Mass Spectrometric Identification of a Novel Factor XIIIa Cross-Linking Site in Fibrinogen
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis of a Model TG Cross-Linked Peptide
2.2. MS Fragmentation Analysis of the Model TG Cross-Linked Peptide
2.3. Software Analysis
2.4. Cross-Linking of Fibrinogen with Factor XIIIa
2.5. Identification of a Transglutaminase Cross-Linking Site in a Fibrinogen Dimer
3. Results
3.1. Synthesis of TXLP
3.2. Fragmentation Analysis of TXLP
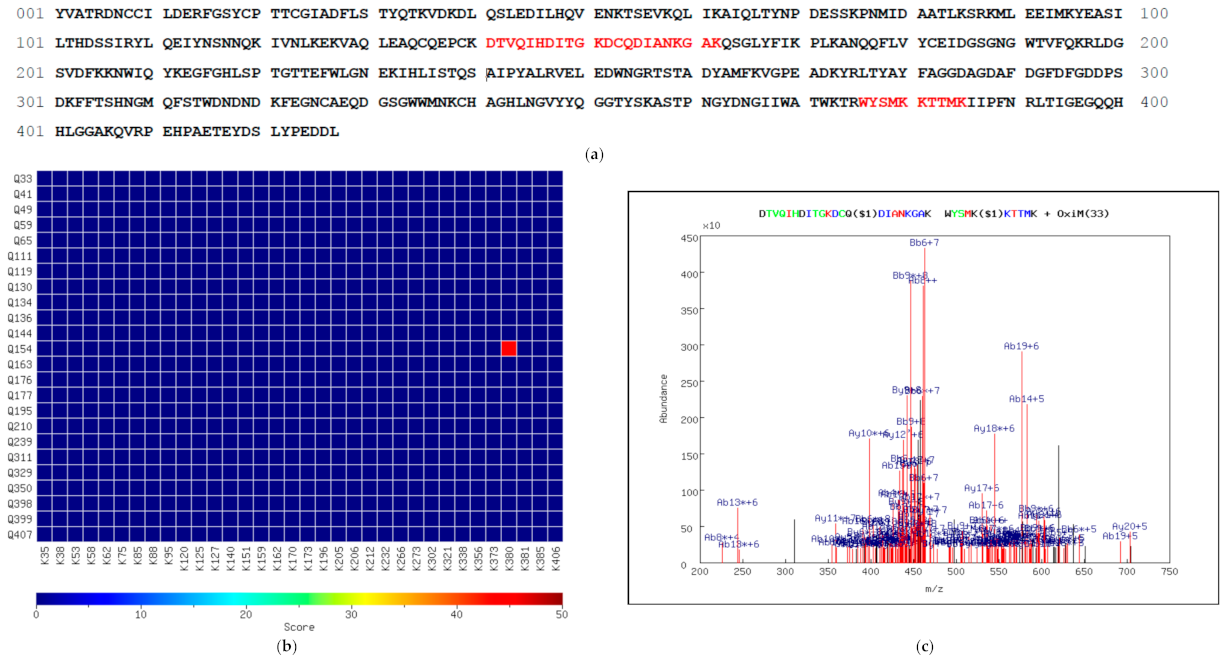
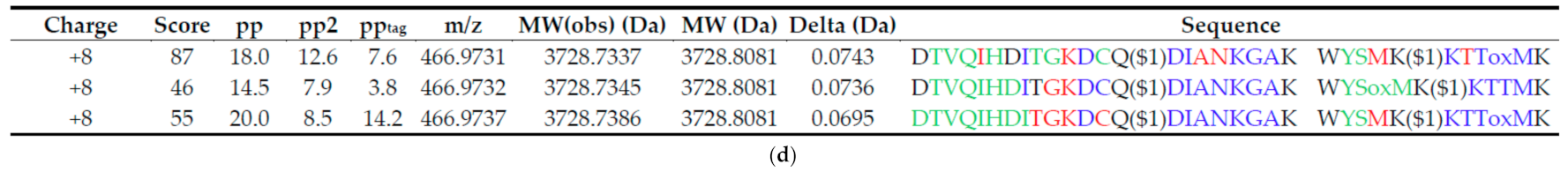
3.3. Identification of TXLP Using MassMatrix
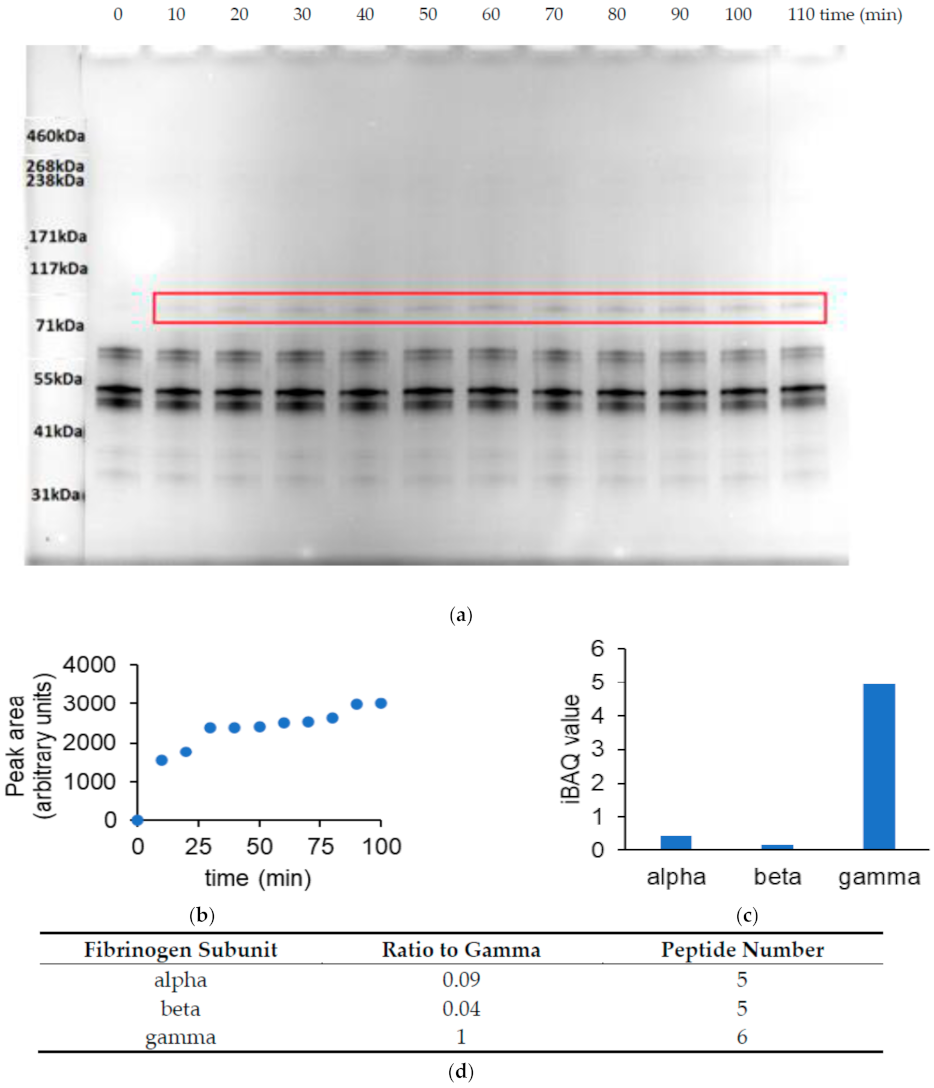
3.4. Identification of TG Cross-Linked Sites in a Fibrinogen γ-γ Dimer
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
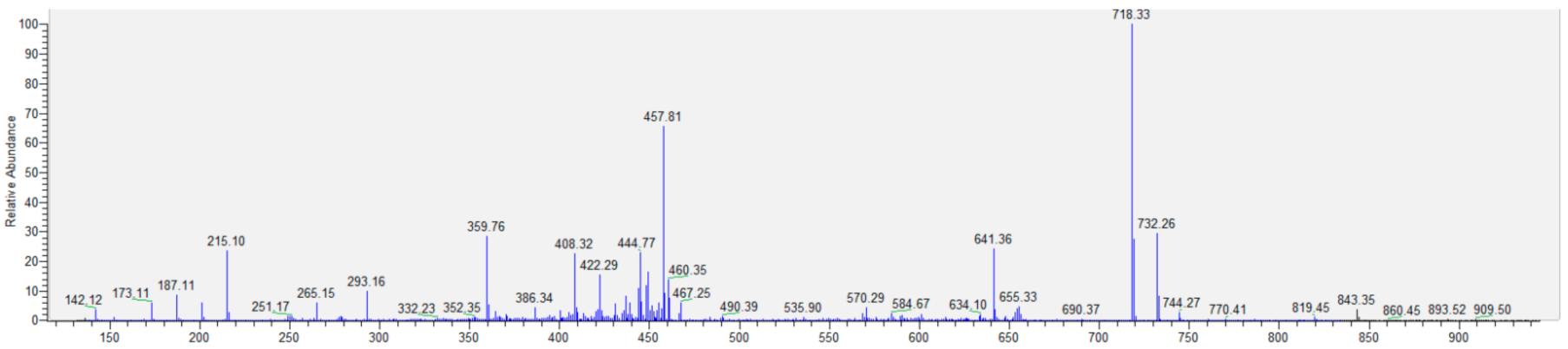
Appendix B
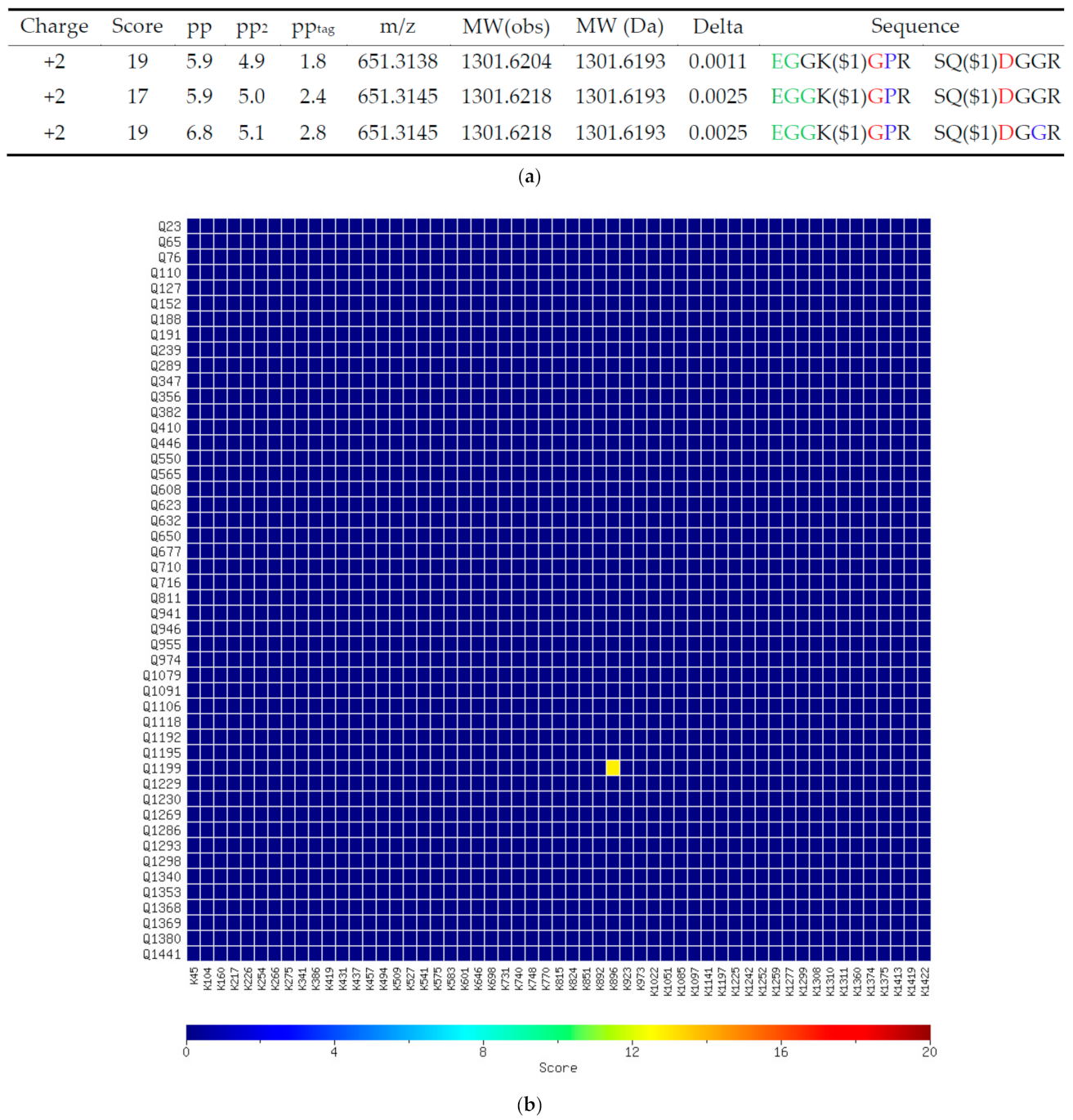
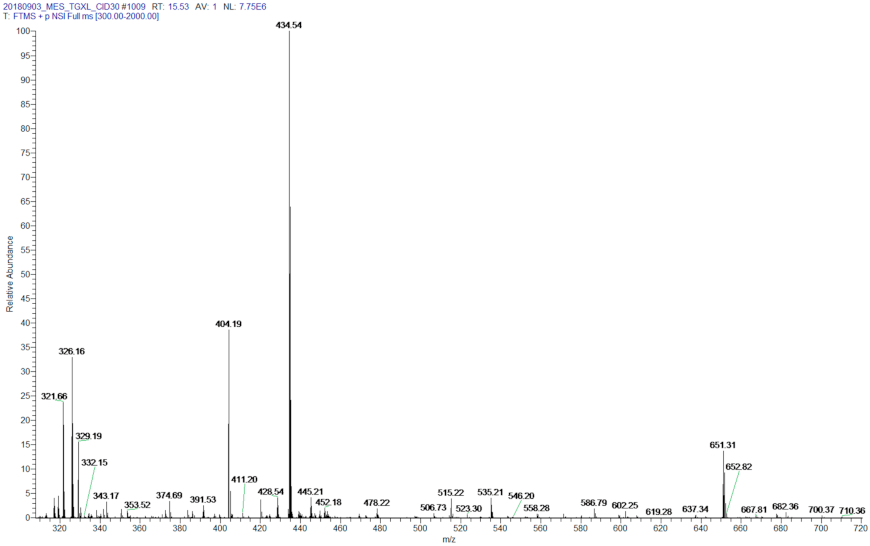
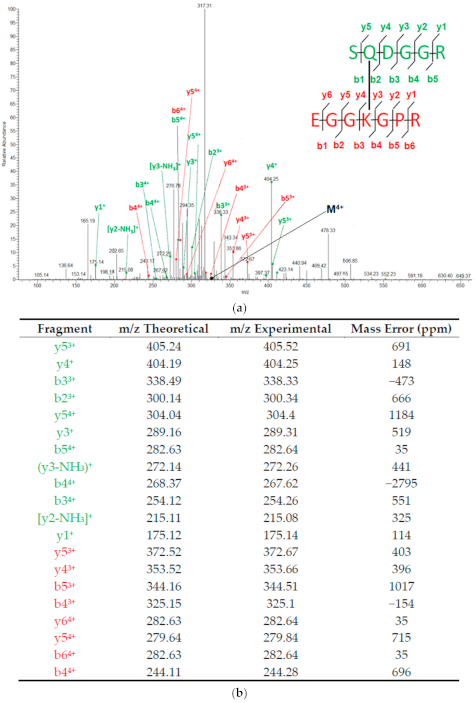
References
- Lorand, L.; Graham, R.M. Transglutaminases: Crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 2003, 4, 140–156. [Google Scholar] [CrossRef]
- Lorand, L. Factor XIII: Structure, Activation, and Interactions with Fibrinogen and Fibrin. Ann. N. Y. Acad. Sci. 2001, 936, 291–311. [Google Scholar] [CrossRef]
- Chen, J.S.K.; Mehta, K. Tissue transglutaminase: An enzyme with a split personality. Int. J. Biochem. Cell Biol. 1999, 31, 817–836. [Google Scholar] [CrossRef]
- Kalinin, A.; Marekov, L.N.; Steinert, P.M. Assembly of the epidermal cornified cell envelope. J. Cell Sci. 2001, 114, 3069–3070. [Google Scholar] [CrossRef]
- Akimov, S.S.; Krylov, D.; Fleischman, L.F.; Belkin, A.M. Tissue Transglutaminase Is an Integrin-binding Adhesion Coreceptor for Fibronectin. J. Cell Biol. 2000, 148, 825–838. [Google Scholar] [CrossRef]
