1. Introduction
Statistics represent an important part of dealing with everyday data, i.e., data that follow a certain distribution. Determining the number of modes in a distribution can be a crucial step in making further decisions based on said data. However, this can prove to be rather difficult, in most cases, and many approaches simply rely on just assigning the best number that fits, via trial and error.
Many have explored ways to test whether a distribution is bimodal and have found better methods than the ones available before them. This field has been growing for a long time and there seems to be no end in sight. An important application for these tests is the binarization of an image, for which worldwide competitions are often held to encourage finding an even better algorithm than the ones before it. Since these sorts of applications just require finding a threshold, we only need to determine whether a distribution is either bimodal or unimodal to find an optimal threshold. Thus, a metric that could ascertain this characteristic would be beneficial. Such metrics have been proposed before, but none are without flaws, so a more robust solution is needed.
The dip test of unimodality is a non-parametric test proposed by Hartigan and Hartigan [
1] that measures multimodality in an empirical distribution by computing, over all points, the maximum difference between the empirical distribution and the unimodal distribution that minimizes said maximum difference. However, this test only determines if a distribution is multimodal or not and does not help with determining how many modes a multimodal distribution has.
Silverman [
2] uses kernel density estimates to investigate multimodality. His test consists of finding the maximal window h for which the kernel density estimate has k modes or less. A large value for this window indicates that the distribution has k modes or less and a small value indicates that it has more than k modes. This test is computationally demanding and must be run for both the number of modes you want to test for, and that number minus one.
Muller and Sawitzki [
3] propose a test based on the excess mass functional, which measures excessive empirical mass in comparison with multiples of uniform distributions. Estimators for the excess mass functional are built iteratively and they converge uniformly towards it.
The MAP test for multimodality proposed by Rozal and Hartigan [
4] uses minimal ascending path spanning trees to compute the MAPk statistic which indicates k-modality if it has a large value. Computing the MAPk statistic is straightforward, but the trees are created with Prim’s algorithm which has a complexity of log-linear order. Creating the trees is time-consuming.
Ashman et al. [
5] use the KMM algorithm to test for multimodality. They fit a user-specified number of Gaussian distributions to the empirical distribution and iteratively adjust the fit until the likelihood function converges to its maximum value. A good fit for a k-modal model indicates k-modality, but multiple values for k must be tested. Fitting a complex model is extremely cumbersome, especially for high values for k, and the test is not reliable for distributions drastically different from the model.
Zhang et al. [
6] present three measures for bimodality after fitting a bimodal normal mixture to a distribution. The first one, bimodal amplitude, is calculated as
, with
being the amplitude of the minimum PDF between the two peaks and
the amplitude of the smaller peak of the two. The second one, bimodal separation, is based on the characteristics of the two normal distributions that form the mixture and has the following formula
. The third one, bimodal ratio, is computed with
, where
and
are the amplitudes of the right and left peaks.
Wang et al. [
7] also fit a bimodal normal mixture to a distribution, but with equal variances, and define their bimodality index as
, where
are the means of the normal distributions,
is their common variance, and
and
are the mixing weights of the normal distributions.
Bayramoglu [
8] defines a bimodality degree
, where
is the probabilty density function of the distribution,
is a local minimum of
, and
are local maxima of
. This bimodality degree takes values from 0 to 1, with 1 indicating unimodality and lower values indicating a more pronounced antimode. In practice, finding local minima and maxima from samples of a distribution requires fitting a model
over the observed samples and solving
.
Jammalamadaka et al. [
9] propose a Bayesian test for the number of modes in a Gaussian mixture, and compute a Bayes factor for a mixture of two Gaussians
, where
is the hypothesis that the mixture is unimodal and
is the hypothesis that the mixture is bimodal. Higher values for this Bayes factor indicate bimodality, but a suitable threshold must be selected.
Chaudhuri and Agrawal [
10] measure the bimodality of a distribution after applying a threshold k with the following function
, where
is the number of samples with values
,
is their variance,
are the analogs for the samples with values
and
for all the samples. Low values for this function indicate bimodality.
Van Der Eijk [
11] presents a way to measure agreement in a discrete distribution with a finite range of possible values. The formula for this agreement is
, where U is a measure of unimodality, S is the number of distinct values present in the sample, and K is the total number of possible values for the distribution. This formula takes values between −1 and 1, with 1 indicating unimodality and -1 indicating multimodality.
