Deep Learning Approaches to Automatic Chronic Venous Disease Classification
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Mining
2.1.1. Scrapy Data Mining
2.1.2. Selenium Data Mining
2.1.3. Datasets
2.2. Neural Networks
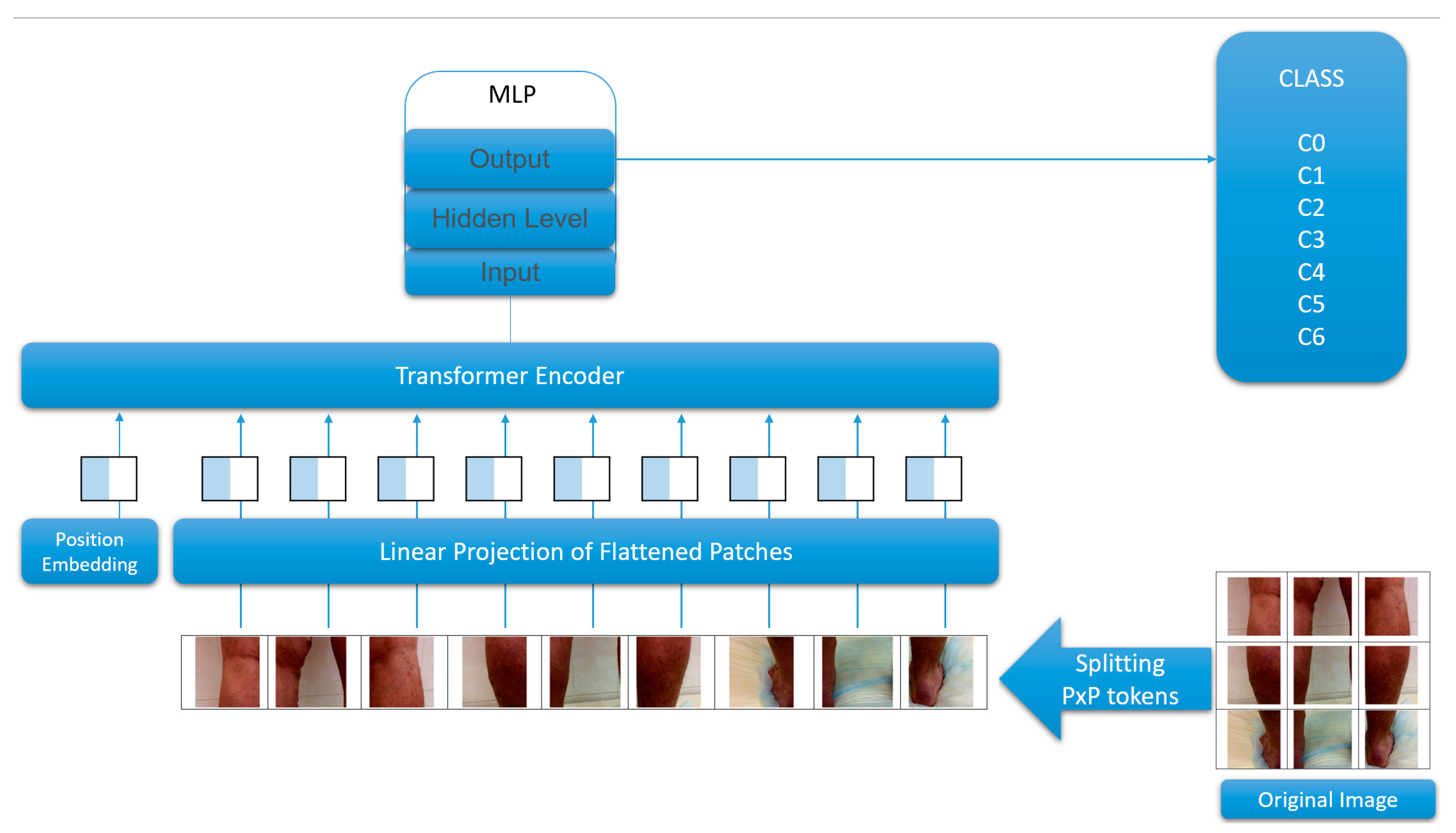
2.2.1. Filter “Legs–No Legs”
2.2.2. Multi-Classification Problem
- Precision = TruePositive/(TruePositive + FalsePositive)
- Recall = TruePositive/(TruePositive + FalseNegative)
- F-Measure = (2 × Precision × Recall)/(Precision + Recall)
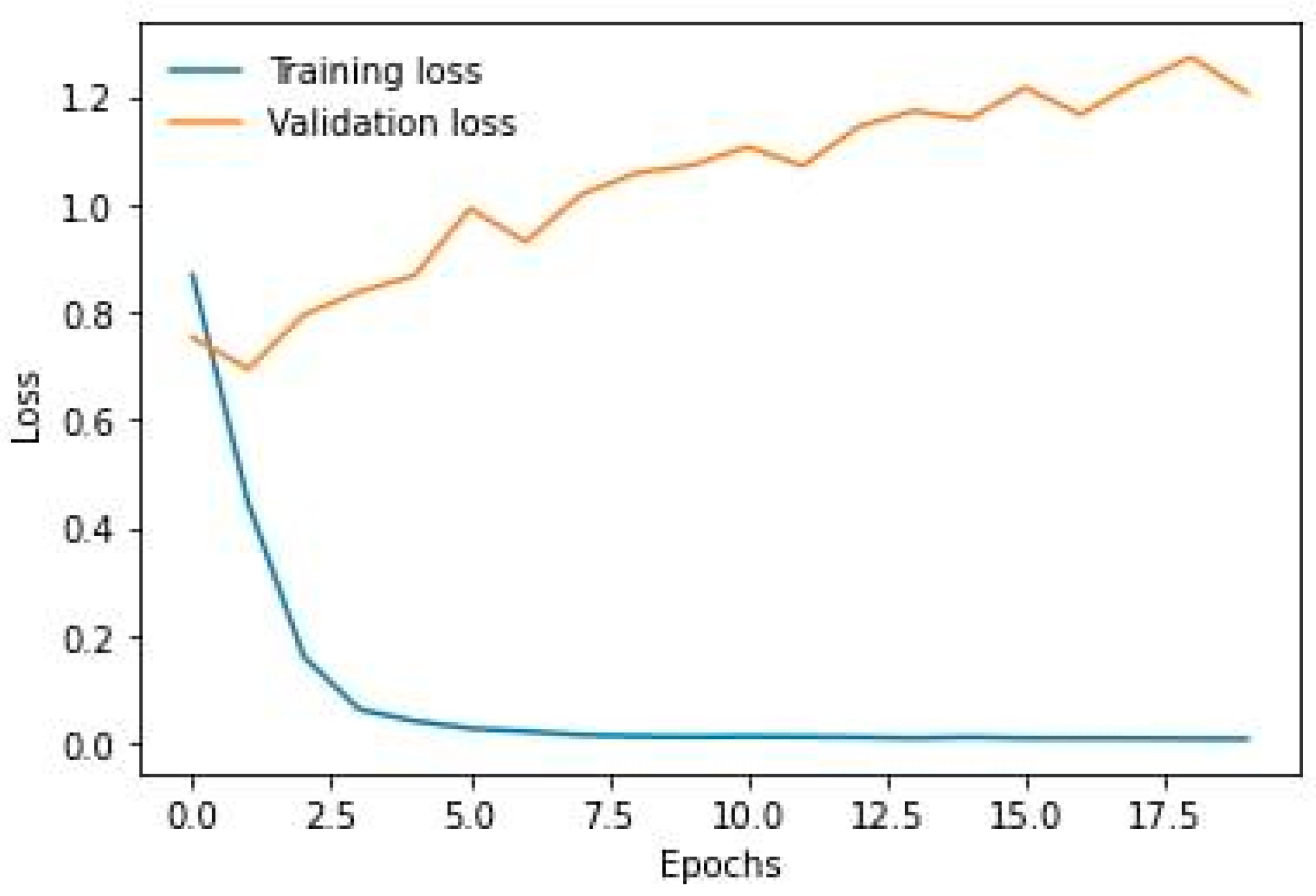
- Logistic Loss curve.
