Subcellular Proteomics to Understand Promotive Effect of Plant-Derived Smoke Solution on Soybean Root
Abstract
1. Introduction
2. Materials and Methods
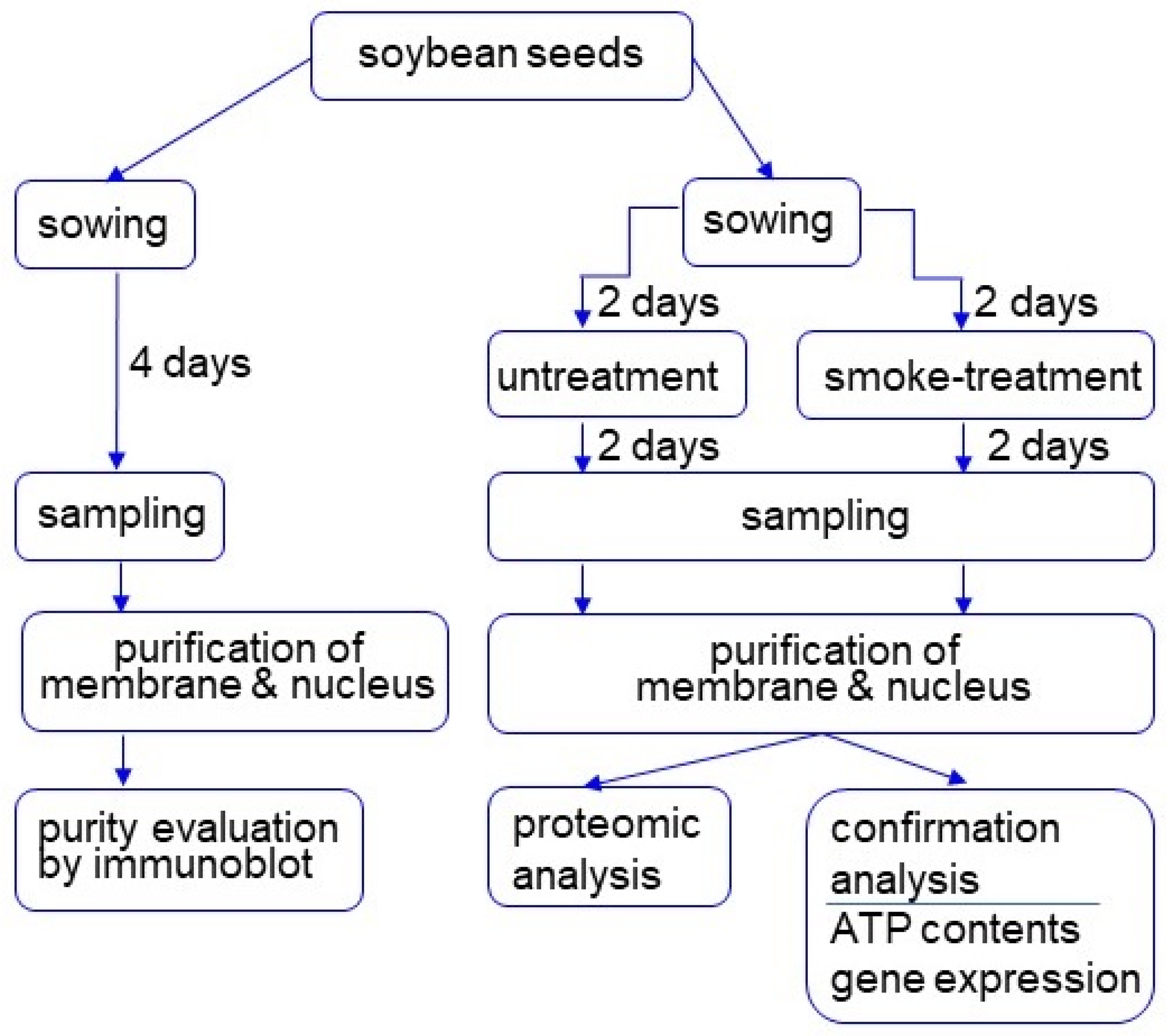
2.1. Plant Material and Treatment
2.2. Isolation of Membrane Fractions
2.3. Isolation of Nuclear Fractions
2.4. Protein Concentration Measurement
2.5. Immunoblot Analysis
2.6. Protein Enrichment, Reduction, Alkylation, and Digestion
2.7. Protein Identification Using Nano-Liquid Chromatography Mass Spectrometry
2.8. MS Data Analysis
2.9. Differential Analysis of Proteins Using MS Data
2.10. Measurement of ATP Contents
2.11. Reverse Transcriptase-Polymerase Chain Reaction Analysis
2.12. Statistical Analysis
3. Results
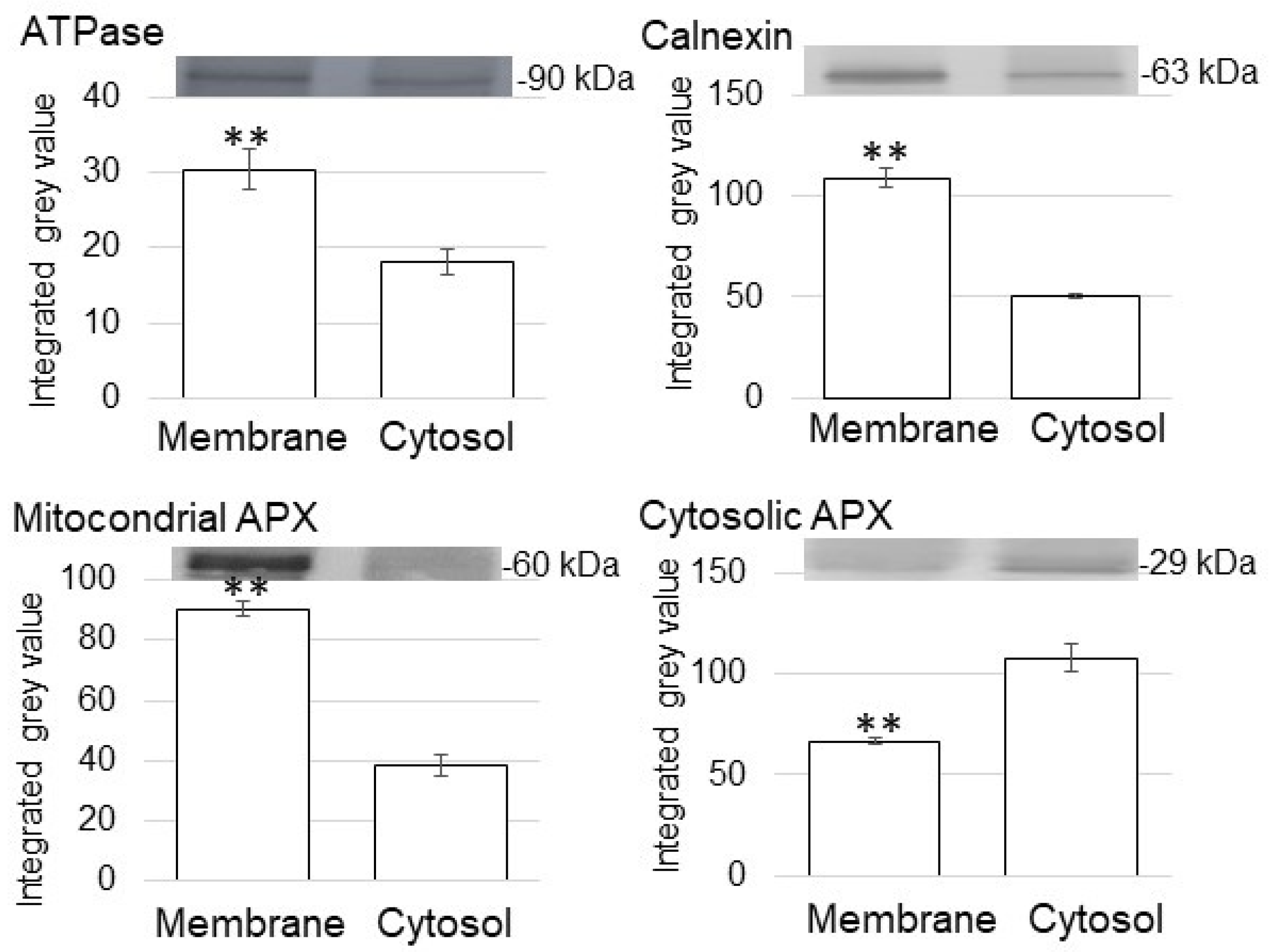
3.1. Purification of Membrane and Nucleus Fractions in Soybean Root Tip
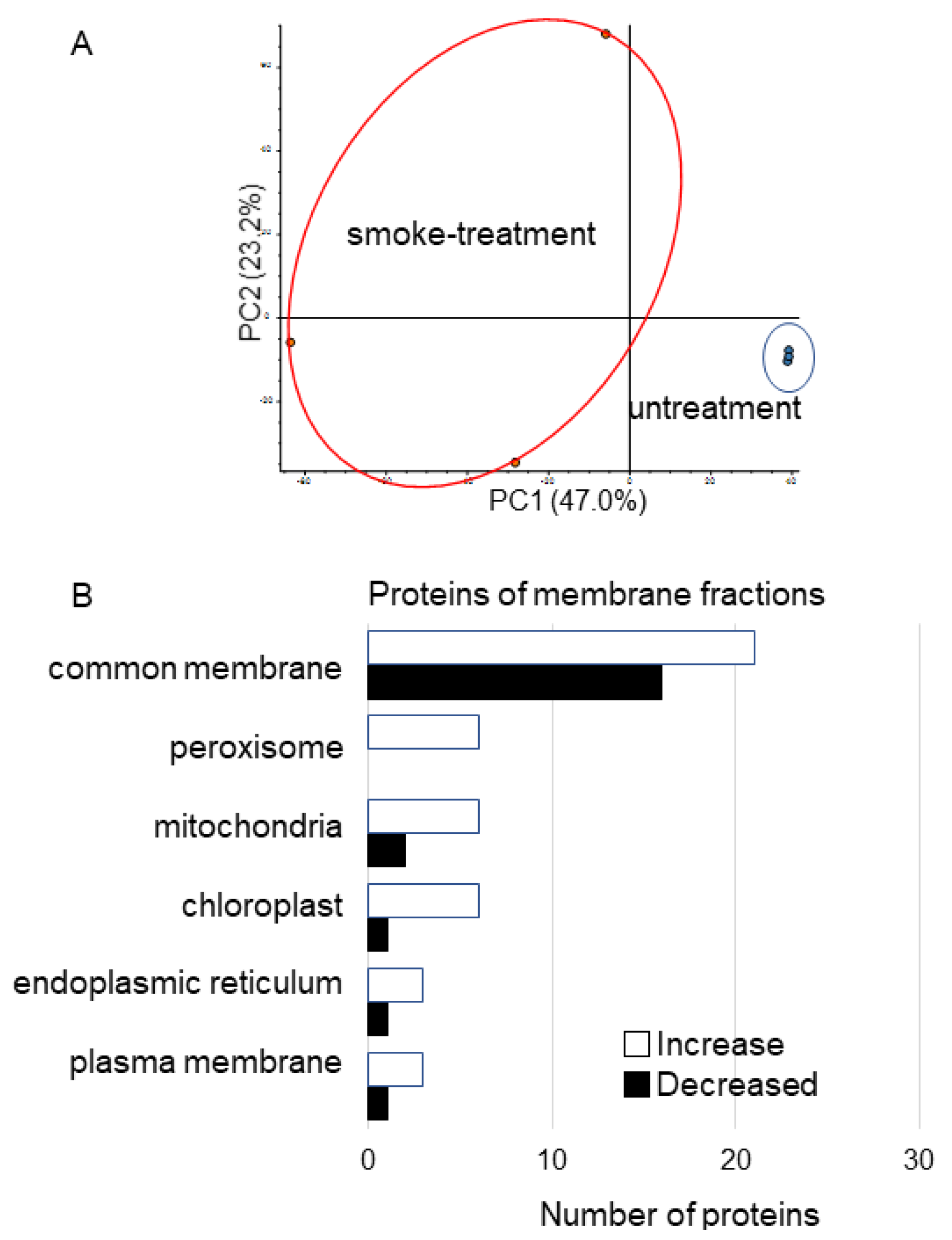
3.2. Membrane Proteomics of Soybean Treated with Plant-Derived Smoke
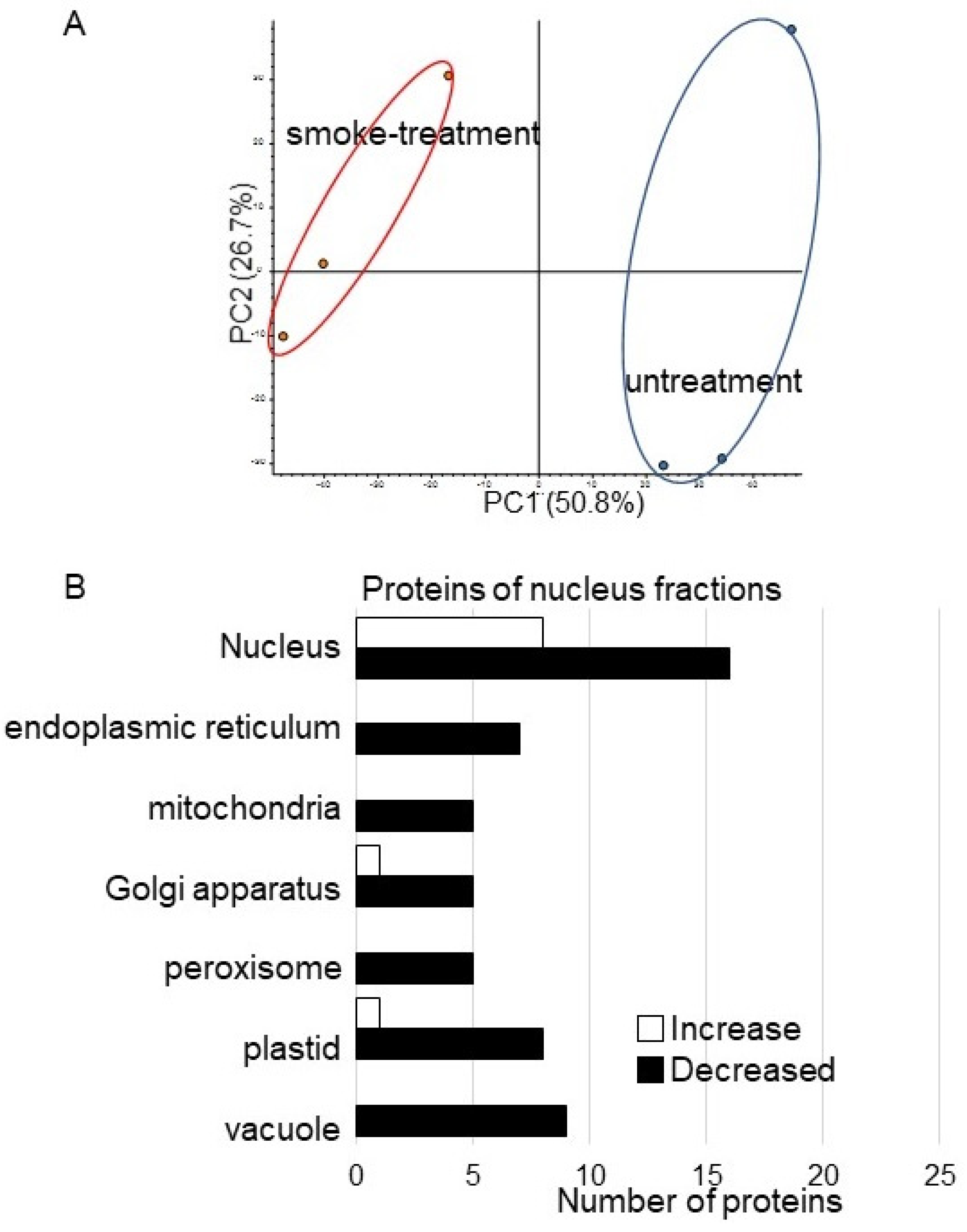
3.3. Nuclear Proteomics of Soybean Treated with Plant-Derived Smoke
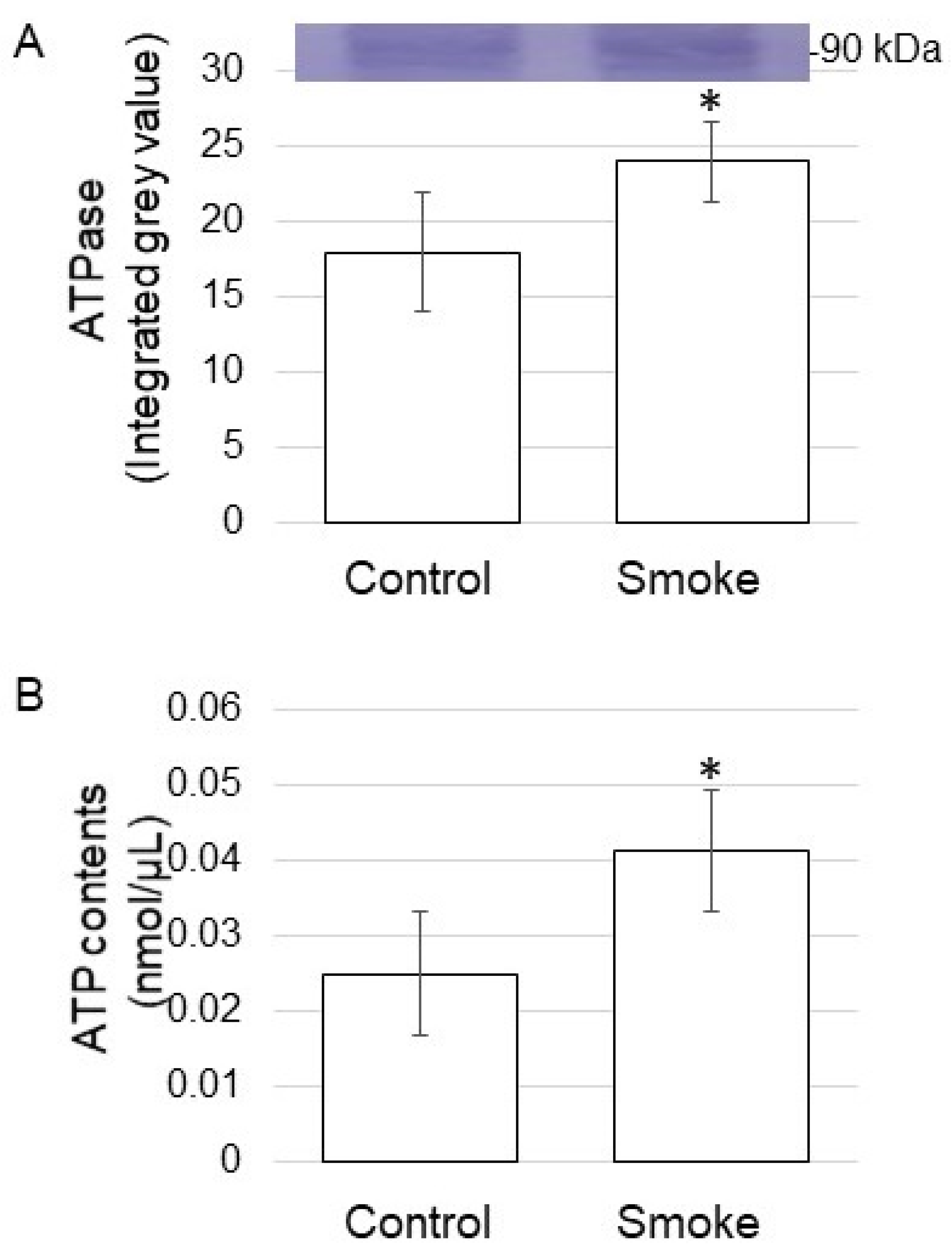
3.4. ATPase Abundance and ATP Contents in Soybean Treated with Plant-Derived Smoke
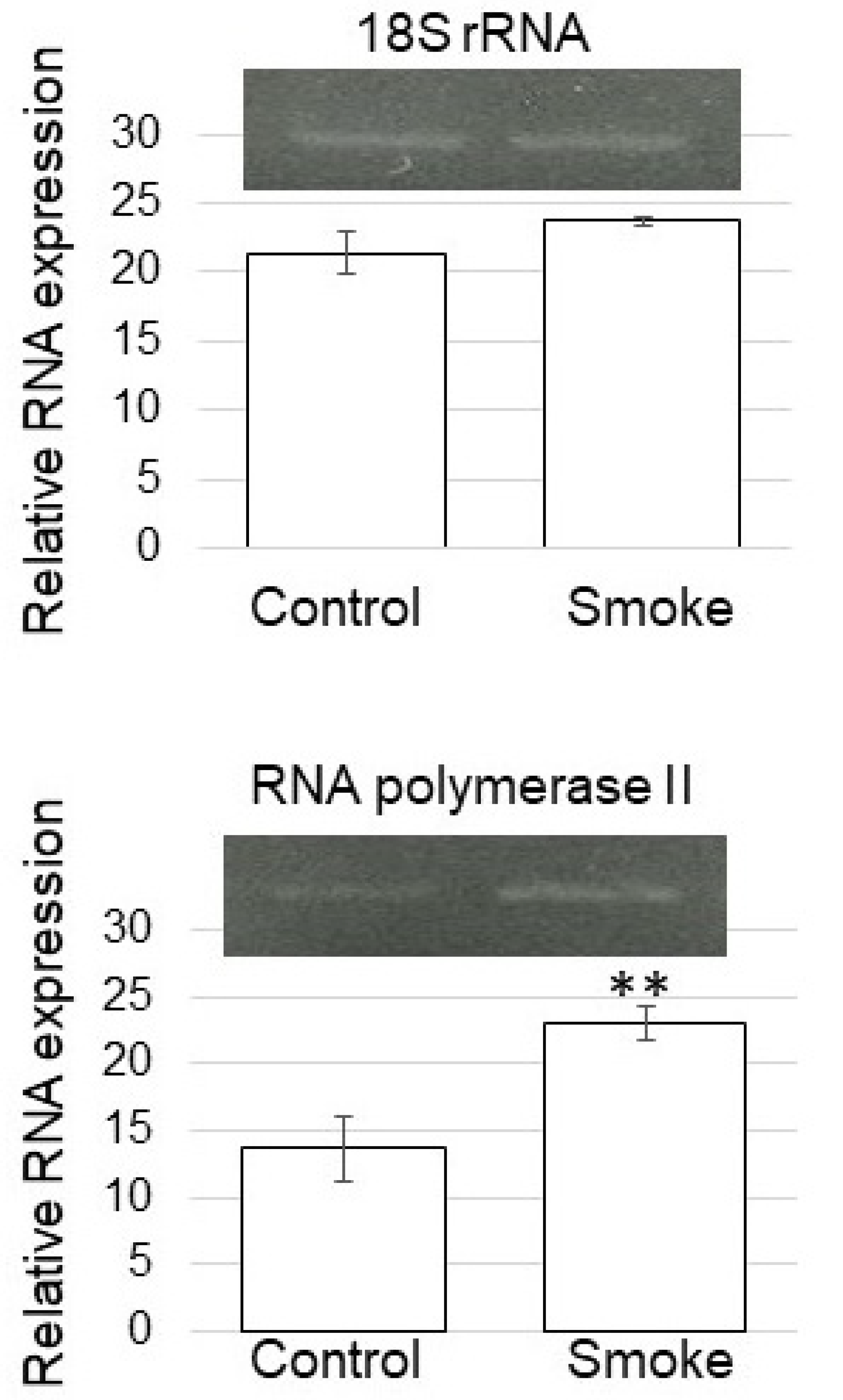
3.5. Expression of RNA Polymerase II in Soybean Treated with Plant-Derived Smoke
4. Discussion
4.1. The Effect of Plant-Derived Smoke Solution on Membrane in Soybean Root
4.2. The Effect of Plant-Derived Smoke Solution on Nucleus in Soybean Root
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khatoon, A.; Rehman, S.U.; Aslam, M.M.; Jamil, M.; Komatsu, S. Plant-derived smoke affects biochemical mechanism on plant growth and seed germination. Int. J. Mol. Sci. 2020, 21, 7760. [Google Scholar] [CrossRef]
