Heat Stress Triggers Differential Protein Accumulation in the Extracellular Matrix of Sorghum Cell Suspension Cultures
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Heat-Stress Treatment
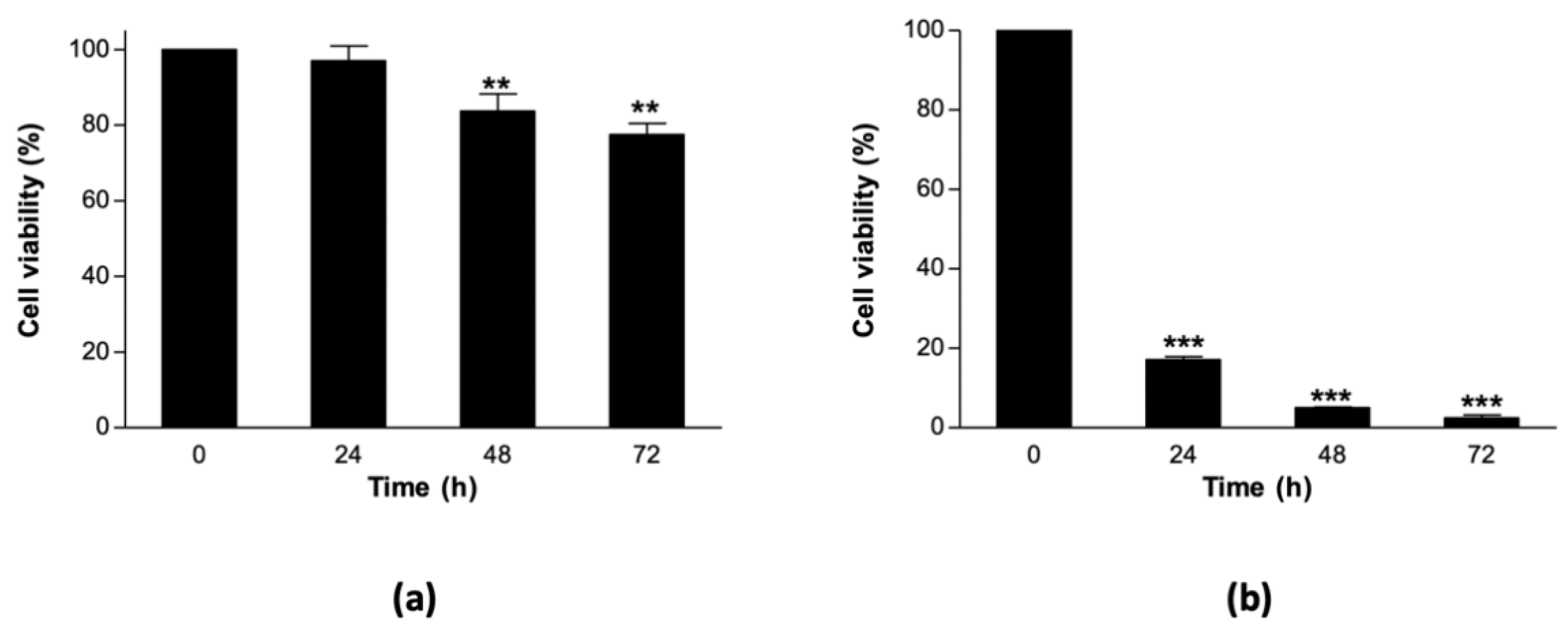
2.2. Cell Viability Estimations Using the MTT Assay
2.3. Protein Extraction, iTRAQ Labelling and Cleaning-Up
2.4. LC-MS/MS and Mass Spectra Data Analyses
2.5. Bioinformatic Analyses
2.6. Gene Expression Analysis
3. Results
3.1. Sorghum Cell Suspension Cultures Are Relatively Thermostable at 40 °C
3.2. Heat Stress Upregulates Expression of Sorghum HSP70 and HSP90 Genes
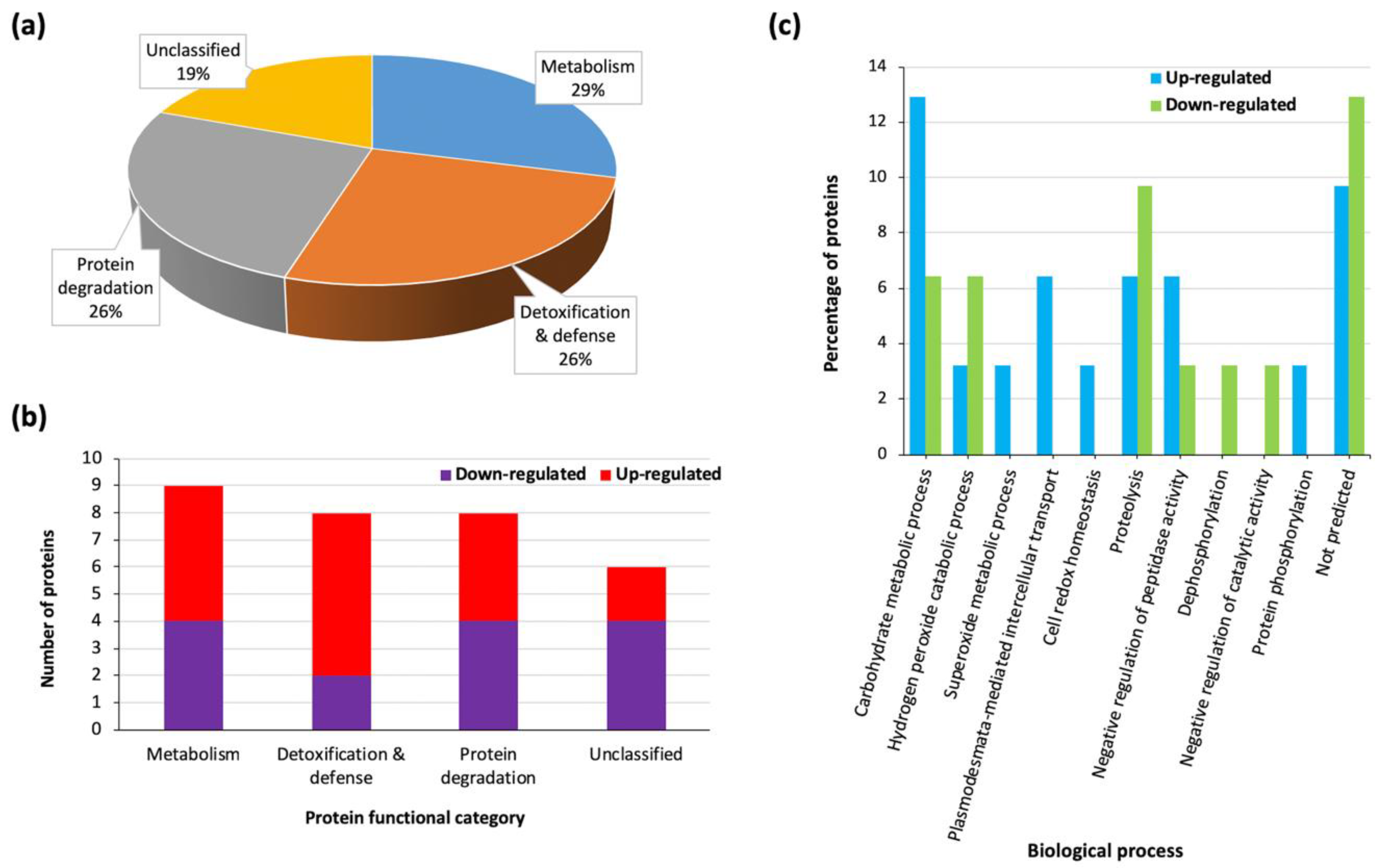
3.3. Heat Stress Triggers Differential Protein Accumulation in the Sorghum Extracellular Matrix (ECM)
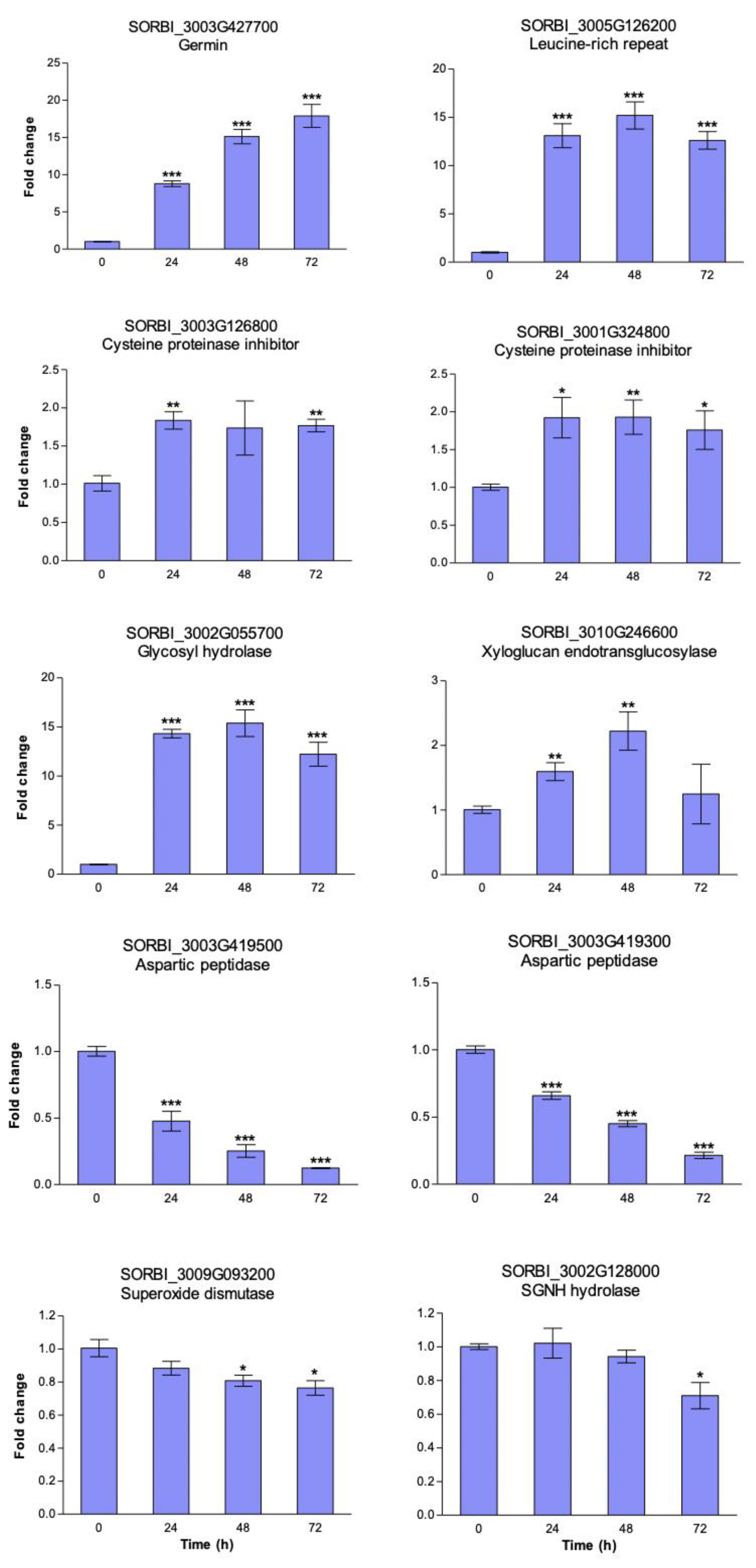
3.4. Heat Induced Gene Expression Patterns in Sorghum Cell Suspension Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hopkins, W.G.; Hüner, N.P.A. Introduction to Plant Physiology, 4th ed.; John Wiley & Sons: New Jersey, NJ, USA, 2009. [Google Scholar]
- Hatfield, J.L.; Boote, K.J.; Kimball, B.A.; Ziska, L.H.; Izaurralde, R.C.; Ort, D.R.; Thomson, A.M.; Wolfe, D. Climate impacts on agriculture: Implications for crop production. Agron. J. 2011, 103, 351–370. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Hughes, M.A.; Dunn, M.A. The effect of temperature on plant growth and development. Biotechnol. Genet. Eng. Rev. 1990, 8, 161–188. [Google Scholar] [CrossRef]
- Levitt, J. Responses of Plants to Environmental Stresses: Chilling, Freezing, and High Temperature Stress, 2nd ed.; Academic Press: New York, NY, USA, 1980; Volume 1. [Google Scholar]
- Tiaz, L.; Zeiger, E. Plant Physiology; Sinauer Associates, Inc.: Sunderland, MA, USA, 2010. [Google Scholar]
- Neilson, K.A.; Gammulla, C.G.; Mirzaei, M.; Imin, N.; Haynes, P.A. Proteomic analysis of temperature stress in plants. Proteomics 2010, 10, 828–845. [Google Scholar] [CrossRef]
- Nelson, G.C.; Rosegrant, M.; Koo, J.; Robertson, R.; Sulser, T.; Zhu, T.; Msangi, S.; Ringler, C.; Palazzo, A.; Batka, M.; et al. Climate Change Impact on Agriculture and Costs of Adapation; International Food Policy Research Institure: Washington, DC, USA, 2009; p. 32. [Google Scholar]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Hale, M.G.; Orcutt, D.M.; Thompson, L.K. The Physiology of Plants Under Stress; Wiley: New York, NY, USA, 1987; p. 206. [Google Scholar]