| Predicted | |||
| Positive | Negative | ||
| Actual | Positive | Rated TP = TruePositive/ActualPositive | Rated FN = FalseNegative/ActualPositive |
| Negative | Rated FP = FalsePositive/ActualNegative | Rated TN = TrueNegative/ActualNegative | |
3. Results
3.1. Resnet50 for Filter “Legs–No Legs”
3.2. Resnet50 for the Multi-Classification Problem
3.3. ViT Transformers
3.4. DeiT Multi-Classification Problem
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vuylsteke, M.E.; Colman, R.; Thomis, S.; Guillaume, G.; van Quickenborne, D.; Staelens, I. An epidemiological survey of venous disease among general practitioner attendees in different geographical regions on the globe: The final results of the vein consult program. Angiology 2018, 69, 779–785. [Google Scholar] [CrossRef]
- Feodor, T.; Baila, S.; Mitea, I.-A.; Branisteanu, D.-E.; Vittos, O. Epidemiology and clinical characteristics of chronic venous disease in Romania. Exp. Ther. Med. 2019, 17, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Carlton, R.; Mallick, R.; Campbell, C.; Raju, A.; O’Donnell, T.; Eaddy, M. Evaluating the expected costs and budget impact of interventional therapies for the treatment of chronic venous disease. Am. Health Drug Benefits 2015, 8, 366. [Google Scholar] [CrossRef] [PubMed]
- Epstein, D.; Gohel, M.; Heatley, F.; Davies, A.H. Cost-effectiveness of treatments for superficial venous reflux in patients with chronic venous ulceration. BJS Open 2018, 2, 203–212. [Google Scholar] [CrossRef]
- Vlajinac, H.D.; Radak, Ð.J.; Marinković, J.M.; Maksimović, M.Ž. Risk factors for chronic venous disease. Phlebology 2012, 27, 416–422. [Google Scholar] [CrossRef]
- Bailey, M.; Solomon, C.; Kasabov, N.; Greig, S. Hybrid systems for medical data analysis and decision making-a case study on varicose vein disorders. In Proceedings of the 1995 Second New Zealand International Two-Stream Conference on Artificial Neural Networks and Expert Systems, Dunedin, New Zealand, 20–23 November 1995; pp. 265–268. [Google Scholar]