- Doherty, L.C.; Cohn, M.A. Seed dormancy in rice (Oryza sativa). Commercial liquid smoke elicits germination. Seed Sci. Res. 2000, 10, 415–421. [Google Scholar] [CrossRef]
- Jamil, M.; Kanwal, M.; Aslam, M.M.; Khan, S.S.; Malook, I.; Tu, J.; Rehman, S.U. Effect of plant-derived smoke priming on physiological and biochemical characteristics of rice under salt stress condition. Aust. J. Crop Sci. 2014, 8, 159–170. [Google Scholar]
- Malook, J.; Shah, G.; Jan, M.; Shinwari, K.I.; Aslam, M.M.; Rehman, S.U.; Jamil, M. Smoke priming regulates growth and the expression of myeloblastosis and zinc-finger genes in rice under salt stress. Arab. J. Sci. Eng. 2017, 42, 2207–2215. [Google Scholar] [CrossRef]
- Aslam, M.M.; Rehman, S.; Khatoon, A.; Jamil, M.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Li, X.; Sunohara, Y.; Matsumoto, H.; et al. Molecular responses of maize shoot to a plant derived smoke solution. Int. J. Mol. Sci. 2019, 20, 1319. [Google Scholar] [CrossRef]
- Aslam, M.M.; Jamil, M.; Khatoon, A.; Hendawy, S.E.; AL-Suhaibani, N.A.; Malook, I.; Rehman, S.U. Physiological and biochemical responses of maize (Zea mays L.) to plant derived smoke solution. Pak. J. Bot. 2017, 49, 435–443. [Google Scholar]
- Li, X.; Rehman, S.U.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Yamaguchi, T.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Proteomic analysis of the effect of plant-derived smoke on soybean during recovery from flooding stress. J. Proteom. 2018, 181, 238–248. [Google Scholar] [CrossRef]