- Lorand, L.; Conrad, S.M. Transglutaminases. Mol. Cell Biochem. 1984, 58, 9–35. [Google Scholar] [CrossRef]
- Konno, T.; Morii, T.; Hirata, A.; Sato, S.; Oiki, S.; Ikura, K. Covalent blocking of fibril formation and aggregation of intracellular amyloidgenic proteins by transglutaminase-catalyzed intramolecular cross-linking. Biochemistry 2005, 44, 2072–2079. [Google Scholar] [CrossRef]
- Mosesson, M.W.; Siebenlist, K.R.; Meh, D.A. The structure and biological features of fibrinogen and fibrin. Ann. N. Y. Acad. Sci. 2001, 936, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Nemes, Z.; Petrovski, G.; Aerts, M.; Sergeant, K.; Devreese, B.; Fesus, L. Transglutaminase-mediated intramolecular cross-linking of membrane-bound alpha-synuclein promotes amyloid formation in Lewy bodies. J. Biol. Chem. 2009, 284, 27252–27264. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Murphy, L.J. Tissue transglutaminase has intrinsic kinase activity: Identification of transglutaminase 2 as an insulin-like growth factor-binding protein-3 kinase. J. Biol. Chem. 2004, 279, 23863–23868. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Molberg, O.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Lee, S.G.; Cho, Y.D. Identification of glycinin in vivo as a polyamine-conjugated protein via a γ-glutamyl linkage. Biochem. J. 1998, 332, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Nemes, Z.; Marekov, L.N.; Fésüs, L.; Steinert, P.M. A novel function for transglutaminase 1: Attachment of long-chain omega-hydroxyceramides to involucrin by ester bond formation. Proc. Natl. Acad. Sci. USA 1999, 96, 8402–8407. [Google Scholar] [CrossRef]
