | 91.06 ± 0.49 |
| DocEmbFastTextSG | 60.22 ± 0.22 | 88.13 ± 1.08 | 20.93 ± 0.41 | 93.41 ± 0.31 | 94.05 ± 0.34 | 92.28 ± 0.58 |
| DocEmbGloVe | 62.22 ± 0.49 | 81.02 ± 1.27 | 29.04 ± 0.91 | 90.63 ± 0.30 | 89.98 ± 0.48 | 90.83 ± 0.36 |
| DocEmbBERT | 70.52 ± 0.47 | 82.03 ± 0.70 | 50.34 ± 1.06 | 92.91 ± 0.34 | 93.58 ± 0.22 | 91.69 ± 0.65 |
| DocEmbRoBERTa | 81.86 ± 0.56 | 87.45 ± 0.78 | 73.18 ± 1.27 | 92.42 ± 0.22 | 92.51 ± 0.33 | 91.82 ± 0.45 |
| DocEmbBART | 90.14 ± 0.42 | 91.69 ± 0.63 | 87.64 ± 0.52 | 99.06 ± 0.11 | 98.82 ± 0.24 | 99.24 ± 0.19 |
| Perceptron | Multi-Layer Perceptron | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 93.76 ± 0.27 | 94.45 ± 0.51 | 92.59 ± 0.56 | 95.36 ± 0.23 | 95.24 ± 0.42 | 95.20 ± 0.44 |
| DocEmbWord2VecCBOW | 90.15 ± 0.35 | 90.84 ± 0.63 | 88.67 ± 0.71 | 91.54 ± 0.40 | 91.67 ± 1.42 | 90.89 ± 1.45 |
| DocEmbWord2VecSG | 88.98 ± 0.46 | 90.48 ± 0.51 | 86.41 ± 0.86 | 92.45 ± 0.32 | 92.59 ± 1.19 | 91.83 ± 1.29 |
| DocEmbFastTextCBOW | 90.10 ± 0.44 | 90.23 ± 0.95 | 89.30 ± 0.62 | 92.00 ± 0.44 | 91.99 ± 1.03 | 91.54 ± 1.68 |
| DocEmbFastTextSG | 88.45 ± 0.59 | 90.65 ± 0.82 | 84.99 ± 1.07 | 92.15 ± 0.43 | 92.78 ± 1.03 | 90.94 ± 0.81 |
| DocEmbGloVe | 83.90 ± 0.52 | 85.26 ± 1.15 | 80.89 ± 1.97 | 87.57 ± 0.57 | 88.79 ± 2.16 | 85.29 ± 2.64 |
| DocEmbBERT | 92.09 ± 0.50 | 92.84 ± 1.01 | 90.74 ± 1.36 | 94.80 ± 0.47 | 94.51 ± 1.95 | 94.89 ± 1.73 |
| DocEmbRoBERTa | 91.62 ± 0.40 | 91.02 ± 2.03 | 91.92 ± 1.94 | 92.74 ± 0.96 | 93.03 ± 3.76 | 92.31 ± 4.31 |
| DocEmbBART | 99.73 ± 0.09 | 99.66 ± 0.11 | 99.78 ± 0.13 | 99.77 ± 0.07 | 99.70 ± 0.14 | 99.82 ± 0.08 |
| LSTM | Bidirectional LSTM | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 95.05 ± 0.25 | 94.83 ± 0.44 | 95.00 ± 0.50 | 95.01 ± 0.25 | 94.75 ± 0.39 | 95.00 ± 0.34 |
| DocEmbWord2VecCBOW | 93.70 ± 0.32 | 94.11 ± 1.11 | 92.87 ± 1.50 | 93.81 ± 0.39 | 93.60 ± 0.82 | 93.68 ± 0.87 |
| DocEmbWord2VecSG | 93.03 ± 0.32 | 93.22 ± 1.35 | 92.40 ± 1.24 | 93.14 ± 0.50 | 94.08 ± 1.74 | 91.71 ± 1.80 |
| DocEmbFastTextCBOW | 93.25 ± 0.65 | 92.83 ± 2.21 | 93.44 ± 2.16 | 93.52 ± 0.30 | 93.04 ± 1.66 | 93.73 ± 2.08 |
| DocEmbFastTextSG | 92.90 ± 0.41 | 93.13 ± 1.17 | 92.21 ± 1.15 | 92.67 ± 0.56 | 94.61 ± 1.65 | 90.12 ± 2.34 |