- Zhong, Z.; Kobayashi, T.; Zhu, W.; Imai, H.; Zhao, R.; Ohno, T.; Rehman, S.U.; Uemura, M.; Tian, J.; Komatsu, S. Plant-derived smoke enhances plant growth through ornithine-synthesis pathway and ubiquitin-proteasome pathway in soybean. J. Proteom. 2020, 221, 103781. [Google Scholar] [CrossRef] [PubMed]
- Otori, M.; Murashita, Y.; Rehman, S.; Komatsu, S. Proteomic study to understand promotive effects of plant-derived smoke on soybean root growth under flooding stress. Plant Mol. Biol. Rep. 2021, 39, 24–33. [Google Scholar] [CrossRef]
- Rehman, A.; Rehman, S.U.; Khatoon, A.; Qasim, M.; Itoh, T.; Iwasaki, Y.; Wang, X.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Proteomic analysis of the promotive effect of plant-derived smoke on plant growth of chickpea. J. Proteom. 2018, 176, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Hrdlička, J.; Ngoroyemoto, N.; Nemahunguni, N.K.; Gucký, T.; Novák, O.; Kulkarni, M.J.; Dolezal, K.; van Staden, J. Preparation and standardisation of smoke-water for seed germination and plant growth stimulation. Plant Growth Regul. 2019, 39, 338–345. [Google Scholar] [CrossRef]
- Bose, U.; Juhász, A.; Broadbent, J.A.; Komatsu, S.; Colgrave, M.L. Multi-omics strategies for decoding smoke-assisted germination pathways and seed vigour. Int. J. Mol. Sci. 2020, 21, 7512. [Google Scholar] [CrossRef]
- Baldrianova, J.; Černy, M.; Novak, J.; Jedelsky, P.L.; Diviškova, E.; Brzobohaty, B. Arabidopsis proteome responses to the smoke-derived growth regulator Karrikin. J. Proteom. 2015, 120, 7–20. [Google Scholar] [CrossRef]
- Struk, S.; De Cuyper, C.; Jacobs, A.; Braem, L.; Walton, A.; De Keyser, A.; Depuydt, S.; Vu, L.D.; De Smet, I.; Boyer, F.D.; et al. Unraveling the MAX2 protein network in Arabidopsis thaliana: Identification of the protein phosphatase PAPP5 as a novel MAX2 interactor. Mol. Cell. Proteom. 2021, 20, 100040. [Google Scholar] [CrossRef] [PubMed]