- Grenard, P.; Bresson-Hadni, S.; El Alaoui, S.; Chevallier, M.; Vuitton, D.A.; Ricard-Blum, S. Transglutaminase-mediated cross-linking is involved in the stabilization of extracellular matrix in human liver fibrosis. J. Hepatol. 2001, 35, 367–375. [Google Scholar] [CrossRef]
- Olsen, K.C.; Sapinoro, R.E.; Kottmann, R.M.; Kulkarni, A.A.; Iismaa, S.E.; Johnson, G.V.; Thatcher, T.H.; Phipps, R.P.; Sime, P.J. Transglutaminase 2 and its role in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2011, 184, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Stuckey, D.J.; Murdoch, C.E.; Camelliti, P.; Lip, G.Y.H.; Griffin, M. Cardiac fibrosis can be attenuated by blocking the activity of transglutaminase 2 using a selective small-molecule inhibitor. Cell Death Dis. 2018, 9, 613. [Google Scholar] [CrossRef]
- Lee, J.; Condello, S.; Yakubov, B.; Emerson, R.; Caperell-Grant, A.; Hitomi, K.; Xie, J.; Matei, D. Tissue Transglutaminase Mediated Tumor-Stroma Interaction Promotes Pancreatic Cancer Progression. Clin. Cancer Res. 2015, 21, 4482–4493. [Google Scholar] [CrossRef]
- Lee, J.; Yakubov, B.; Ivan, C.; Jones, D.R.; Caperell-Grant, A.; Fishel, M.; Cardenas, H.; Matei, D. Tissue Transglutaminase Activates Cancer-Associated Fibroblasts and Contributes to Gemcitabine Resistance in Pancreatic Cancer. Neoplasia 2016, 18, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Martucciello, S.; Paolella, G.; Esposito, C.; Lepretti, M.; Caputo, I. Anti-type 2 transglutaminase antibodies as modulators of type 2 transglutaminase functions: A possible pathological role in celiac disease. Cell Mol. Life Sci. 2018, 75, 4107–4124. [Google Scholar] [CrossRef]