- Schoffl, F.; Prandl, R.; Reindl, A. Regulation of the heat-shock response. Plant Physiol. 1998, 117, 1135–1141. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Sachs, M.M.; Ho, T.-H.D. Alteration of gene expression during environmental stress in plants. Ann. Rev. Plant Physiol. 1986, 37, 363–376. [Google Scholar] [CrossRef]
- Howarth, C.J. Heat shock proteins in sorghum and pearl millet; ethanol, sodium arsenite, sodium malonate and the development of thermotolerance. J. Exp. Bot. 1990, 41, 877–883. [Google Scholar] [CrossRef]
- Vierling, E. The roles of heat shock proteins in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed]
- Nievola, C.C.; Carvalho, C.P.; Carvalho, V.; Rodrigues, E. Rapid responses of plants to temperature changes. Temperature 2017, 4, 371–405. [Google Scholar] [CrossRef]
- Al-Whaibi, M.M. Plant heat-shock proteins: A mini review. J. King Saud Univ. Sci. 2011, 23, 139–150. [Google Scholar] [CrossRef]
- Wahid, A. Physiological implications of metabolite biosynthesis for net assimilation and heat-stress tolerance of sugarcane (Saccharum officinarum) sprouts. J. Plant Res. 2007, 120, 219–228. [Google Scholar] [CrossRef]
- Ruelland, E.; Zachowski, A. How plants sense temperature. Environ. Exp. Bot. 2010, 69, 225–232. [Google Scholar] [CrossRef]
- Mittler, R.; Finka, A.; Goloubinoff, P. How do plants feel the heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Janni, M.; Gulli, M.; Maestri, E.; Marmiroli, M.; Valliyodan, B.; Nguyen, H.T.; Marmiroli, N. Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. J. Exp. Bot. 2020, 71, 3780–3802. [Google Scholar] [CrossRef]
- Doggett, H. Sorghum, 2nd. ed.; Longman Scientific & Technical: New York, NY, USA, 1988; p. 512. [Google Scholar]
- Rosenow, D.T.; Quisenberry, J.E.; Wendt, C.W.; Clark, L.E. Drought tolerant sorghum and cotton germplasm. Agr. Water Manag. 1983, 7, 207–222. [Google Scholar] [CrossRef]
- Kimber, C.T.; Dahlberg, J.A.; Kresovich, S. The gene pool of Sorghum bicolor and its improvement. In Genomics of the Saccharinae; Paterson, A.H., Ed.; Springer: New York, NY, USA, 2013; Volume 11, pp. 23–41. [Google Scholar]
- Motlhaodi, T.; Geleta, M.; Chite, S.; Fatih, M.; Ortiz, R.; Bryngelsson, T. Genetic diversity in sorghum (Sorghum bicolor (L.) Moench) germplasm from Southern Africa as revealed by microsatellite markers and agro-morphological traits. Gen. Resour. Crop Evol. 2017, 64, 599–610. [Google Scholar] [CrossRef]
- Goche, T.; Shargie, N.G.; Cummins, I.; Brown, A.P.; Chivasa, S.; Ngara, R. Comparative physiological and root proteome analyses of two sorghum varieties responding to water limitation. Sci. Rep. 2020, 10, 11835. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [PubMed]