- Tieu, A.; Dixon, K.A.; Sivasithamparam, K.; Plummer, J.A. Germination of four species of native western Australian plant using plant-derived smoke. Aust. J. Bot. 1999, 47, 207–219. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Yamamoto, A.; Nakamura, T.; Nouri, M.Z.; Nanjo, Y.; Nishizawa, K.; Furukawa, K. Comprehensive analysis of mitochondria in roots and hypocotyls of soybean under flooding stress using proteomics and metabolomics techniques. J. Proteom. Res. 2011, 10, 3993–4004. [Google Scholar] [CrossRef]
- Nouri, M.Z.; Hiraga, S.; Yanagawa, Y.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Characterization of calnexin in soybean roots and hypocotyls under osmotic stress. Phytochemistry 2012, 74, 20–29. [Google Scholar] [CrossRef]
- Komatsu, S.; Han, C.; Nanjo, Y.; Altaf-Un-Nahar, M.; Wang, K.; He, D.; Yang, P. Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteom. Res. 2013, 12, 4769–4784. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Bailly, C.; El-Maarouf-Bouteau, H.; Corbineau, F. From intracellular signaling networks to cell death: The dual role of reactive oxygen species in seed physiology. Comptes Rendus Biol. 2008, 331, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Sun, J.; Wang, K.Y.; Liu, D.; Li, Z.Y.; Zhang, J. Spermidine enhances waterlogging tolerance via regulation of antioxidant defence, heat shock protein expression and plasma membrane H+-ATPase activity in Zea mays. J. Agron. Crop Sci. 2014, 200, 199–211. [Google Scholar] [CrossRef]
- Yiu, J.-C.; Liu, C.-W.; Yi-Tan Fang, D.; Lai, Y.-S. Waterlogging tolerance of Welsh onion (Allium fistulosum L.) enhanced by exogenous spermidine and spermine. Plant Physiol. Biochem. 2009, 47, 710–716. [Google Scholar] [CrossRef]