| DocEmbGloVe | 88.84 ± 0.82 | 88.36 ± 2.96 | 88.98 ± 2.98 | 88.83 ± 0.79 | 89.23 ± 3.27 | 87.93 ± 4.70 |
| DocEmbBERT | 96.31 ± 0.72 | 96.15 ± 2.41 | 96.36 ± 1.84 | 96.36 ± 1.32 | 96.08 ± 3.10 | 96.59 ± 1.13 |
| DocEmbRoBERTa | 93.52 ± 0.78 | 94.41 ± 2.02 | 92.24 ± 2.94 | 93.12 ± 1.29 | 94.87 ± 2.53 | 90.93 ± 4.62 |
| DocEmbBART | 99.79 ± 0.06 | 99.74 ± 0.11 | 99.84 ± 0.08 | 99.80 ± 0.12 | 99.72 ± 0.18 | 99.87 ± 0.11 |
| GRU | Bidirectional GRU | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 94.99 ± 0.22 | 94.64 ± 0.34 | 95.08 ± 0.45 | 94.98 ± 0.27 | 94.71 ± 0.38 | 94.98 ± 0.38 |
| DocEmbWord2VecCBOW | 93.67 ± 0.41 | 93.66 ± 1.02 | 93.32 ± 1.19 | 93.68 ± 0.30 | 93.48 ± 0.67 | 93.54 ± 1.20 |
| DocEmbWord2VecSG | 93.05 ± 0.36 | 93.28 ± 1.48 | 92.40 ± 1.35 | 92.91 ± 0.77 | 93.55 ± 2.49 | 91.87 ± 2.32 |
| DocEmbFastTextCBOW | 93.18 ± 0.54 | 92.94 ± 2.21 | 93.14 ± 1.85 | 93.34 ± 0.59 | 92.43 ± 1.26 | 94.02 ± 1.01 |
| DocEmbFastTextSG | 92.73 ± 0.76 | 92.94 ± 2.54 | 92.16 ± 2.16 | 92.86 ± 0.45 | 94.07 ± 1.14 | 91.09 ± 2.10 |
| DocEmbGloVe | 88.83 ± 0.76 | 89.73 ± 3.25 | 87.26 ± 3.54 | 89.19 ± 0.64 | 90.81 ± 1.47 | 86.59 ± 2.85 |
| DocEmbBERT | 96.25 ± 0.52 | 96.81 ± 1.95 | 95.50 ± 1.87 | 96.37 ± 0.81 | 97.71 ± 0.90 | 94.77 ± 2.28 |
| DocEmbRoBERTa | 92.32 ± 1.93 | 92.10 ± 5.31 | 92.80 ± 5.35 | 92.93 ± 1.27 | 95.36 ± 2.48 | 90.01 ± 4.51 |
| DocEmbBART | 99.77 ± 0.07 | 99.72 ± 0.14 | 99.82 ± 0.09 | 99.78 ± 0.07 | 99.72 ± 0.12 | 99.83 ± 0.09 |
| Accuracy | Precision | Recall | ||||
| MisRoBÆRTa [23] | 97.57 ± 0.29 | 97.58 ± 0.28 | 97.57 ± 0.31 | |||
| C-CNN [24] | 99.90 | 99.90 | 99.90 | |||
| Accuracy | ||||||
| FNDNet [12] | 98.36 | |||||
| FakeBERT [18] | 98.90 | |||||
| Naïve Bayes | Gradient Boosted Trees | |||||
|---|---|---|---|---|---|---|
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 | 78.29 ± 0.99 | 70.70 ± 2.62 | 78.29 ± 0.99 |
| DocEmbWord2VecCBOW | 55.26 ± 1.52 | 72.29 ± 2.16 | 55.26 ± 1.52 | 78.72 ± 0.88 | 72.97 ± 2.71 | 78.72 ± 0.88 |
| DocEmbWord2VecSG | 54.42 ± 2.99 | 72.24 ± 2.05 | 54.42 ± 2.99 | 78.97 ± 0.91 | 73.30 ± 2.66 | 78.97 ± 0.91 |
| DocEmbFastTextCBOW | 49.13 ± 2.26 | 71.76 ± 1.86 | 49.13 ± 2.26 | 78.54 ± 0.57 | 72.26 ± 1.50 | 78.54 ± 0.57 |