- Shad, R.; Cunningham, J.P.; Ashley, E.A.; Langlotz, C.P.; Hiesinger, W. Designing clinically translatable artificial intelligence systems for high-dimensional medical imaging. Nat. Mach. Intell. 2021, 3, 929–935. [Google Scholar] [CrossRef]
- Bharati, S.; Mondal, M.R.H.; Podder, P.; Prasath, V.B.S. Deep learning for medical image registration: A comprehensive review. Int. J. Comput. Inf. Syst. Ind. Manag. Appl. 2022, 14, 173–190. [Google Scholar]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; Ginneken, B.V.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A review of deep learning in medical imaging: Imaging traits, technology trends, case studies with progress highlights, and future promises. Proc. IEEE 2021, 109, 820–838. [Google Scholar] [CrossRef]
- Gergenreter, Y.S.; Zakharova, N.B.; Barulina, M.A.; Maslyakov, V.V.; Fedorov, V.E. Analysis of the cytokine profile of blood serum and tumor supernatants in breast cancer. Acta Biomed. Sci. 2022, 7, 134–146. [Google Scholar] [CrossRef]
- Wang, Y.B.; You, Z.H.; Yang, S.; Yi, H.-C.; Chen, Z.-H.; Zheng, K. A deep learning-based method for drug-target interaction prediction based on long short-term memory neural network. BMC Med. Inf. Decis. Mak. 2020, 20 (Suppl. S2), 49. [Google Scholar] [CrossRef]
- Qu, R.; Wang, Y.; Yang, Y. COVID-19 detection using CT image based on YOLOv5 network. In Proceedings of the 2021 3rd International Academic Exchange Conference on Science and Technology Innovation (IAECST), Guangzhou, China, 10–12 December 2021. [Google Scholar]
- Saxena, A.; Singh, S.P. A deep learning approach for the detection of COVID-19 from chest X-Ray images using convolutional neural networks. arXiv 2022, arXiv:2201.09952. [Google Scholar]
- Gromov, M.S.; Rogacheva, S.M.; Barulina, M.A.; Reshetnikov, A.A.; Prokhozhev, D.A.; Fomina, A.Y. Analysis of some physiological and biochemical indices in patients with COVID-19 pneumonia using mathematical methods. J. Evol. Biochem. Physiol. 2021, 57, 1394–1407. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.A.; Al-Khateeb, B.; Yousif, M.; Mostafa, S.A.; Kadry, S.; Abdulkareem, K.H.; Garcia-Zapirain, B. Novel crow swarm optimization algorithm and selection approach for optimal deep learning COVID-19 diagnostic model. Comput. Intell. Neuralsci. 2022, 2022, 1307944. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-scale Chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar]