- Bajerski, F.; Stock, J.; Hanf, B.; Darienko, T.; Heine-Dobbernack, E.; Lorenz, M.; Naujox, L.; Keller, E.R.J.; Schumacher, H.M.; Friedl, T.; et al. ATP content and cell viability as indicators for cryostress across the diversity of life. Front. Physiol. 2018, 9, 921. [Google Scholar] [CrossRef]
- De Col, V.; Fuchs, P.; Nietzel, T.; Elsasser, M.; Voon, C.P.; Candeo, A.; Seeliger, I.; Fricker, M.D.; Grefen, C.; Moller, I.M.; et al. ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. eLife 2017, 6, 26770. [Google Scholar] [CrossRef]
- Vlachonasios, K.; Poulios, S.; Mougiou, N. The histone acetyltransferase GCN5 and the associated coactivators ADA2: From evolution of the SAGA complex to the biological roles in plants. Plants 2021, 10, 308. [Google Scholar] [CrossRef]
- Hajheidari, M.; Koncz, C.; Eick, D. Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 2013, 18, 633–643. [Google Scholar] [CrossRef]
- Hu, Z.; Song, N.; Zheng, M.; Liu, X.; Liu, Z.; Xing, J.; Ma, J.; Guo, W.; Yao, Y.; Peng, H.; et al. Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J. 2015, 84, 1178–1191. [Google Scholar] [CrossRef]
- Duan, J.; Cai, W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE 2012, 7, e45117. [Google Scholar] [CrossRef]
- Ndong, C.; Danyluk, J.; Wilson, K.E.; Pocock, T.; Huner, N.P.A.; Sarhan, F. Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiol. 2002, 129, 1368–1381. [Google Scholar] [CrossRef]
- Tolleter, D.; Hincha, D.K.; Macherel, D. A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim. Biophys. Acta 2010, 1798, 1926–1933. [Google Scholar] [CrossRef]