| DocEmbFastTextSG | 55.58 ± 2.35 | 73.95 ± 1.76 | 55.58 ± 2.35 | 78.94 ± 0.66 | 73.46 ± 2.97 | 78.94 ± 0.66 |
| DocEmbGloVe | 48.63 ± 8.40 | 69.79 ± 3.13 | 48.63 ± 8.40 | 78.16 ± 1.35 | 71.89 ± 4.01 | 78.16 ± 1.35 |
| DocEmbBERT | 59.91 ± 1.79 | 76.22 ± 0.85 | 59.91 ± 1.79 | 78.44 ± 0.73 | 71.31 ± 1.95 | 78.44 ± 0.73 |
| DocEmbRoBERTa | 62.02 ± 7.65 | 70.54 ± 1.52 | 62.02 ± 7.65 | 77.98 ± 0.70 | 69.83 ± 4.17 | 77.98 ± 0.70 |
| DocEmbBART | 61.56 ± 1.29 | 80.57 ± 1.17 | 61.56 ± 1.29 | 79.28 ± 1.18 | 73.92 ± 2.50 | 79.28 ± 1.18 |
| Perceptron | Multi-Layer Perceptron | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 | 78.29 ± 0.50 | 70.91 ± 6.32 | 78.29 ± 0.50 |
| DocEmbWord2VecCBOW | 77.98 ± 0.31 | 63.63 ± 3.26 | 77.98 ± 0.31 | 78.10 ± 0.50 | 65.94 ± 5.52 | 78.10 ± 0.50 |
| DocEmbWord2VecSG | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 |
| DocEmbFastTextCBOW | 77.79 ± 0.62 | 66.38 ± 5.91 | 77.79 ± 0.62 | 77.91 ± 0.65 | 66.63 ± 3.36 | 77.91 ± 0.65 |
| DocEmbFastTextSG | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 |
| DocEmbGloVe | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 |
| DocEmbBERT | 77.76 ± 1.02 | 68.20 ± 3.39 | 77.76 ± 1.02 | 77.60 ± 1.65 | 70.76 ± 3.70 | 77.60 ± 1.65 |
| DocEmbRoBERTa | 77.85 ± 0.33 | 63.79 ± 5.23 | 77.85 ± 0.33 | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 |
| DocEmbBART | 79.78 ± 0.84 | 75.84 ± 1.30 | 79.78 ± 0.84 | 79.75 ± 1.75 | 75.40 ± 2.30 | 79.75 ± 1.75 |
| LSTM | Bidirectional LSTM | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 78.91 ± 0.98 | 73.90 ± 1.40 | 78.91 ± 0.98 | 78.63 ± 0.80 | 73.77 ± 1.63 | 78.63 ± 0.80 |
| DocEmbWord2VecCBOW | 77.88 ± 1.40 | 70.88 ± 3.53 | 77.88 ± 1.40 | 77.23 ± 1.55 | 70.58 ± 2.30 | 77.23 ± 1.55 |
| DocEmbWord2VecSG | 78.04 ± 0.16 | 63.35 ± 1.97 | 78.04 ± 0.16 | 77.88 ± 0.59 | 67.35 ± 2.43 | 77.88 ± 0.59 |
| DocEmbFastTextCBOW | 78.07 ± 0.59 | 71.74 ± 2.23 | 78.07 ± 0.59 | 78.10 ± 0.79 | 73.05 ± 1.58 | 78.10 ± 0.79 |
| DocEmbFastTextSG | 77.98 ± 0.20 | 61.64 ± 1.69 | 77.98 ± 0.20 | 77.85 ± 0.74 | 67.51 ± 5.94 | 77.85 ± 0.74 |
| DocEmbGloVe | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 | 77.85 ± 0.17 | 62.74 ± 3.44 | 77.85 ± 0.17 |