- Ali, O.; Ali, H.; Shah, S.A.A.; Shahzad, A. Implementation of a modified U-Net for medical image segmentation on edge devices. IEEE Trans. Circuits Syst. II Express Briefs 2022, in press. [CrossRef]
- Intel® Neural Compute Stick 2 (Intel® NCS2). Available online: https://www.intel.com/content/www/us/en/developer/tools/neural-compute-stick/overview.html (accessed on 16 September 2022).
- Antonelli, M.; Reinke, A.; Bakas, S.; Farahani, K.; Landman, B.A.; Litjens, G.; Menze, B.; Ronneberger, O.; Summers, R.M.; Ginneken, B.; et al. The medical segmentation decathlon. arXiv 2021, arXiv:2106.05735. [Google Scholar] [CrossRef]
- Shah, M.I.; Mishra, S.; Yadav, V.K.; Chauhan, A.; Sarkar, M.; Sharma, S.K.; Rout, C. Ziehl–neelsen sputum smear microscopy image database: A resource to facilitate automated bacilli detection for tuberculosis diagnosis. J. Med. Imaging 2017, 4, 027503. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Wang, Y. Medical image segmentation with limited supervision: A review of deep network models. IEEE Access 2021, 9, 36827–36851. [Google Scholar] [CrossRef]
- Chi, W.; Ma, L.; Wu, J.; Chen, M.; Lu, W.; Gu, X. Deep learning-based medical image segmentation with limited labels. Phys. Med. Biol. 2020, 65, 235001. [Google Scholar] [CrossRef]
- Butova, X.; Shayakhmetov, S.; Fedin, M.; Zolotukhin, I.; Gianesini, S. Artificial Intelligence Evidence-Based Current Status and Potential for Lower Limb Vascular Management. J. Pers. Med. 2021, 11, 1280. [Google Scholar] [CrossRef]
- Ryan, L.; Mataraso, S.; Siefkas, A.; Pellegrini, E.; Barnes, G.; Green-Saxena, A. A machine learning approach to predict deep venous thrombosis among hospitalized patients. Clin. Appl. Thromb. Hemost. 2021, 27, 1076029621991185. [Google Scholar] [CrossRef]