- Ukaji, N.; Kuwabara, C.; Takezawa, D.; Arakawa, K.; Fujikawa, S. Cold acclimation-induced WAP27 localized in endoplasmic reticulum in cortical parenchyma cells of mulberry tree was homologous to group 3 late-embryogenesis abundant proteins. Plant Physiol. 2001, 126, 1588–1597. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heyen, B.J.; Alsheikh, M.K.; Smith, E.A.; Torvik, C.F.; Seals, D.F.; Randall, S.K. The calcium-binding activity of a vacuole-associated, dehydrin-like protein is regulated by phosphorylation. Plant Physiol. 2002, 130, 675–687. [Google Scholar] [CrossRef]
- Goyal, K.; Pinelli, C.; Maslen, S.L.; Rastogi, R.K.; Stephens, E.; Tunnacliffe, A. Dehydration-regulated processing of late embryogenesis abundant protein in a desiccation-tolerant nematode. FEBS Lett. 2005, 57, 4093–4098. [Google Scholar] [CrossRef] [PubMed]
- Danyluk, J.; Perron, A.; Houde, M.; Limin, A.; Fowler, B.; Benhamou, N.; Sarhan, F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 1998, 10, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.S.; Reddy, G.M.; Pandey, P.; Chandrasekhar, K.; Reddy, M.K. Cloning and molecular characterization of a gene encoding late embryogenesis abundant protein from Pennisetum glaucum: Protection against abiotic stresses. Mol. Biol. Rep. 2012, 39, 7163–7174. [Google Scholar] [CrossRef]
- Borrell, A.; Cutanda, M.C.; Lumbreras, V.; Pujal, J.; Goday, A.; Culiáñez-Macià, F.A.; Pagès, M. Arabidopsis thaliana atrab28: A nuclear targeted protein related to germination and toxic cation tolerance. Plant Mol. Biol. 2002, 50, 249–259. [Google Scholar] [CrossRef]