| DocEmbBERT | 77.57 ± 2.95 | 73.20 ± 2.13 | 77.57 ± 2.95 | 77.51 ± 2.22 | 74.46 ± 2.19 | 77.51 ± 2.22 |
| DocEmbRoBERTa | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 |
| DocEmbBART | 78.07 ± 2.16 | 76.31 ± 3.01 | 78.07 ± 2.16 | 77.35 ± 2.39 | 75.97 ± 1.89 | 77.35 ± 2.39 |
| GRU | Bidirectional GRU | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 78.94 ± 1.21 | 73.99 ± 2.19 | 78.94 ± 1.21 | 78.60 ± 1.13 | 73.69 ± 1.40 | 78.60 ± 1.13 |
| DocEmbWord2VecCBOW | 77.57 ± 1.26 | 70.72 ± 2.80 | 77.57 ± 1.26 | 77.32 ± 1.31 | 71.04 ± 2.30 | 77.32 ± 1.31 |
| DocEmbWord2VecSG | 78.29 ± 0.37 | 66.63 ± 2.52 | 78.29 ± 0.37 | 78.22 ± 1.00 | 68.24 ± 2.56 | 78.22 ± 1.00 |
| DocEmbFastTextCBOW | 78.19 ± 0.78 | 72.06 ± 2.37 | 78.19 ± 0.78 | 77.73 ± 1.51 | 74.22 ± 1.06 | 77.73 ± 1.51 |
| DocEmbFastTextSG | 77.88 ± 0.37 | 64.47 ± 3.68 | 77.88 ± 0.37 | 77.76 ± 0.79 | 67.54 ± 2.82 | 77.76 ± 0.79 |
| DocEmbGloVe | 77.82 ± 0.31 | 61.81 ± 2.43 | 77.82 ± 0.31 | 77.66 ± 0.52 | 64.45 ± 3.17 | 77.66 ± 0.52 |
| DocEmbBERT | 78.54 ± 1.39 | 72.23 ± 3.39 | 78.54 ± 1.39 | 74.86 ± 4.28 | 72.99 ± 2.21 | 74.86 ± 4.28 |
| DocEmbRoBERTa | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 | 77.88 ± 0.01 | 60.66 ± 0.01 | 77.88 ± 0.01 |
| DocEmbBART | 77.48 ± 2.08 | 75.96 ± 2.38 | 77.48 ± 2.08 | 76.45 ± 4.56 | 77.31 ± 3.14 | 76.45 ± 4.56 |
| Accuracy | Precision | Recall | ||||
| MisRoBÆRTa [23] | 77.39 ± 0.83 | 77.39 ± 0.83 | 77.39 ± 0.83 | |||
| Accuracy | ||||||
| SVM [52] | 78.00 | |||||
| UFD [29] | 67.90 | |||||
| Naïve Bayes | Gradient Boosted Trees | |||||
|---|---|---|---|---|---|---|
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 92.08 ± 0.15 | 92.11 ± 0.15 | 92.08 ± 0.15 | 98.05 ± 0.12 | 98.05 ± 0.12 | 98.05 ± 0.12 |
| DocEmbWord2VecCBOW | 70.06 ± 0.50 | 73.01 ± 0.36 | 70.06 ± 0.50 | 95.64 ± 0.26 | 95.63 ± 0.26 | 95.64 ± 0.26 |
| DocEmbWord2VecSG | 55.33 ± 0.63 | 68.18 ± 0.33 | 55.33 ± 0.63 | 95.76 ± 0.21 | 95.75 ± 0.21 | 95.76 ± 0.21 |
| DocEmbFastTextCBOW | 62.83 ± 0.47 | 70.17 ± 0.66 | 62.83 ± 0.47 | 94.36 ± 0.27 | 94.34 ± 0.27 | 94.36 ± 0.27 |
| DocEmbFastTextSG | 59.72 ± 0.52 | 69.49 ± 0.63 | 59.72 ± 0.52 | 95.66 ± 0.28 | 95.65 ± 0.28 | 95.66 ± 0.28 |
| DocEmbGloVe | 52.99 ± 0.56 | 65.84 ± 0.43 | 52.99 ± 0.56 | 96.19 ± 0.28 | 96.19 ± 0.28 | 96.19 ± 0.28 |