- Jones, R.A.; Kotsakis, P.; Johnson, T.S.; Chau, D.Y.; Ali, S.; Melino, G.; Griffin, M. Matrix changes induced by transglutaminase 2 lead to inhibition of angiogenesis and tumor growth. Cell Death Differ. 2006, 13, 1442–1453. [Google Scholar] [CrossRef]
- Hitomi, K.; Kitamura, M.; Sugimura, Y. Preferred substrate sequences for transglutaminase 2: Screening using a phage-displayed peptide library. Amino Acids 2009, 36, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Song, C.; Kim, D.H.; Park, I.H.; Lee, S.G.; Lee, Y.S.; Kim, B.G. Glutamine (Q)-peptide screening for transglutaminase reaction using mRNA display. Biotechnol. Bioeng. 2013, 110, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Malesevic, M.; Migge, A.; Hertel, T.C.; Pietzsch, M. A fluorescence-based array screen for transglutaminase substrates. Chembiochem 2015, 16, 1169–1174. [Google Scholar] [CrossRef] [PubMed]
- Bains, W. Transglutaminse 2 and EGGL, the protein cross-link formed by transglutaminse 2, as therapeutic targets for disabilities of old age. Rejuvenation Res. 2013, 16, 495–517. [Google Scholar] [CrossRef] [PubMed]
- Groenen, P.J.; Smulders, R.H.; Peters, R.F.; Grootjans, J.J.; van den Ijssel, P.R.; Bloemendal, H.; de Jong, W.W. The amine-donor substrate specificity of tissue-type transglutaminase. Influence of amino acid residues flanking the amine-donor lysine residue. Eur. J. Biochem. 1994, 220, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Csosz, E.; Bagossi, P.; Nagy, Z.; Dosztanyi, Z.; Simon, I.; Fesus, L. Substrate preference of transglutaminase 2 revealed by logistic regression analysis and intrinsic disorder examination. J. Mol. Biol. 2008, 383, 390–402. [Google Scholar] [CrossRef]