- Nafee, T.; Gibson, C.M.; Travis, R.; Yee, M.K.; Kerneis, M.; Chi, G. Machine learning to predict venous thrombosis in acutely ill medical patients. Res. Pract. Thromb. Haemost. 2020, 4, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Fong-Mata, M.B.; García-Guerrero, E.E.; Mejía-Medina, D.A.; López-Bonilla, O.R.; Villarreal-Gómez, L.J.; Zamora-Arellano, F.; Inzunza-González, E. An artificial neural network approach and a data augmentation algorithm to systematize the diagnosis of deep-vein thrombosis by using wells’ criteria. Electronics 2020, 9, 1810. [Google Scholar] [CrossRef]
- Rosenberg, D.; Eichorn, A.; Alarcon, M.; McCullagh, L.; McGinn, T.; Spyropoulos, A.C. External validation of the risk assessment model of the International Medical Prevention Registry on Venous Thromboembolism (IMPROVE) for medical patients in a tertiary health system. J. Am. Heart Assoc. 2014, 3, e001152. [Google Scholar] [CrossRef] [PubMed]
- Barbar, S.; Noventa, F.; Rossetto, V.; Ferrari, A.; Brandolin, B.; Perlati, M. A risk assessment model for the identification of hospitalized medical patients at risk for venous thromboembolism: The Padua Prediction Score. J. Thromb. Haemost. 2010, 8, 2450–2457. [Google Scholar] [CrossRef]
- Hippisley-Cox, J.; Coupland, C. Development and validation of risk prediction algorithm (QThrombosis) to estimate future risk of venous thromboembolism: Prospective cohort study. BMJ 2011, 343, d4656. [Google Scholar] [CrossRef]
- Van Es, N.; Di Nisio, M.; Cesarman, G.; Kleinjan, A.; Otten, H.M.; Mahé, I.; Büller, H.R. Comparison of risk prediction scores for venous thromboembolism in cancer patients: A prospective cohort study. Haematologica 2017, 102, 1494. [Google Scholar] [CrossRef]
- Ferroni, P.; Zanzotto, F.M.; Scarpato, N.; Riondino, S.; Guadagni, F.; Roselli, M. Validation of a machine learning approach for venous thromboembolism risk prediction in oncology. Dis. Mrk. 2017, 2017, 8781379. [Google Scholar] [CrossRef]