| DocEmbBERT | 84.65 ± 0.63 | 87.60 ± 0.42 | 84.65 ± 0.63 | 98.18 ± 0.10 | 98.18 ± 0.10 | 98.18 ± 0.10 |
| DocEmbRoBERTa | 52.15 ± 0.33 | 65.77 ± 0.64 | 52.15 ± 0.33 | 79.15 ± 0.36 | 79.11 ± 0.38 | 79.15 ± 0.36 |
| DocEmbBART | 94.08 ± 0.28 | 94.66 ± 0.26 | 94.08 ± 0.28 | 99.01 ± 0.10 | 99.01 ± 0.10 | 99.01 ± 0.10 |
| Perceptron | Multi-Layer Perceptron | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 97.64 ± 0.13 | 97.64 ± 0.13 | 97.64 ± 0.13 | 97.54 ± 0.16 | 97.54 ± 0.16 | 97.54 ± 0.16 |
| DocEmbWord2VecCBOW | 94.24 ± 0.23 | 94.22 ± 0.24 | 94.24 ± 0.23 | 96.11 ± 0.22 | 96.11 ± 0.22 | 96.11 ± 0.22 |
| DocEmbWord2VecSG | 90.14 ± 0.29 | 90.09 ± 0.29 | 90.14 ± 0.29 | 93.91 ± 0.24 | 93.89 ± 0.24 | 93.91 ± 0.24 |
| DocEmbFastTextCBOW | 93.14 ± 0.30 | 93.14 ± 0.30 | 93.14 ± 0.30 | 95.63 ± 0.24 | 95.64 ± 0.24 | 95.63 ± 0.24 |
| DocEmbFastTextSG | 90.00 ± 0.20 | 89.94 ± 0.21 | 90.00 ± 0.20 | 93.64 ± 0.30 | 93.64 ± 0.29 | 93.64 ± 0.30 |
| DocEmbGloVe | 90.97 ± 0.27 | 90.95 ± 0.27 | 90.97 ± 0.27 | 94.04 ± 0.30 | 94.05 ± 0.29 | 94.04 ± 0.30 |
| DocEmbBERT | 98.44 ± 0.15 | 98.44 ± 0.15 | 98.44 ± 0.15 | 98.78 ± 0.14 | 98.78 ± 0.13 | 98.78 ± 0.14 |
| DocEmbRoBERTa | 77.79 ± 1.85 | 78.89 ± 0.65 | 77.79 ± 1.85 | 80.30 ± 1.55 | 81.29 ± 0.61 | 80.30 ± 1.55 |
| DocEmbBART | 99.54 ± 0.05 | 99.54 ± 0.05 | 99.54 ± 0.05 | 99.55 ± 0.06 | 99.55 ± 0.06 | 99.55 ± 0.06 |
| LSTM | Bidirectional LSTM | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 97.10 ± 0.17 | 97.10 ± 0.17 | 97.10 ± 0.17 | 97.03 ± 0.22 | 97.03 ± 0.22 | 97.03 ± 0.22 |
| DocEmbWord2VecCBOW | 96.93 ± 0.16 | 96.94 ± 0.15 | 96.93 ± 0.16 | 96.88 ± 0.09 | 96.88 ± 0.09 | 96.88 ± 0.09 |
| DocEmbWord2VecSG | 95.37 ± 0.17 | 95.40 ± 0.17 | 95.37 ± 0.17 | 95.55 ± 0.32 | 95.56 ± 0.30 | 95.55 ± 0.32 |
| DocEmbFastTextCBOW | 96.24 ± 0.30 | 96.26 ± 0.29 | 96.24 ± 0.30 | 96.37 ± 0.23 | 96.38 ± 0.23 | 96.37 ± 0.23 |
| DocEmbFastTextSG | 95.06 ± 0.13 | 95.07 ± 0.13 | 95.06 ± 0.13 | 95.10 ± 0.37 | 95.11 ± 0.32 | 95.10 ± 0.37 |
| DocEmbGloVe | 95.17 ± 0.34 | 95.22 ± 0.28 | 95.17 ± 0.34 | 95.31 ± 0.42 | 95.35 ± 0.36 | 95.31 ± 0.42 |
| DocEmbBERT | 98.86 ± 0.24 | 98.87 ± 0.22 | 98.86 ± 0.24 | 98.89 ± 0.17 | 98.89 ± 0.16 | 98.89 ± 0.17 |