- Paterson, A.H. The sorghum genome sequence: A core resource for Saccharinae genomics. In Genomics of the Saccharinae; Paterson, A.H., Ed.; Springer: New York, NY, USA, 2013; Volume 11, pp. 105–117. [Google Scholar]
- Alexandersson, E.; Ali, A.; Resjo, S.; Andreasson, E. Plant secretome proteomics. Front Plant Sci. 2013, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M. Structure and function of plant cell wall proteins. Plant Cell 1993, 5, 9–23. [Google Scholar] [PubMed]
- Hoson, T. Apoplast as the site of response to environmental signals. J. Plant Res. 1998, 111, 167–177. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Jwa, N.S.; Lebrun, M.H.; Job, D.; Rakwal, R. Plant secretome: Unlocking secrets of the secreted proteins. Proteomics 2010, 10, 799–827. [Google Scholar] [CrossRef]
- Ramulifho, E.; Goche, T.; Van As, J.; Tsilo, T.J.; Chivasa, S.; Ngara, R. Establishment and characterization of callus and cell suspension cultures of selected Sorghum bicolor (L.) Moench varieties: A resource for gene discovery in plant stress biology. Agronomy 2019, 9, 218. [Google Scholar] [CrossRef]
- Satish, L.; Shilpha, J.; Pandian, S.; Rency, A.S.; Rathinapriya, P.; Ceasar, S.A.; Largia, M.J.; Kumar, A.A.; Ramesh, M. Analysis of genetic variation in sorghum (Sorghum bicolor (L.) Moench) genotypes with various agronomical traits using SPAR methods. Gene 2016, 576, 581–585. [Google Scholar] [CrossRef]
- Ngara, R.; Ramulifho, E.; Movahedi, M.; Shargie, N.G.; Brown, A.P.; Chivasa, S. Identifying differentially expressed proteins in sorghum cell cultures exposed to osmotic stress. Sci. Rep. 2018, 8, 8671. [Google Scholar] [CrossRef]
- May, M.J.; Leaver, C.J. Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 1993, 103, 621–627. [Google Scholar] [CrossRef]
- Ngara, R. A Proteomic Analysis of Drought and Salt Stress Responsive Proteins of Different Sorghum Varieties. Ph.D. Thesis, University of the Western Cape, Cape Town, South Africa, 2009. [Google Scholar]
- Ngara, R.; Ndimba, B.K. Mapping and characterisation of the sorghum cell suspension culture secretome. Afr. J. Biotechnol. 2011, 10, 253–266. [Google Scholar]
- Smith, S.J.; Kroon, J.T.; Simon, W.J.; Slabas, A.R.; Chivasa, S. A novel function for Arabidopsis CYCLASE1 in programmed cell death revealed by isobaric tags for relative and absolute quantitation (iTRAQ) analysis of extracellular matrix proteins. Mol. Cell Proteom. 