- Murthy, S.N.P.; Lorand, L. Cross-linked Aalpha.gamma chain hybrids serve as unique markers for fibrinogen polymerized by tissue transglutaminase. Proc. Natl. Acad. Sci. USA 1990, 87, 9679–9682. [Google Scholar] [CrossRef]
- Tatsukawa, H.; Tani, Y.; Otsu, R.; Nakagawa, H.; Hitomi, K. Global identification and analysis of isozyme-specific possible substrates crosslinked by transglutaminases using substrate peptides in mouse liver fibrosis. Sci. Rep. 2017, 7, 45049. [Google Scholar] [CrossRef]
- Purves, L.; Purves, M.; Brandt, W. Cleavage of Fibrin-Derived D-Dimer into Monomers by Endopeptidase from Puff Adder Venom (Bitis arietans) Acting at Cross-Linked Sites of the gamma-Chain. Sequence of Carboxy-Terminal Cyanogen Bromide gamma-Chain Fragments. Biochemistry 1987, 26, 4640–4646. [Google Scholar] [CrossRef] [PubMed]
- Recktenwald, C.V.; Hansson, G.C. The Reduction-insensitive Bonds of the MUC2 Mucin Are Isopeptide Bonds. J. Biol. Chem. 2016, 291, 13580–13590. [Google Scholar] [CrossRef]
- Velez, V.F.; Romano, J.A.; McKown, R.L.; Green, K.; Zhang, L.; Raab, R.W.; Ryan, D.S.; Hutnik, C.M.; Frierson, H.F., Jr.; Laurie, G.W. Tissue transglutaminase is a negative regulator of monomeric lacritin bioactivity. Investig. Ophthalmol. Vis. Sci. 2013, 54, 2123–2132. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, L.R.; Henderson, R.; Barrett, A.; Darula, Z.; Issaian, A.; D’Alessandro, A.; Clendenen, N.; Hansen, K.C. Mass spectrometry-based molecular mapping of native FXIIIa cross-links in insoluble fibrin clots. J. Biol. Chem. 2019, 294, 8773–8778. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J. The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 2011, 173, 530–540. [Google Scholar] [CrossRef]