Wilcock [
12] defines a bimodality parameter based on the amplitudes and probabilities of the modes. The formula is
, where
are the amplitudes of the left and right mode and
are the probabilities. High values for B indicate bimodality.
Smith et al. [
13] propose an alternative bimodality index with the formula
, where
are the modal sizes in phi units, subscript 1 refers to the primary mode, and subscript 2 to the secondary mode. If the modes have equal amplitudes, then 1 refers to the right mode and 2 to the left one. The two indices are numerically similar in the range of
, which covers most of the bimodal distributions presented in [
13], but the index presented in [
13] has an asymptotic behavior of tending to zero as the separation of modes vanishes.
Contreras-Reyes [
14] constructs an asymptotic test for bimodality for a bimodal skew-symmetric normal (BSSN) distribution from the Kullback–Leibler divergence. For the
parameter of the BSSN distribution, he selects a threshold
and tests the hypothesis
versus the alternative
.
indicates bimodality and
unimodality.
Highly specialized tests are much more accurate than the more general ones; however, they should only be used for the specific distributions they are specialized for. For example, Vichitlimaporn et al. [
15] present highly specialized bimodality criterions for the molecular weight distributions in copolymers. Hassanian-Moghaddam et al. [
16] present a similar criterion for the bivariate distribution of chain length and chemical composition of copolymers. Voříšek [
17] uses Cardan’s discriminant to determine whether or not a cusp distribution is bimodal.
Sarle’s bimodality coefficient [
18] uses a formula based on skewness and kurtosis that takes values between 0 and 1, with the value of 1 corresponding only to the perfect bimodal distribution. Values closer to 1 usually indicate bimodality, but not necessarily. Hildebrand [
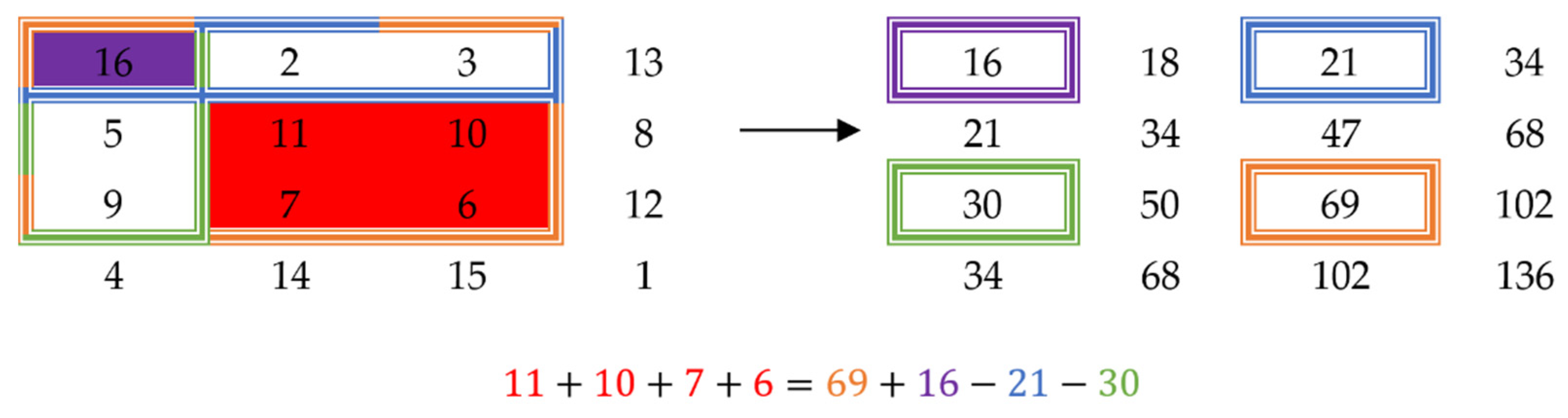
19] shows that kurtosis can be used as a bimodality indicator for some distributions but says that using it uncritically is hazardous and proves that for some distributions it cannot be used as such. Even though this test is not reliable enough to prove that a certain distribution is bimodal, it is quite easy to compute, especially when using summed-area tables to quickly compute the values of skewness and kurtosis.
In this paper, we discuss the strengths and weaknesses of Sarle’s bimodality coefficient and improve upon it by proposing generalized bimodality coefficients and combining several of them to create and fine-tune a composite bimodality coefficient that offers better results on the datasets we tested, while still being able to run in constant time on variable-sized datasets.