2015, 14, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- McDonald, J.H. Handbook of Biological Statistics, 3rd ed.; Sparky House Publishing: Baltimore, MD, USA, 2014; pp. 254–260. [Google Scholar]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- UniProt Consortium. Uniprot: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- Mulder, N.J.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Biswas, M.; Bradley, P.; Bork, P.; Bucher, P.; et al. Interpro: An integrated documentation resource for protein families, domains and functional sites. Brief. Bioinform. 2002, 3, 225–235. [Google Scholar] [PubMed]
- Li, J.; Fan, F.; Wang, L.; Zhan, Q.; Wu, P.; Du, J.; Yang, X.; Liu, Y. Cloning and expression analysis of cinnamoyl-CoA reductase (CCR) genes in sorghum. PeerJ. 2016, 4, e2005. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Lim, F.L.; Finkler, A.; Fromm, H.; Slabas, A.R.; Knight, M.R. Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genom. 2014, 15, 456. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-blast: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Pavli, O.I.; Ghikas, D.K.; Katsiotis, A.; Skaracis, G.N. Differential expression of heat shock protein genes in sorghum (Sorghum bicolor L.) genotypes under heat stress. Aust. J. Crop Sci. 2011, 5, 511–515. [Google Scholar]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Echevarría-Zomeño, S.; Fernández-Calvino, L.; Castro-Sanz, A.B.; López, J.A.; Vázquez, J.; Castellano, M.M. Dissecting the proteome dynamics of the early heat stress response leading to plant survival or death in Arabidopsis. Plant Cell Environ. 2016, 39, 1264–1278. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, J.; Wang, J.; Stierhof, Y.D.; Robinson, D.G.; Jiang, L. Unconventional protein secretion. Trends Plant Sci. 2012, 17, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Robinson, D.G.; Jiang, L. Unconventional protein secretion (UPS) pathways in plants. Curr. Opin. Cell Biol. 2014, 29, 107–115. [Google Scholar] [CrossRef]
- Robinson, D.G.; Ding, Y.; Jiang, L. Unconventional protein secretion in plants: A critical assessment. Protoplasma 2016, 253, 31–43. [Google Scholar] [CrossRef]
- Vincent, D.; Rafiqi, M.; Job, D. The multiple facets of plant-fungal interactions revealed through plant and fungal secretomics. Front Plant Sci. 2020, 10, 1626. [Google Scholar] [CrossRef]
- Mita, G.; Nocco, G.; Leuci, C.; Greco, V.; Rampino, P.; Perrotta, C. Secreted heat shock proteins in sunflower suspension cell cultures. Plant Cell Rep. 1997, 16, 792–796. [Google Scholar] [CrossRef]
- Chivasa, S.; Goodman, H.L. Stress-adaptive gene discovery by exploiting collective decision-making of decentralized plant response systems. New Phytol. 2020, 225, 2307–2313. [Google Scholar] [CrossRef]