- Belsom, A.; Schneider, M.; Fischer, L.; Brock, O.; Rappsilber, J. Serum Albumin Domain Structures in Human Blood Serum by Mass Spectrometry and Computational Biology. Mol. Cell Proteom. 2016, 15, 1105–1116. [Google Scholar] [CrossRef]
- Barysz, H.M.; Malmstrom, J. Development of Large-scale Cross-linking Mass Spectrometry. Mol. Cell. Proteomics 2018, 17, 1055–1066. [Google Scholar] [CrossRef]
- Ohtsuka, T.; Ota, M.; Nio, N.; Motoki, M. Comparison of Substrate Specificities of Transglutaminases Using Synthetic Peptides as Acyl donors. Biosci. Biotechnol. Biochem. 2000, 64, 2608–2613. [Google Scholar] [CrossRef]
- Xu, H.; Hsu, P.; Zhang, L.; Tsai, M.; Freitas, M.A. Database Search Algorithm for Identification of Intact Cross-Links in Proteins and Peptides Using Tandem Mass Spectrometry. J. Proteome Res. 2010, 9, 3384–3393. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J.H.; Gawinowicz, M.A. Identification of the alpha Chain Lysine Donor Sites Involved in Factor XIIIa Fibrin Cross-linking. J. Biol. Chem. 1996, 271, 19288–19297. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Barrett, A.S.; Wither, M.J.; Hill, R.C.; Dzieciatkowska, M.; D’Alessandro, A.; Reisz, J.A.; Hansen, K.C. Hydroxylamine Chemical Digestion for Insoluble Extracellular Matrix Characterization. J. Proteome Res. 2017, 16, 4177–4184. [Google Scholar] [CrossRef] [PubMed]
- Frese, C.K.; Altelaar, A.F.; Hennrich, M.L.; Nolting, D.; Zeller, M.; Griep-Raming, J.; Heck, A.J.; Mohammed, S. Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 2011, 10, 2377–2388. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Schwartz, J.C.; Griep-Raming, J.; Nielsen, M.L.; Damoc, E.; Denisov, E.; Lange, O.; Remes, P.; Taylor, D.; Splendore, M.; et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 2009, 8, 2759–2769. [Google Scholar] [CrossRef] [PubMed]