- Pabinger, I.; van Es, N.; Heinze, G.; Posch, F.; Riedl, J.; Reitter, E.M.; Ay, C. A clinical prediction model for cancer-associated venous thromboembolism: A development and validation study in two independent prospective cohorts. Lancet Haematol. 2018, 5, e289–e298. [Google Scholar] [CrossRef]
- Zhu, R.; Niu, H.; Yin, N.; Wu, T.; Zhao, Y. Analysis of Varicose Veins of Lower Extremities Based on Vascular Endothelial Cell Inflammation Images and Multi-Scale Deep Learning. IEEE Access 2019, 7, 174345–174358. [Google Scholar] [CrossRef]
- Atreyapurapu, V.; Vyakaranam, M.I.; Atreya III, S.; Gupta, P.I.; Atturu, G.V. Assessment of Anatomical Changes in Advanced Chronic Venous Insufficiency Using Artificial Intelligence ) and Machine Learning Techniques. J. Vasc. Surg. Venous Lymphat. Disord. 2022, 10, 571–572. [Google Scholar] [CrossRef]
- Ortega, M.A.; Fraile-Martínez, O.; García-Montero, C.; Álvarez-Mon, M.A.; Chaowen, C.; Ruiz-Grande, F.; Pekarek, L.; Monserrat, J.; Asúnsolo, A.; García-Honduvilla, N.; et al. Understanding chronic venous disease: A critical overview of its pathophysiology and medical management. J. Clin. Med. 2021, 10, 3239. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Chen, W.; Pan, Y.; Yin, S.; Fu, Y.; Mei, J.; Xue, Z. An Automatic Classification Method on Chronic Venous Insufficiency Images. Sci. Rep. 2018, 8, 17952. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Song, Y.; Cai, W.; Hauptmann, A.G.; Liu, S.; Pujol, S.; Kikinis, R.; Fulham, M.J.; Feng, D.D.; Chen, M. Dictionary pruning with visual word significance for medical image retrieval. Neuralcomputing 2016, 177, 75–88. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Houlsby, N. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- DeiT: Data-Efficient Image Transformers. Available online: https://github.com/facebookresearch/deit/blob/main/README_deit.md (accessed on 17 September 2022).
- Patrício, C.; Neves, J.C.; Teixeira, L.F. Explainable Deep Learning Methods in Medical Imaging Diagnosis: A Survey. arXiv 2022, arXiv:2205.04766. [Google Scholar]
- Wang, R.; Lei, T.; Cui, R.; Zhang, B.; Meng, H.; Nandi, A.K. Medical Image Segmentation Using Deep Learning: A Survey. arXiv 2022, arXiv:2009.13120. [Google Scholar] [CrossRef]