- Ngara, R.; Ndimba, B.K. Model plant systems in salinity and drought stress proteomics studies: A perspective on Arabidopsis and Sorghum. Plant Biol. 2014, 16, 1029–1032. [Google Scholar]
- Tran, H.T.; Plaxton, W.C. Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics 2008, 8, 4317–4326. [Google Scholar] [CrossRef] [PubMed]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.; Cheong, H.; Park, O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef]
- Wu, M.T.; Wallner, S.J. Heat stress responses in cultured plant cells: Development and comparison of viability tests. Plant Physiol. 1983, 72, 817–820. [Google Scholar] [CrossRef] [PubMed]
- Moisyadi, S.; Harrington, H.M. Characterization of the heat shock response in cultured sugarcane cells: I. Physiology of the heat shock response and heat shock protein synthesis. Plant Physiol. 1989, 90, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Harrington, H.M.; Alm, D.M. Interaction of heat and salt shock in cultured tobacco cells. Plant Physiol. 1988, 88, 618–625. [Google Scholar] [CrossRef]
- Gammulla, C.G.; Pascovici, D.; Atwell, B.J.; Haynes, P.A. Differential metabolic response of cultured rice (Oryza sativa) cells exposed to high- and low-temperature stress. Proteomics 2010, 10, 3001–3019. [Google Scholar] [CrossRef]
- Qu, A.L.; Ding, Y.F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef]
- Wahid, A.; Close, T.J. Expression of dehydrins under heat stress and their relationship with water relations of sugarcane leaves. Biol. Plant 2007, 51, 104–109. [Google Scholar] [CrossRef]
- Cheng, F.Y.; Williamson, J.D. Is there leaderless protein secretion in plants? Plant Signal Behav. 2010, 5, 129–131. [Google Scholar] [CrossRef]
- Caliskan, M. Germin, an oxalate oxidase, has a function in many aspects of plant life. Turk. J. Biol. 2000, 24, 717–724. [Google Scholar]
- Lane, B.G.; Dunwell, J.M.; Ray, J.A.; Schmitt, M.R.; Cuming, A.C. Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 1993, 268, 12239–12242. [Google Scholar]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.; Sasaki, K.; Ito, H.; Ohashi, Y.; Matsui, H. A large family of class III plant peroxidases. Plant Cell Physiol. 2001, 42, 462–468. [Google Scholar] [CrossRef]
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol. 1993, 125, 27–58. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Wojtaszek, P. Oxidative burst: An early plant response to pathogen infection. Biochem. J. 1997, 322, 681–692. [Google Scholar] [CrossRef]