- Riera, M.; Figueras, M.; López, C.; Goday, A.; Pagès, M. Protein kinase CK2 modulates developmental functions of the abscisic acid responsive protein Rab17 from maize. Proc. Natl. Acad. Sci. USA 2004, 101, 9879–9884. [Google Scholar] [CrossRef] [PubMed]
- Goday, A.; Jensen, A.B.; Culiáñez-Macià, F.A.; Mar Albà, M.; Figueras, M.; Serratosa, J.; Torrent, M.; Pagès, M. The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell 1994, 6, 351–360. [Google Scholar]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar] [CrossRef] [PubMed]
- Tolleter, D.; Jaquinod, M.; Mangavel, C.; Passirani, C.; Saulnier, P.; Manon, S.; Teyssier, E.; Payet, N.; Avelange-Macherel, M.H.; Macherel, D. Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell 2007, 19, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Côté, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 41, D1063–D1069. [Google Scholar] [CrossRef] [PubMed]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. jPOSTTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 2017, 45, D1107–D1111. [Google Scholar] [CrossRef] [PubMed]

| No | Accession Number | Description | Matched Peptides | Ratio |
|---|---|---|---|---|
| Increased | ||||
| 1 | I1L3V3 | Root hair initiation protein root hairless | 3 | 100 |
| 2 | I1LDU4 | RNA polymerases II | 2 | 100 |
| 3 | C6TGY7 | Proliferating cell nuclear antigen | 3 | 29.341 |
| 4 | I1LHP2 | Tyrosyl-tRNA synthetase/Nucleotidylyl transferase | 2 | 5.495 |
| 5 | I1JJS2 | WD repeats region domain-containing protein | 3 | 5.458 |
| 6 | C6TAF1 | Nucleolar essential protein-like protein | 2 | 5.391 |
| 7 | I1JHW9 | CBF domain-containing protein | 10 | 5.143 |
| 8 | C6TGI7 | Leucine-rich repeat (LRR) family protein | 2 | 5.688 |
| Decreased | ||||
| 1 | I1LP68 | Bet v 1 domain-containing protein | 2 | 0.174 |