- Scrapy. Available online: https://scrapy.org/ (accessed on 25 August 2022).
- Selenium Automates Browsers. That’s it! Available online: https://www.selenium.dev/ (accessed on 25 August 2022).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Flickr Image Dataset. Available online: https://www.kaggle.com/datasets/hsankesara/flickr-image-dataset (accessed on 17 September 2022).
- Available online: https://pytorch.org/hub/nvidia_deeplearningexamples_resnet50/ (accessed on 26 August 2022).
- PyTorch. Available online: https://pytorch.org/ (accessed on 25 August 2022).
- Hoens, T.R.; Chawla, N.V. Imbalanced Datasets: From Sampling to Classifiers. In Book Imbalanced Learning: Foundations, Algorithms, and Applications, 1st ed.; Ma, Y., He, H., Eds.; Wiley: Hoboken, NJ, USA, 2013; pp. 43–60. [Google Scholar]
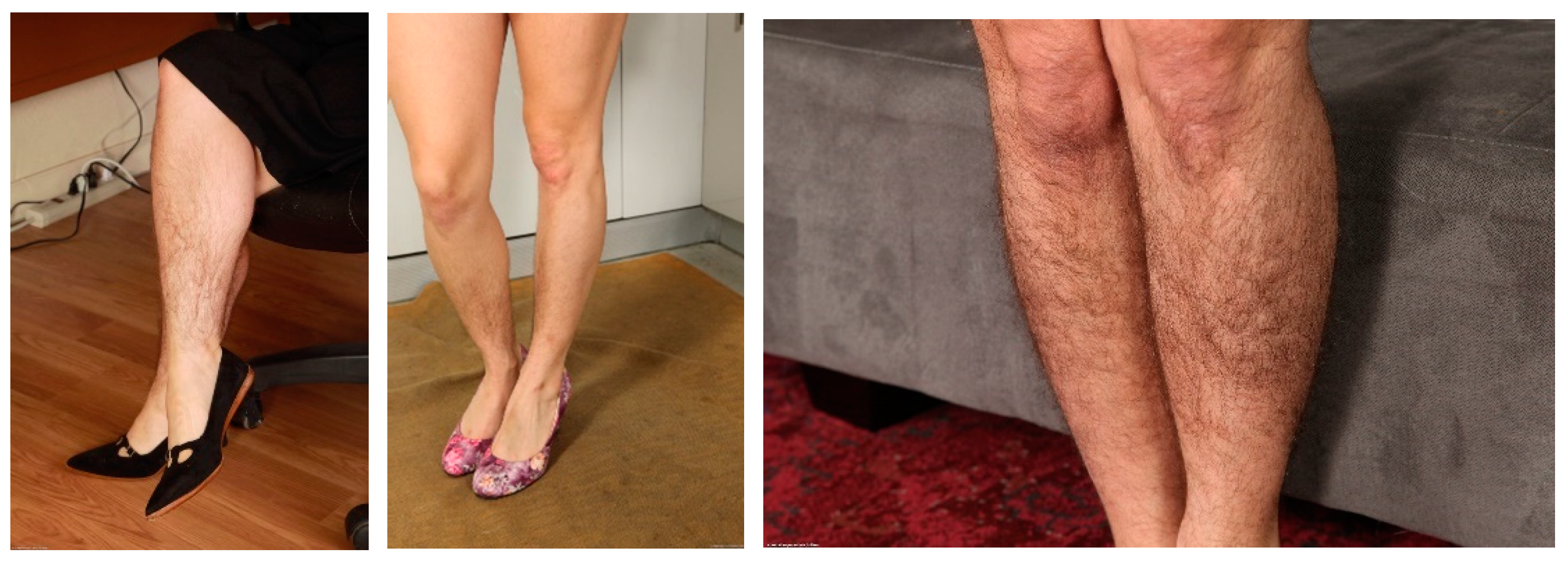



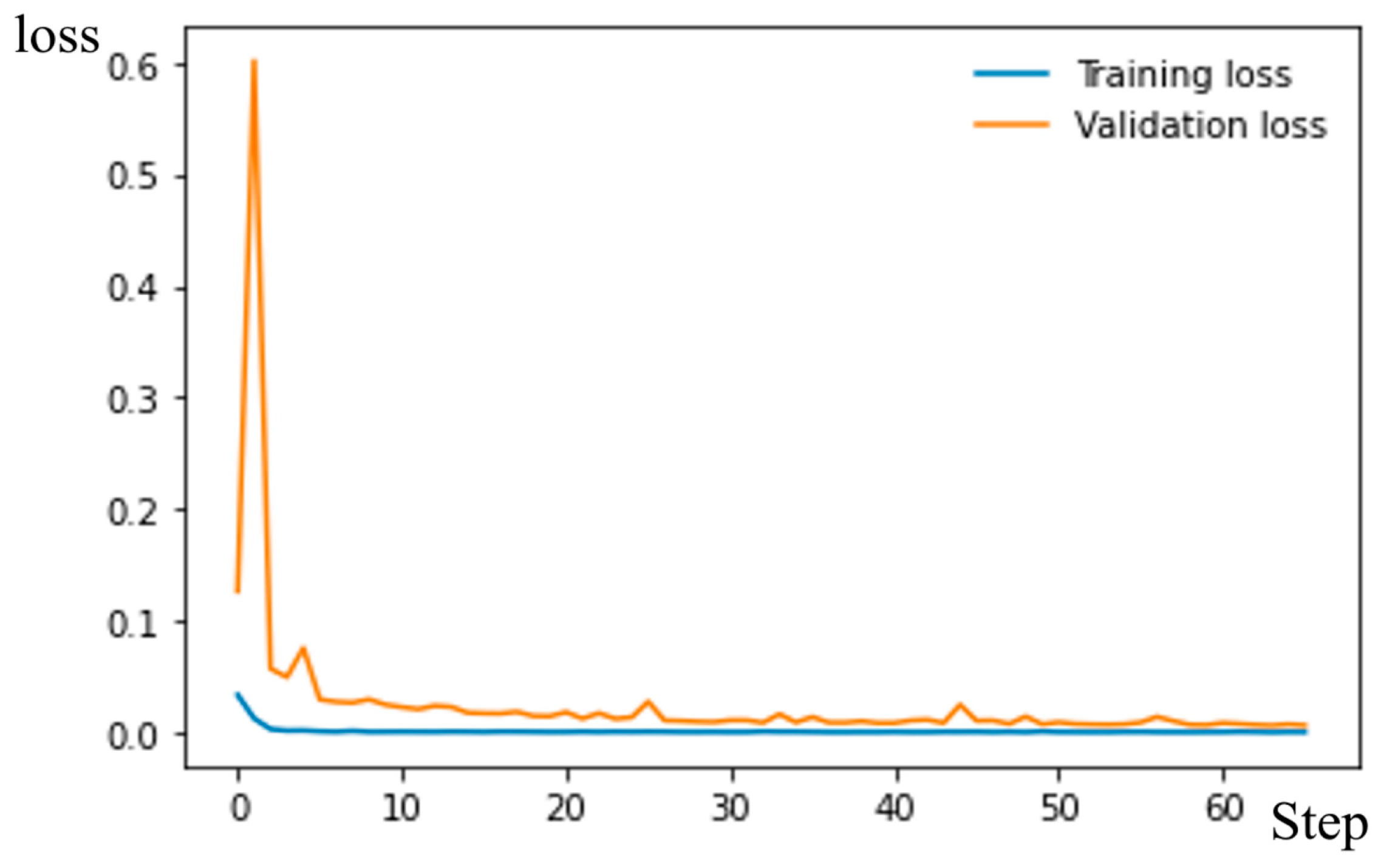
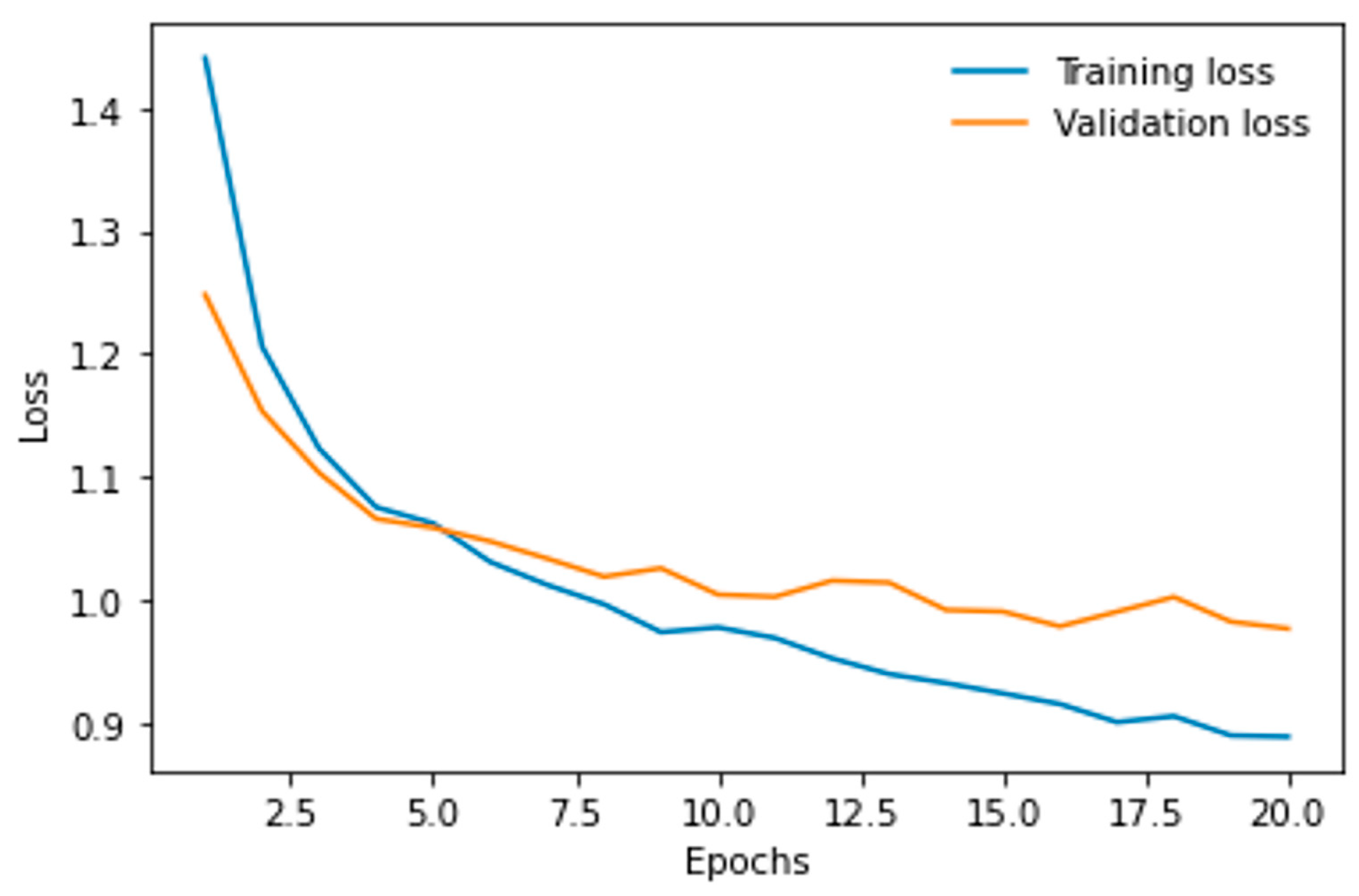
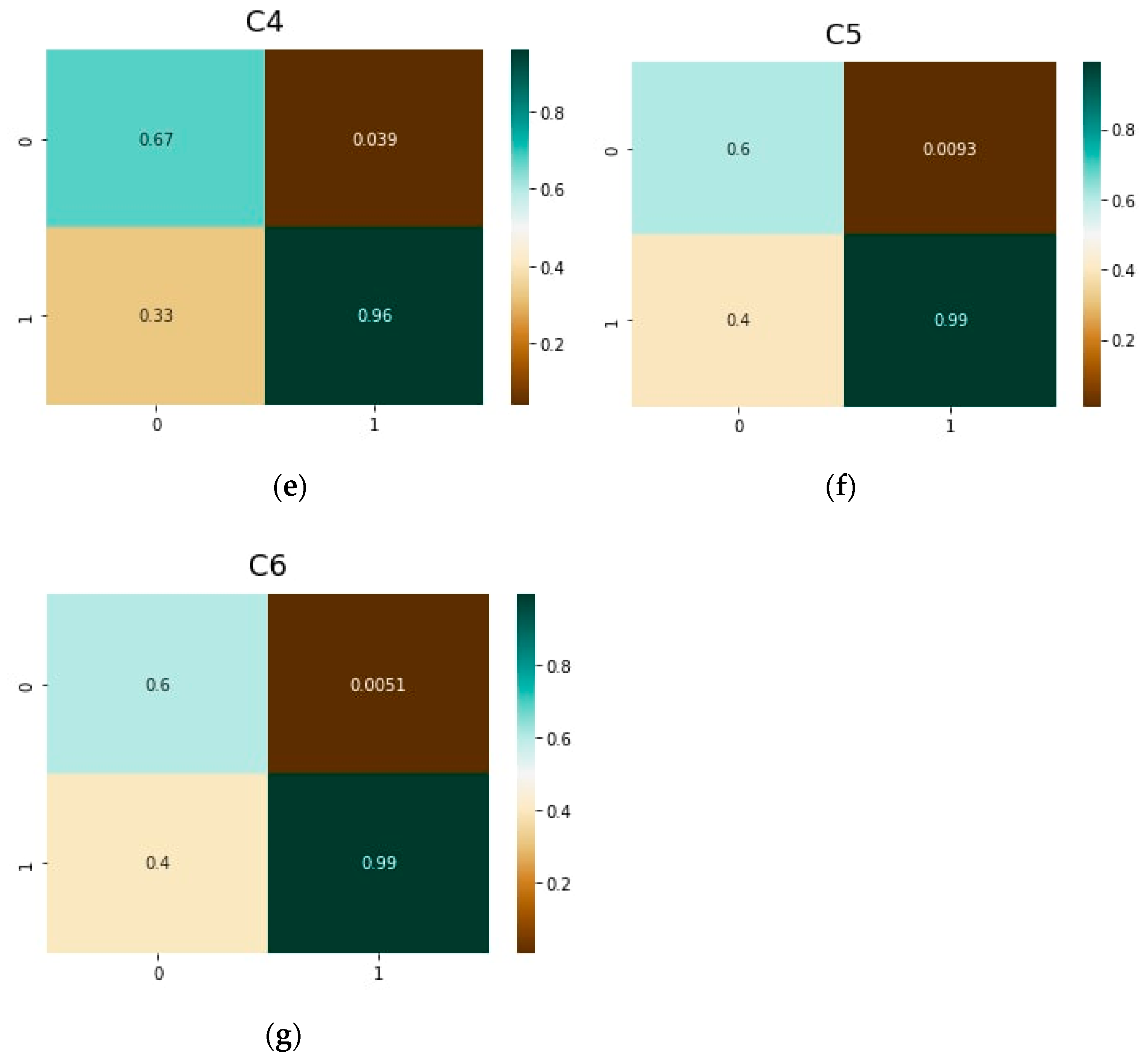
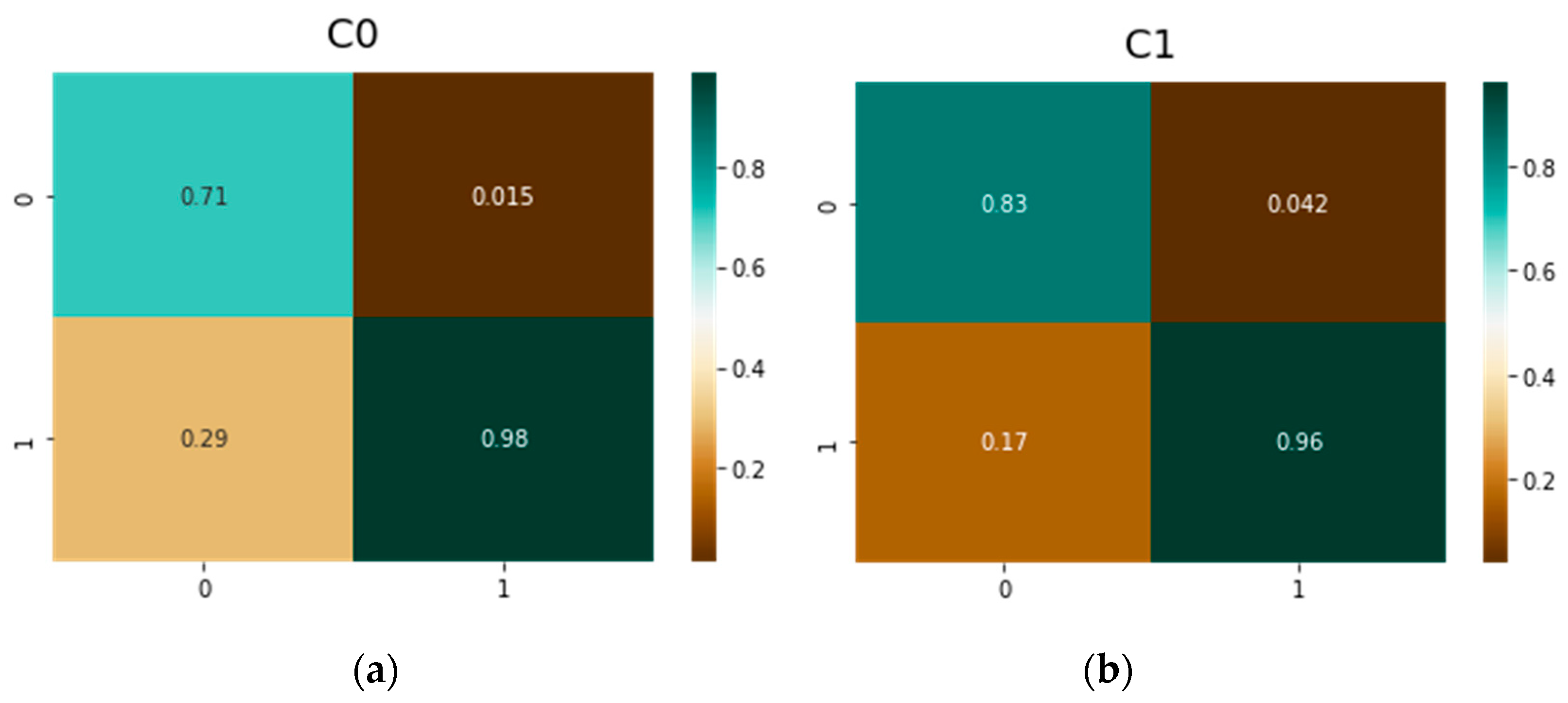
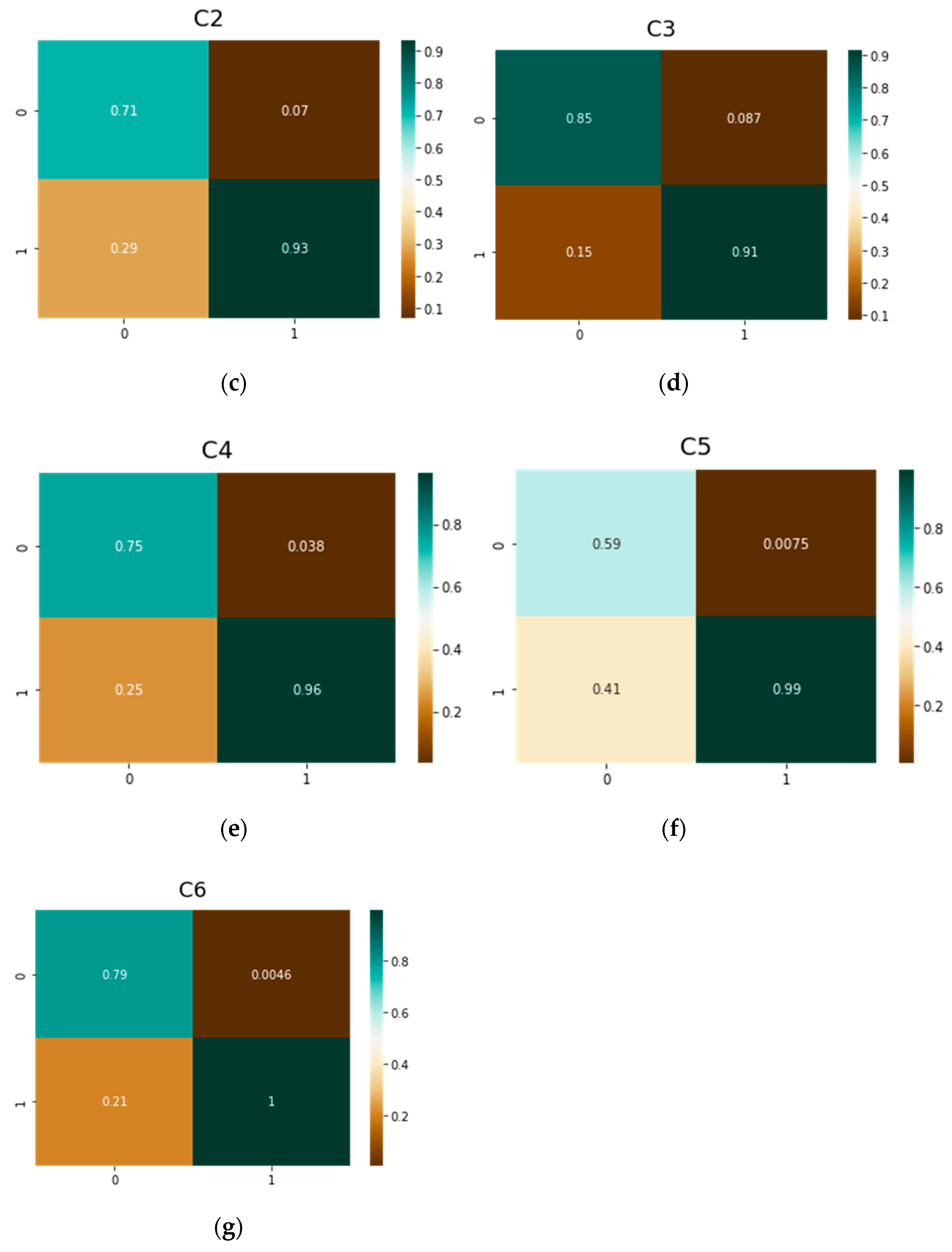

| Parameter | vit-base-patch16-224 | vit-base-patch16-384 |
|---|---|---|
| hidden_size | 768 | 768 |
| image_size | 224 | 384 |
| num_hidden_layers | 12 | 12 |
| patch_size | 16 | 16 |
| NN | Precision | Recall | F1 Score |
|---|---|---|---|
| Resnet50 | 0.62 | 0.61 | 0.61 |
| vit-base-patch16-224 | 0.75 | 0.75 | 0.75 |
| vit-base-patch16-384 | 0.79 | 0.79 | 0.79 |
| DeiT | 0.77 | 0.77 | 0.77 |
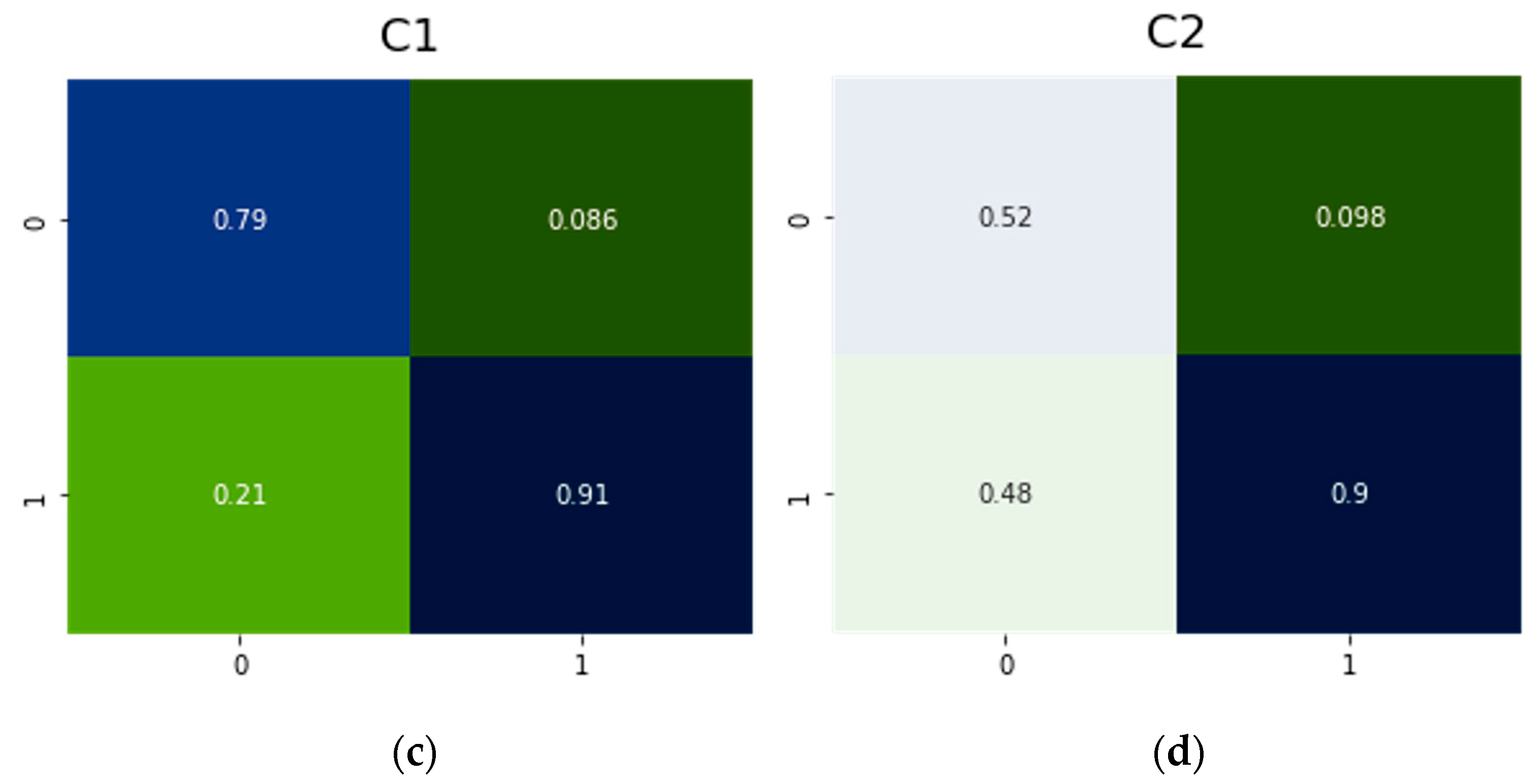
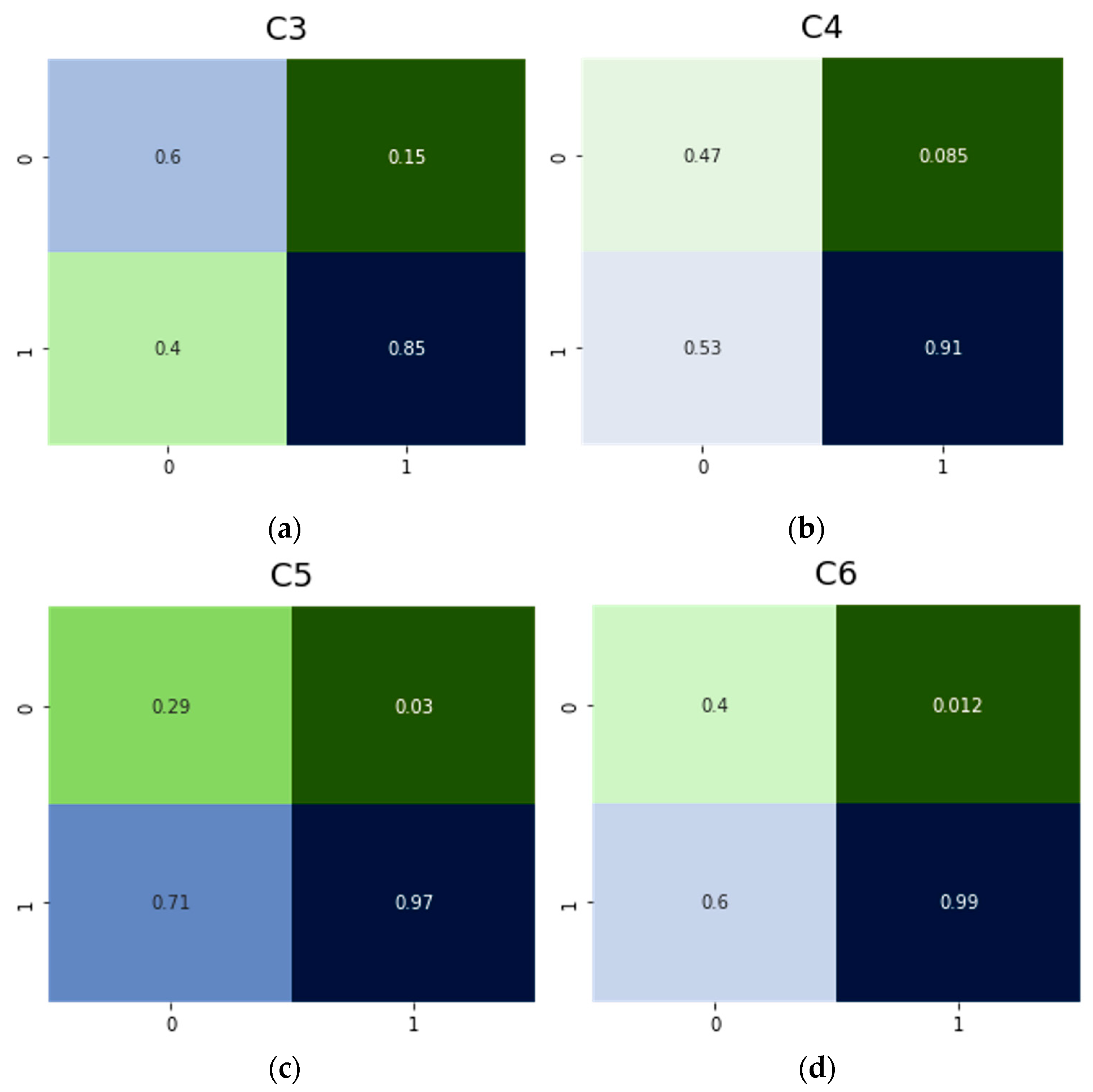
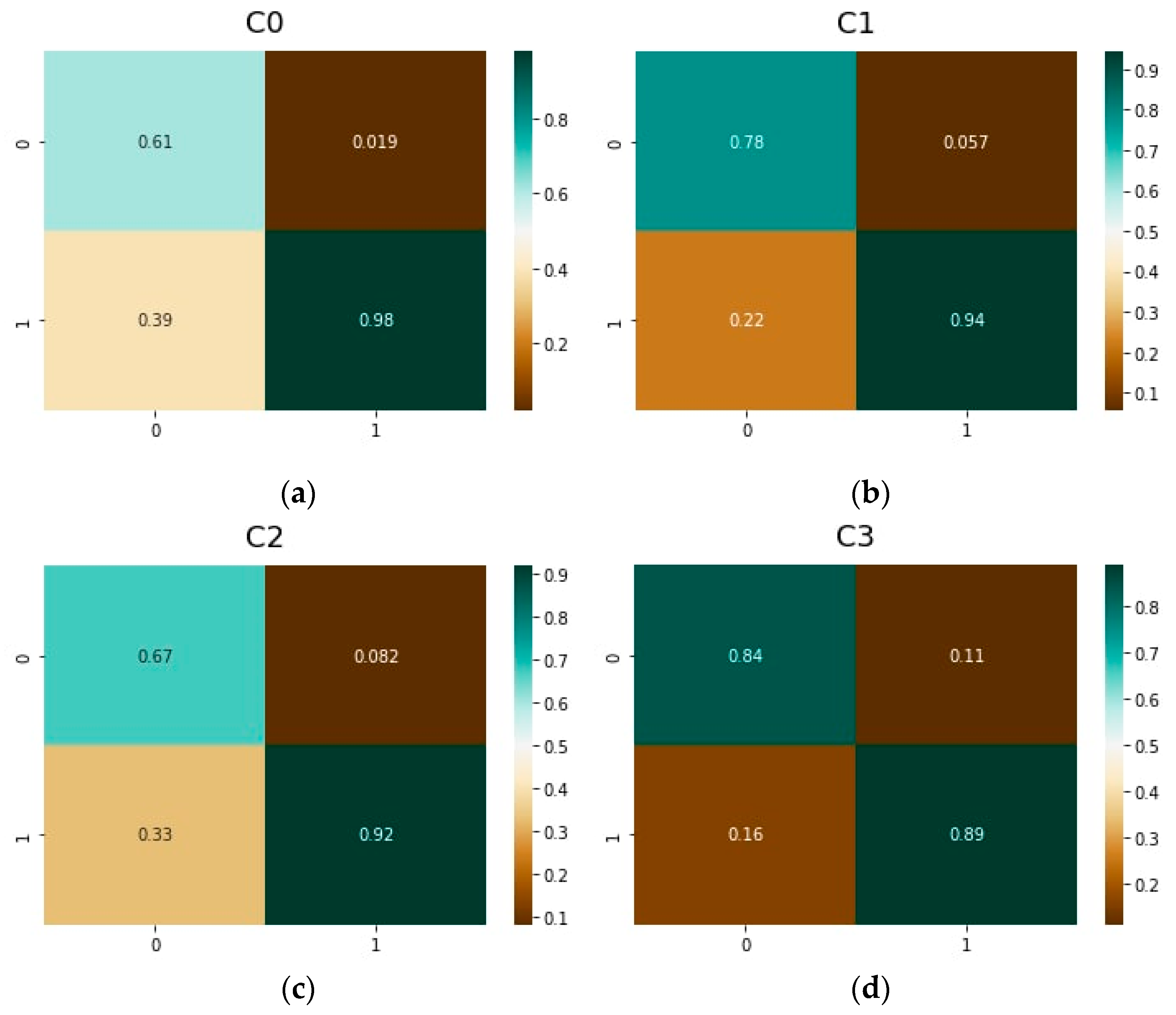
| Model | NN | Rated TP | Rated TN | Rated FP | Rated FN |
|---|---|---|---|---|---|
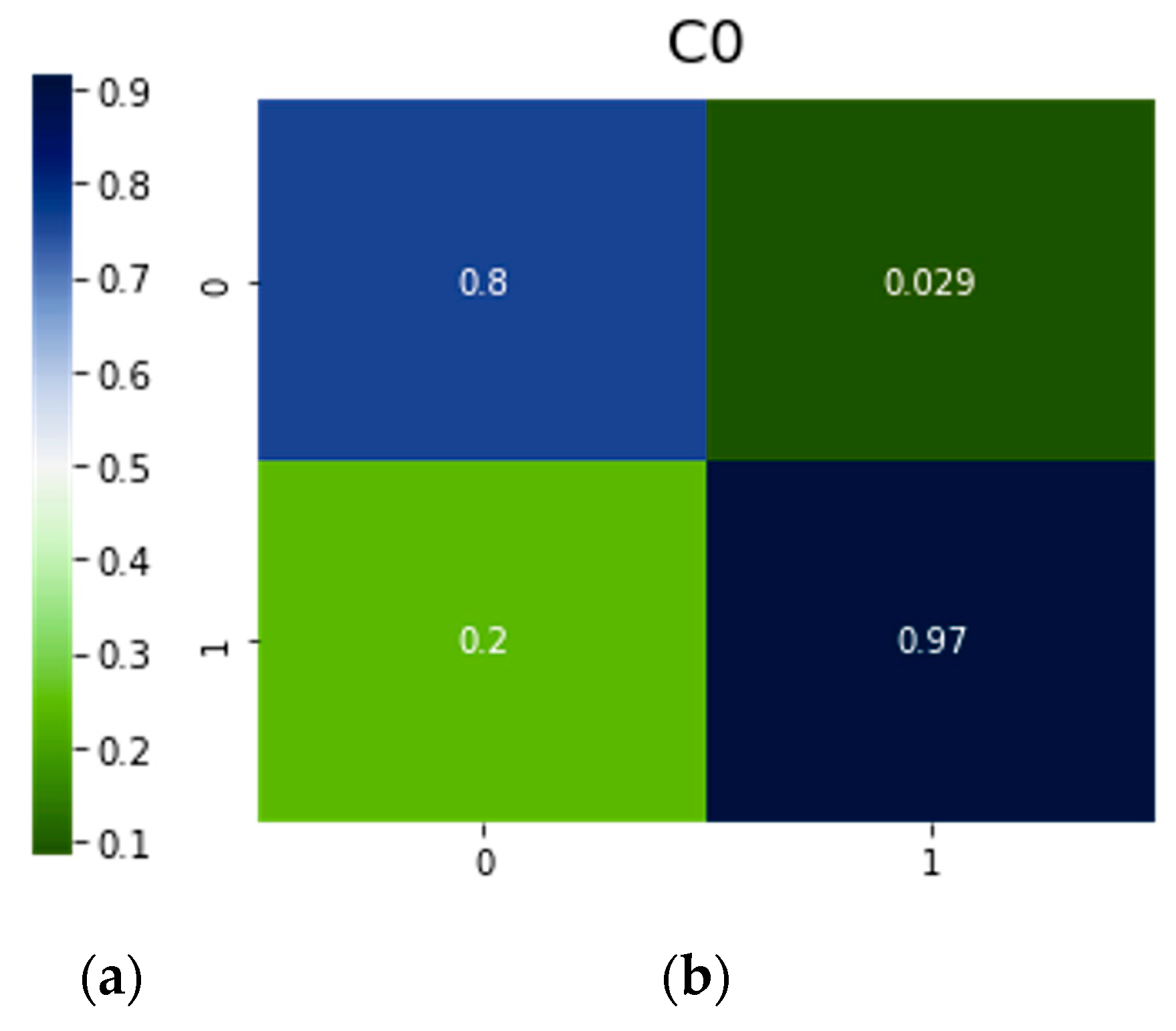
| C0 | Resnet50 | 0.80 | 0.97 | 0.20 | 0.029 |
| vit-base-patch16-224 | 0.61 | 0.98 | 0.39 | 0.019 | |
| vit-base-patch16-384 | 0.71 | 0.98 | 0.29 | 0.015 | |
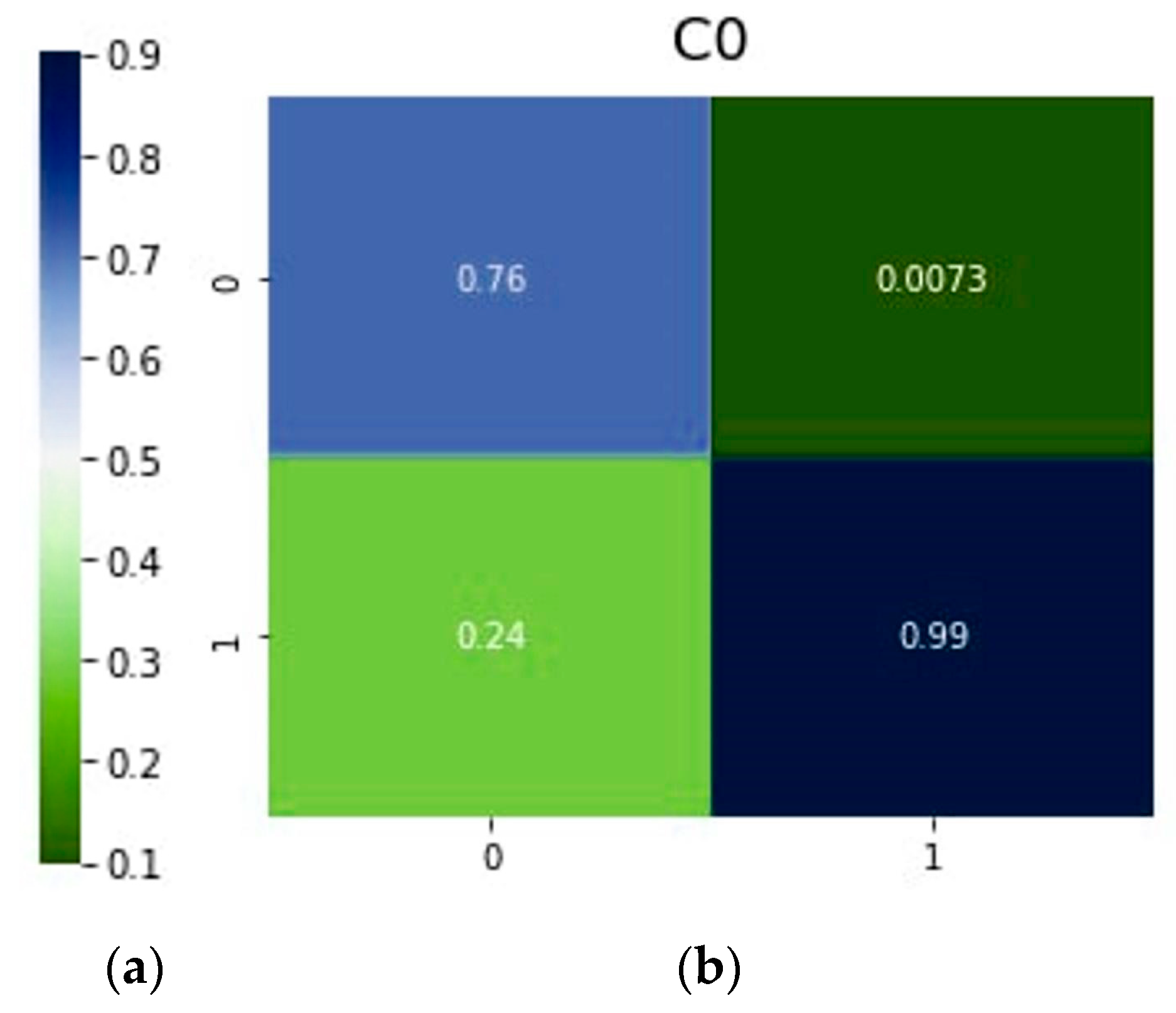
| DeiT | 0.76 | 0.99 | 0.24 | 0.001 | |
| C1 | Resnet50 | 0.79 | 0.91 | 0.21 | 0.086 |