- Davies, G.; Henrissat, B. Structures and mechanisms of glycosyl hydrolases. Structure 1995, 3, 853–859. [Google Scholar] [CrossRef]
- Okazaki, Y.; Saito, K. Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 2014, 79, 584–596. [Google Scholar] [CrossRef]
- Ding, L.-N.; Li, M.; Wang, W.-J.; Cao, J.; Wang, Z.; Zhu, K.-M.; Yang, Y.-H.; Li, Y.-L.; Tan, X.-L. Advances in plant GDSL lipases: From sequence to functional mechanisms. Acta Physiol. Plant 2019, 41, 151. [Google Scholar] [CrossRef]
- Ramulifho, E. Proteomic Mapping of Sorghum Bicolor (L.) Moench Cell Suspension Culture Secretome and Identification of its Drought Stress Responsive Proteins. Master’s Thesis, University of the Free State, Bloemfontein, South Africa, 2017. [Google Scholar]
- Kidrič, M.; Kos, J.; Sabotič, J. Proteases and their endogenous inhibitors in the plant response to abiotic stress. Bot. SERB 2014, 38, 139–158. [Google Scholar]
- Cooke, R.J. Protein degradation in plants. Sci. Prog. Oxf. 1981, 67, 461–480. [Google Scholar]
- Vierstra, R.D. Proteolysis in plants: Mechanisms and functions. Plant Mol. Biol. 1996, 32, 275–302. [Google Scholar] [CrossRef] [PubMed]
- Franssen, H.J.; Bisseling, T. Peptide signaling in plants. Proc. Natl Acad. Sci. USA 2001, 98, 12855–12856. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, S.; Fernandez, A.; Hilson, P.; Beeckman, T. Signaling peptides in plants. Cell Dev. Biol. 2014, 3. Available online: https://biblio.ugent.be/publication/5779157 (accessed on 27 August 2020). [CrossRef]
- Chen, Y.L.; Fan, K.T.; Hung, S.C.; Chen, Y.R. The role of peptides cleaved from protein precursors in eliciting plant stress reactions. New Phytol. 2020, 225, 2267–2282. [Google Scholar] [CrossRef]
- De Sousa Abreu, R.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Maier, T.; Guell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef]

| Prot No. a | Accession b | Protein Name | Ratio c | SD d | p-Value e | Signal Peptide f | Family Name g |
|---|---|---|---|---|---|---|---|
| Metabolism | |||||||
| 11 | A0A1B6QCB0 * | Alpha-amylase OS = Sorghum bicolor GN = SORBI_3002G190500 | 2.84 | 0.22 | 4.89 10−5 | + | Alpha-amylase, plant |
| 36 | C5XB39 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3002G055700 | 2.41 | 0.20 | 3.80 10−4 | + | Glycosyl hydrolase superfamily |
| 92 | C5XD22 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3002G356800 | −2.32 | 0.05 | 2.54 10−3 | + | Lipase_GDSL domain |
| 106 | A0A1W0VY92 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3003G205900 | −2.31 | 0.08 | 4.37 10−4 | + | Lipase_GDSL domain |