- Kolbowski, L.; Mendes, M.L.; Rappsilber, J. Optimizing the Parameters Governing the Fragmentation of Cross-Linked Peptides in a Tribrid Mass Spectrometer. Anal. Chem. 2017, 89, 5311–5318. [Google Scholar] [CrossRef] [PubMed]
- Stieger, C.E.; Doppler, P.; Mechtler, K. Optimized Fragmentation Improves the Identification of Peptides Cross-Linked by MS-Cleavable Reagents. J. Proteome Res. 2019, 18, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Chen, R.; Doolittle, R.F. γ-γ Cross-linking sites in human and bovine fibrin. Biochemistry 2002, 10, 4486–4491. [Google Scholar] [CrossRef]
- Schopfer, L.M.; Onder, S.; Lockridge, O. Evaluation of mass spectrometry MS/MS spectra for the presence of isopeptide crosslinked peptides. PLoS ONE 2021, 16, e0254450. [Google Scholar] [CrossRef]
- Trnka, M.J.; Baker, P.R.; Robinson, P.J.; Burlingame, A.L.; Chalkley, R.J. Matching Cross-linked Peptide Spectra: Only as Good as the Worse Identification. Mol. Cell Proteom. 2014, 13, 420–434. [Google Scholar] [CrossRef]
- Dongré, A.R.; Jones, J.L.; Somogyi, A.; Wysocki, V.H. Influence of Peptide Composition, Gas-Phase Basicity, and Chemical Modification on Fragmentation Efficiency: Evidence for the Mobile Proton Model. J. Am. Chem. Soc. 1996, 118, 8365–8374. [Google Scholar] [CrossRef]
- Sun, F.; Liu, R.; Zong, W.; Tian, Y.; Wang, M.; Zhang, P. A Unique Approach to the Mobile Proton Model: Influence of Charge Distribution on Peptide Fragmentation. J. Phys. Chem. B 2010, 114, 6350–6353. [Google Scholar] [CrossRef]