| vit-base-patch16-224 | 0.78 | 0.94 | 0.22 | 0.057 | |
| vit-base-patch16-384 | 0.83 | 0.96 | 0.17 | 0.042 | |
| DeiT | 0.86 | 0.94 | 0.14 | 0.055 | |
| C2 | Resnet50 | 0.52 | 0.90 | 0.48 | 0.098 |
| vit-base-patch16-224 | 0.67 | 0.92 | 0.33 | 0.082 | |
| vit-base-patch16-384 | 0.71 | 0.93 | 0.29 | 0.07 | |
| DeiT | 0.63 | 0.95 | 0.37 | 0.055 | |
| C3 | Resnet50 | 0.60 | 0.85 | 0.40 | 0.150 |
| vit-base-patch16-224 | 0.84 | 0.89 | 0.16 | 0.11 | |
| vit-base-patch16-384 | 0.85 | 0.91 | 0.15 | 0.087 | |
| DeiT | 0.83 | 0.90 | 0.17 | 0.099 | |
| C4 | Resnet50 | 0.47 | 0.91 | 0.53 | 0.085 |
| vit-base-patch16-224 | 0.67 | 0.96 | 0.33 | 0.039 | |
| vit-base-patch16-384 | 0.75 | 0.96 | 0.25 | 0.038 | |
| DeiT | 0.70 | 0.94 | 0.30 | 0.058 | |
| C5 | Resnet50 | 0.29 | 0.91 | 0.71 | 0.030 |
| vit-base-patch16-224 | 0.6 | 0.99 | 0.40 | 0.009 | |
| vit-base-patch16-384 | 0.59 | 0.99 | 0.41 | 0.007 | |
| DeiT | 0.40 | 0.99 | 0.60 | 0.014 | |
| C6 | Resnet50 | 0.40 | 0.99 | 0.60 | 0.012 |
| vit-base-patch16-224 | 0.60 | 0.99 | 0.40 | 0.005 | |
| vit-base-patch16-384 | 0.79 | 1.00 | 0.21 | 0.004 | |
| DeiT | 0.55 | 0.99 | 0.45 | 0.009 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barulina, M.; Sanbaev, A.; Okunkov, S.; Ulitin, I.; Okoneshnikov, I. Deep Learning Approaches to Automatic Chronic Venous Disease Classification. Mathematics 2022, 10, 3571. https://doi.org/10.3390/math10193571
Barulina M, Sanbaev A, Okunkov S, Ulitin I, Okoneshnikov I. Deep Learning Approaches to Automatic Chronic Venous Disease Classification. Mathematics. 2022; 10(19):3571. https://doi.org/10.3390/math10193571
Chicago/Turabian StyleBarulina, Marina, Askhat Sanbaev, Sergey Okunkov, Ivan Ulitin, and Ivan Okoneshnikov. 2022. "Deep Learning Approaches to Automatic Chronic Venous Disease Classification" Mathematics 10, no. 19: 3571. https://doi.org/10.3390/math10193571
APA StyleBarulina, M., Sanbaev, A., Okunkov, S., Ulitin, I., & Okoneshnikov, I. (2022). Deep Learning Approaches to Automatic Chronic Venous Disease Classification. Mathematics, 10(19), 3571. https://doi.org/10.3390/math10193571