| 113 | A0A1B6Q537 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3003G244600 | 2.02 | 0.47 | 5.86 10−3 | - | Glycosyl hydrolase, family 18 |
| 125 | C5WSY5 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3001G014700 | −2.51 | 0.07 | 4.89 10−3 | + | Glycoside hydrolase, family 17 |
| 127 | C5Z8T4 * | Xyloglucan endotransglucosylase/hydrolase OS = Sorghum bicolor GN = SORBI_3010G246600 | 2.47 | 0.30 | 4.69 10−4 | + | Xyloglucan endotransglucosylase/hydrolase |
| 250 | C5 × 578 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3002G128000 | 2.31 | 0.33 | 1.32 10−2 | + | SGNH hydrolase superfamily |
| 279 | A0A1B6QEM0 | Xyloglucan endotransglucosylase/hydrolase OS = Sorghum bicolor GN = SORBI_3002G324100 | −2.50 | 0.10 | 2.61 10 −2 | + | Xyloglucan endotransglucosylase/hydrolase |
| Detoxification & Defence | |||||||
| 15 | C5X5K6 | Peroxidase OS = Sorghum bicolor GN = SORBI_3002G416700 | −2.17 | 0.02 | 1.34 10−2 | + | Plant peroxidase |
| 50 | C5X040 * | Peroxidase OS = Sorghum bicolor GN = SORBI_3001G080300 | 2.06 | 0.27 | 5.18 10−4 | + | Plant peroxidase |
| 58 | C5YVR0 * | Superoxide dismutase OS = Sorghum bicolor GN = SORBI_3009G093200 | 2.35 | 0.38 | 8.83 10−4 | - | Manganese/iron superoxide dismutase |
| 60 | C5XHF1 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3003G136200 | 2.08 | 0.24 | 2.38 10−4 | + | Germin |
| 108 | C5XL59 * | Peroxidase OS = Sorghum bicolor GN = SORBI_3003G024700 | −3.23 | 0.07 | 2.70 10−5 | - | Plant peroxidase |
| 137 | C5XHX2 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3003G427700 | 2.85 | 0.43 | 4.85 10−4 | + | Germin |
| 154 | C5XJT8 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3003G156400 | 2.17 | 0.29 | 4.45 10−4 | + | Thioredoxin-like superfamily |
| 166 | C5YD83 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3006G031200 | 2.09 | 0.45 | 7.25 10−3 | + | Thioredoxin-like superfamily |
| Protein Degradation | |||||||
| 47 | C5XHP9 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3003G419500 | 2.18 | 0.13 | 5.94 10−5 | + | Aspartic peptidase A1 family |
| 49 | C5XHP7 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3003G419300 | 2.74 | 0.20 | 3.77 10−5 | - | Aspartic peptidase A1 family |
| 74 | C5WVG9 * | Cysteine proteinase inhibitor OS = Sorghum bicolor GN = SORBI_3001G324800 | 2.19 | 0.17 | 8.33 10−5 | + | Cysteine proteinase inhibitor |
| 153 | C5Z1X3 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3010G268400 | −2.10 | 0.03 | 5.84 10−3 | + | Aspartic peptidase A1 family |