- Diedrich, J.K.; Pinto, A.F.; Yates, J.R., 3rd. Energy dependence of HCD on peptide fragmentation: Stepped collisional energy finds the sweet spot. J. Am. Soc. Mass Spectrom. 2013, 24, 1690–1699. [Google Scholar] [CrossRef] [PubMed]
- Baker, E.S.; Tang, K.; Danielson, W.F.; Prior, D.C.; Smith, R.D. Simultaneous fragmentation of multiple ions using IMS drift time dependent collision energies. J. Am. Soc. Mass Spectrom. 2008, 19, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Giese, S.H.; Belsom, A.; Rappsilber, J. Optimized Fragmentation Regime for Diazirine Photo-Cross-Linked Peptides. Anal. Chem. 2016, 88, 8239–8247. [Google Scholar] [CrossRef]
- Xu, H.; Freitas, M.A. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinform. 2007, 8, 133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, H.; Freitas, M.A. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 2009, 9, 1548–1555. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semkova, M.E.; Hsuan, J.J. Mass Spectrometric Identification of a Novel Factor XIIIa Cross-Linking Site in Fibrinogen. Proteomes 2021, 9, 43. https://doi.org/10.3390/proteomes9040043
Semkova ME, Hsuan JJ. Mass Spectrometric Identification of a Novel Factor XIIIa Cross-Linking Site in Fibrinogen. Proteomes. 2021; 9(4):43. https://doi.org/10.3390/proteomes9040043
Chicago/Turabian StyleSemkova, Mariya E., and J. Justin Hsuan. 2021. "Mass Spectrometric Identification of a Novel Factor XIIIa Cross-Linking Site in Fibrinogen" Proteomes 9, no. 4: 43. https://doi.org/10.3390/proteomes9040043
APA StyleSemkova, M. E., & Hsuan, J. J. (2021). Mass Spectrometric Identification of a Novel Factor XIIIa Cross-Linking Site in Fibrinogen. Proteomes, 9(4), 43. https://doi.org/10.3390/proteomes9040043