| DocEmbRoBERTa | 80.25 ± 1.38 | 81.31 ± 0.73 | 80.25 ± 1.38 | 80.17 ± 1.66 | 81.30 ± 0.86 | 80.17 ± 1.66 |
| DocEmbBART | 99.62 ± 0.05 | 99.62 ± 0.05 | 99.62 ± 0.05 | 99.65 ± 0.05 | 99.65 ± 0.05 | 99.65 ± 0.05 |
| GRU | Bidirectional GRU | |||||
| Vectorization | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| TFIDF | 97.00 ± 0.13 | 97.00 ± 0.13 | 97.00 ± 0.13 | 96.85 ± 0.20 | 96.86 ± 0.20 | 96.85 ± 0.20 |
| DocEmbWord2VecCBOW | 96.85 ± 0.16 | 96.86 ± 0.16 | 96.85 ± 0.16 | 96.86 ± 0.14 | 96.87 ± 0.14 | 96.86 ± 0.14 |
| DocEmbWord2VecSG | 95.29 ± 0.28 | 95.31 ± 0.26 | 95.29 ± 0.28 | 95.53 ± 0.14 | 95.56 ± 0.14 | 95.53 ± 0.14 |
| DocEmbFastTextCBOW | 96.28 ± 0.17 | 96.29 ± 0.17 | 96.28 ± 0.17 | 96.25 ± 0.28 | 96.26 ± 0.26 | 96.25 ± 0.28 |
| DocEmbFastTextSG | 94.72 ± 0.31 | 94.75 ± 0.28 | 94.72 ± 0.31 | 94.77 ± 0.52 | 94.85 ± 0.44 | 94.77 ± 0.52 |
| DocEmbGloVe | 95.06 ± 0.34 | 95.10 ± 0.31 | 95.06 ± 0.34 | 95.09 ± 0.29 | 95.15 ± 0.25 | 95.09 ± 0.29 |
| DocEmbBERT | 98.91 ± 0.27 | 98.92 ± 0.25 | 98.91 ± 0.27 | 98.99 ± 0.10 | 98.99 ± 0.10 | 98.99 ± 0.10 |
| DocEmbRoBERTa | 80.44 ± 1.26 | 81.44 ± 0.55 | 80.44 ± 1.26 | 80.04 ± 1.69 | 81.36 ± 0.76 | 80.04 ± 1.69 |
| DocEmbBART | 99.62 ± 0.09 | 99.62 ± 0.09 | 99.62 ± 0.09 | 99.64 ± 0.07 | 99.64 ± 0.07 | 99.64 ± 0.07 |
| Accuracy | Precision | Recall | ||||
| MisRoBÆRTa [23] | 99.52 ± 0.12 | 99.52 ± 0.12 | 99.52 ± 0.12 | |||
| Accuracy | ||||||
| Proppy [70] | 98.36 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Truică, C.-O.; Apostol, E.-S. It’s All in the Embedding! Fake News Detection Using Document Embeddings. Mathematics 2023, 11, 508. https://doi.org/10.3390/math11030508
Truică C-O, Apostol E-S. It’s All in the Embedding! Fake News Detection Using Document Embeddings. Mathematics. 2023; 11(3):508. https://doi.org/10.3390/math11030508
Chicago/Turabian StyleTruică, Ciprian-Octavian, and Elena-Simona Apostol. 2023. "It’s All in the Embedding! Fake News Detection Using Document Embeddings" Mathematics 11, no. 3: 508. https://doi.org/10.3390/math11030508
APA StyleTruică, C.-O., & Apostol, E.-S. (2023). It’s All in the Embedding! Fake News Detection Using Document Embeddings. Mathematics, 11(3), 508. https://doi.org/10.3390/math11030508