| 215 | C5XAQ7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3002G326100 | −2.05 | 0.08 | 4.50 10−2 | + | Aspartic peptidase A1 family |
| 219 | A0A1B6Q242 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3003G085300 | −2.00 | 0.03 | 5.30 10−3 | + | Bowman-Birk type proteinase inhibitor |
| 225 | C5XGM0 | Cysteine proteinase inhibitor OS = Sorghum bicolor GN = SORBI_3003G126800 | 2.16 | 0.42 | 9.39 10−3 | + | Cysteine proteinase inhibitor |
| 259 | A0A1B6Q6G6 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3003G314800 | −2.00 | 0.11 | 2.50 10−2 | + | Aspartic peptidase A1 family |
| Unclassified | |||||||
| 26 | A0A1Z5R915 * | Purple acid phosphatase OS = Sorghum bicolor GN = SORBI_3007G091100 | −4.18 | 0.04 | 2.42 10−4 | - | Purple acid phosphatase-like |
| 33 | C5XBP7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3002G343600 | −2.55 | 0.06 | 3.77 10−3 | + | Leucine-rich repeat domain superfamily |
| 34 | A0A1B6PLT5 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3006G133000 | −2.83 | 0.05 | 5.29 10−4 | + | Domain of unknown function DUF642 |
| 101 | C5Y2R8 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3005G126200 | 2.48 | 0.37 | 6.26 10−4 | + | Leucine-rich repeat domain superfamily |
| 268 | C5X4M5 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3002G255000 | −6.40 | 0.10 | 8.97 10−3 | + | Not predicted |
| 273 | C5WPH2 * | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_3001G130400 | 2.34 | 0.35 | 6.69 10−4 | + | Protein of unknown function DUF538 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngcala, M.G.; Goche, T.; Brown, A.P.; Chivasa, S.; Ngara, R. Heat Stress Triggers Differential Protein Accumulation in the Extracellular Matrix of Sorghum Cell Suspension Cultures. Proteomes 2020, 8, 29. https://doi.org/10.3390/proteomes8040029
Ngcala MG, Goche T, Brown AP, Chivasa S, Ngara R. Heat Stress Triggers Differential Protein Accumulation in the Extracellular Matrix of Sorghum Cell Suspension Cultures. Proteomes. 2020; 8(4):29. https://doi.org/10.3390/proteomes8040029
Chicago/Turabian StyleNgcala, Mamosa G., Tatenda Goche, Adrian P. Brown, Stephen Chivasa, and Rudo Ngara. 2020. "Heat Stress Triggers Differential Protein Accumulation in the Extracellular Matrix of Sorghum Cell Suspension Cultures" Proteomes 8, no. 4: 29. https://doi.org/10.3390/proteomes8040029
APA StyleNgcala, M. G., Goche, T., Brown, A. P., Chivasa, S., & Ngara, R. (2020). Heat Stress Triggers Differential Protein Accumulation in the Extracellular Matrix of Sorghum Cell Suspension Cultures. Proteomes, 8(4), 29. https://doi.org/10.3390/proteomes8040029