In
Section 2 we present Sarle’s bimodality coefficient in greater detail, the summed-area table technique and how we use it to compute the coefficient in constant time, a generalization of Sarle’s bimodality coefficient, and how we combine multiple generalized bimodality coefficients to create a composite bimodality coefficient. In
Section 3, we describe the testing methodology, and in
Section 4 we present the numerical results. In
Section 5, we discuss the benefits and risks of using the generalized bimodality coefficients and the composite bimodality coefficient. In
Section 6, we conclude that the composite bimodality coefficient can be a step-up from Sarle’s bimodality coefficient with the correct ratios.
3. Preparing the Dataset for Training and Testing
For our training dataset, we chose to generate it from a variety of distributions, for two simple reasons. First, since we know the distributions involved in generating the data, we know ahead of time if a sample should follow a one or two-point focused distribution. Second, due to controlling the distributions involved in generating our samples, we can limit some of the caveats that were previously mentioned that negatively impact our GBC. For this purpose, we chose to generate random data points using twelve different distributions (see
Appendix B), respectively a mixture of them, aimed at a variety of potential unimodal and bimodal results, generating a total of approximately 25,000 distributions. In a similar fashion, to vary the potential values of the GBCs pertaining to each distribution, we generated sets with a varying number of points, between 500 and 10,000. Because we wanted to test the veracity of the CBC in a binarization application, all values were generated in the interval
, for an easier overall approach; however, this does not alter the result, as any distribution could be shifted and scaled to any other interval of choice, and the standardized central moments would not change.
After we generated the data, we computed the following for each set of points pertaining to a distribution: the moments, respectively the GBC from them, as well as a value that we would like our CBC to tend to for that set of points. We denoted these values as CBC’ and they will be used as the output of the network during the training step. Since we know the original distributions that generated our sets of points, we can compute CBC′ in two ways, depending on whether or not the original distribution was unimodal or bimodal:
For a bimodal distribution
we denote with
the position of the two modes of the distribution.
Theorem 3. Let. Then.
Proof. The last case for is similar to (14). From (11)–(14) we can see that . □
In the above equations, represents a correction factor based on the distance between the two modes of the distribution, such that two close modes do not give a high value for the CBC’. Similarly, is a normalization factor, typically chosen in the interval to ensure we always raise to subunit powers, and that can raise exponentially closer to 1 when the modes start spreading apart. The reason for using a correction factor is because a set of points generated by a bimodal distribution with two close modes would be almost indistinguishable from one generated by a unimodal distribution with a similar shape and with its mode lying somewhere between the two modes of the bimodal one. In a similar fashion, we used when the value of the PDF differs between the two nodes so that modes that are very spread vertically, but not horizontally, do not result in high CBC’ values.
3.1. Training the Network
Based on the previously computed values, we could train our network using the natural logarithms of GBCs as inputs and CBC’ as the output. However, before training our network, we also considered a second potential remap of our inputs, before applying our logarithmic function on the GBCs, one based on polynomial fitting (see
Appendix C). This approach did lead to increased overall accuracy; thus, it was kept both for later training and testing purposes.
Since the training of the network was based on gradient descent, and we used a very simple network configuration, we had no certainty that our search would hit a global optimum, and, even if it did, we could not be sure of the general applicability of such an optimum. Consequently, since we did not know which set of powers might behave best in practice, we needed to obtain multiple sets and compare them. Thus, we trained different networks, with different constraints, such as using a different dataset when training, or constraining the weights, either to a set interval or simply making them strictly positive, to obtain our possible CBC parameters.