| 2 | K7LQ69 | Poly (ADP-ribose) polymerase | 4 | 0.155 |
| 3 | Q9SWB4 | Protein ADP-ribosyltransferase PARP3 | 3 | 0.126 |
| 4 | Q9XET0 | Seed maturation protein PM30 | 3 | 0.123 |
| 5 | I1L957 | Late embryogenesis abundant protein, | 6 | 0.051 |
| 6 | I1JFX0 | Usp domain-containing protein | 7 | 0.022 |
| 7 | I1JFX0 | Usp domain-containing protein | 7 | 0.022 |
| 8 | I1M3M9 | Late embryogenesis abundant protein | 11 | 0.010 |
| 9 | I1L849 | Late embryogenesis abundant protein D-34 | 10 | 0.010 |
| 10 | I1LE41 | Seed maturation protein | 7 | 0.010 |
| 11 | C6SWV3 | Histone acetyltransferase | 4 | 0.010 |
| 12 | I1MDN4 | Importin N-terminal domain-containing protein | 2 | 0.010 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murashita, Y.; Nishiuchi, T.; Rehman, S.U.; Komatsu, S. Subcellular Proteomics to Understand Promotive Effect of Plant-Derived Smoke Solution on Soybean Root. Proteomes 2021, 9, 39. https://doi.org/10.3390/proteomes9040039
Murashita Y, Nishiuchi T, Rehman SU, Komatsu S. Subcellular Proteomics to Understand Promotive Effect of Plant-Derived Smoke Solution on Soybean Root. Proteomes. 2021; 9(4):39. https://doi.org/10.3390/proteomes9040039
Chicago/Turabian StyleMurashita, Yusuke, Takumi Nishiuchi, Shafiq Ur Rehman, and Setsuko Komatsu. 2021. "Subcellular Proteomics to Understand Promotive Effect of Plant-Derived Smoke Solution on Soybean Root" Proteomes 9, no. 4: 39. https://doi.org/10.3390/proteomes9040039
APA StyleMurashita, Y., Nishiuchi, T., Rehman, S. U., & Komatsu, S. (2021). Subcellular Proteomics to Understand Promotive Effect of Plant-Derived Smoke Solution on Soybean Root. Proteomes, 9(4), 39. https://doi.org/10.3390/proteomes9040039