3.2. Testing the CBC
Tests were conducted against an image dataset generated by combining the following datasets: the DIBCO datasets [
27,
28,
29,
30,
31,
32,
33,
34,
35], the PHIBD 2012 [
36] dataset, the Nabuco dataset [
37], and the Rahul Sharma dataset [
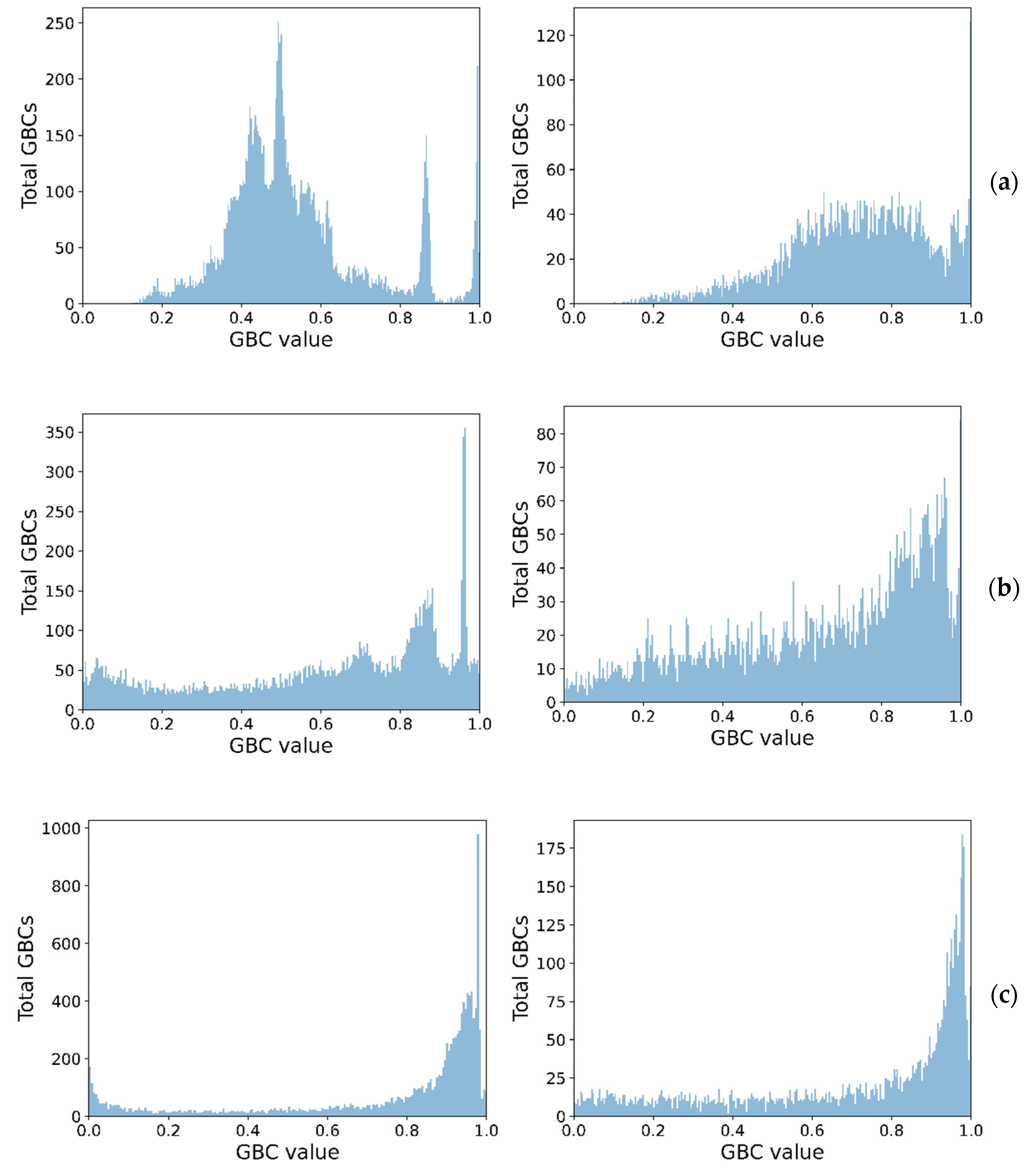
38], the combination of which will be further denoted simply as the Images dataset. On this dataset, we ran a binarization algorithm starting from multiple possible choices, with various window sizes. We decided to settle on one based on Otsu’s method with a fixed 8 × 8 window size centered on each pixel. There were also separate tests conducted against a synthetic dataset obtained from random data pertaining to multiple distributions and mixtures of them.
6. Conclusions
Determining whether some data follow a unimodal or bimodal distribution is an important step in figuring out a proper approach, in order to deal with said data, in a variety of situations. Thus, a proper way to ascertain this property is desired. A bimodality coefficient is a good starting point if it is robust.
In our case, the CBC performs better than any GBC on its own, for most of the composition ratios we tested, on average obtaining F-measure increases of 1.5–2%. However, finding an optimal ratio is no easy task, and even then, we have no certainty on how a CBC based on such a ratio might behave in more generic scenarios. For this reason, we also recommend two possible ratios to be considered: 3:2:1 and 3:0:1.
A certain downside of using the bimodality coefficient on its own is the fact that we do not know how data that follow a multimodal distribution with more than two modes behave. Thus, a multimodality coefficient could be considered for a better approach, and in future works, we could investigate ways to compute one.