Short-Chain Fatty Acids Modulate Metabolic Pathways and Membrane Lipids in Prevotella bryantii B14
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation in Medium M2-A for Proteome Analysis
2.2. Cultivation for Long-Chain Fatty Acid Determination
2.3. Glucose Based Medium M2-B
2.4. Short-Chain Fatty Acid Determination
2.5. Long-Chain Fatty Acid Determination
2.6. Protein Extraction
2.7. In Gel Protein Digestion
2.8. LC-ESI-MS/MS
2.9. Data Analysis for Proteomics
3. Results
3.1. Cultivation of P. bryantii B14 Using Single SCFAs
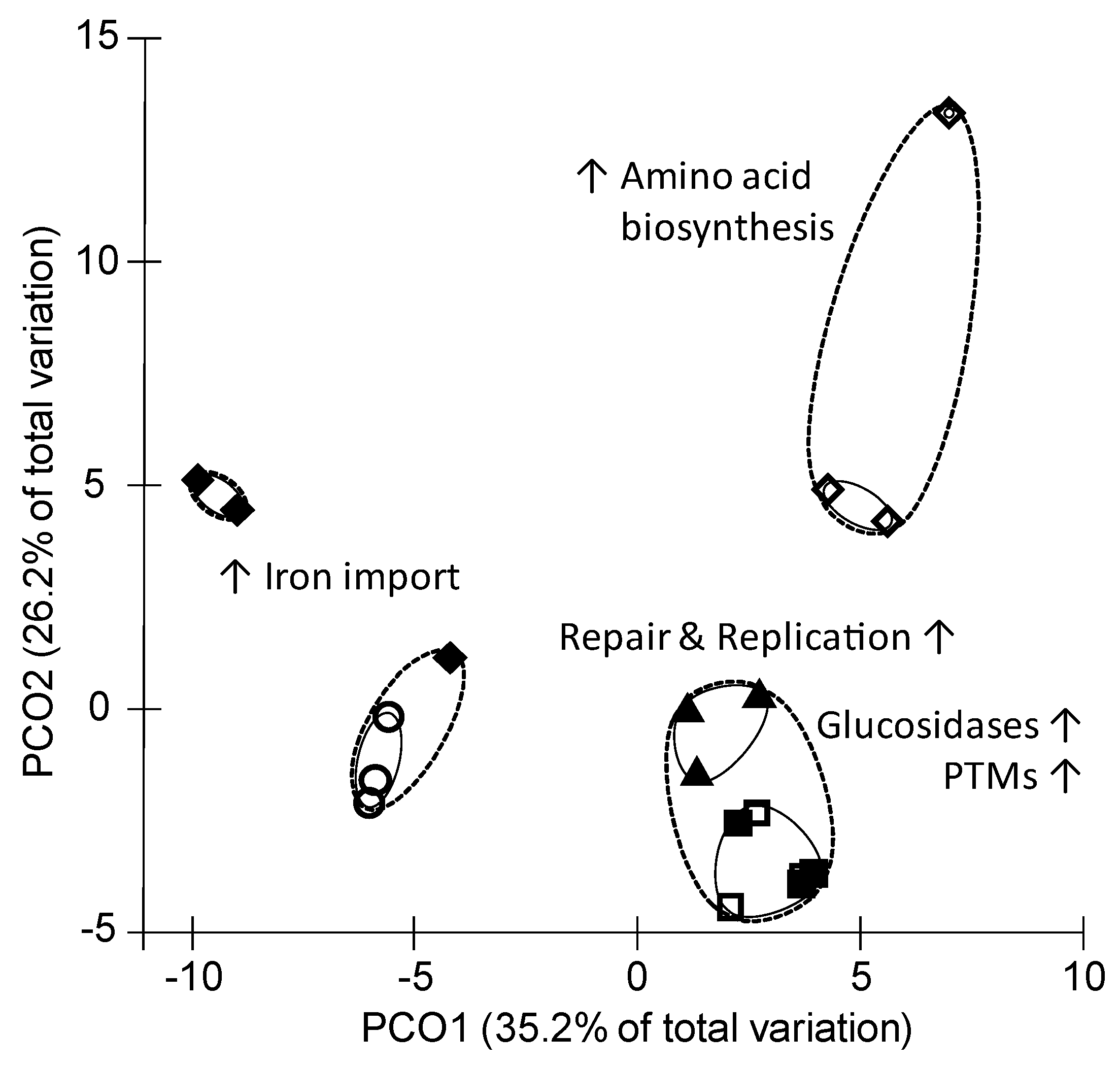
3.2. Proteome Inventory of P. bryantii B14
3.3. Effect of SCFA Exposure on Long-Chain Fatty Acids in Membranes from P. bryantii
3.4. Effect of Carbon Source on Growth of P. bryantii
4. Discussion
4.1. SCFAs Replace Rumen Fluid for Cultivation of P. bryantii B14
4.2. SCFA Influence the Abundance of Iron Transport Proteins
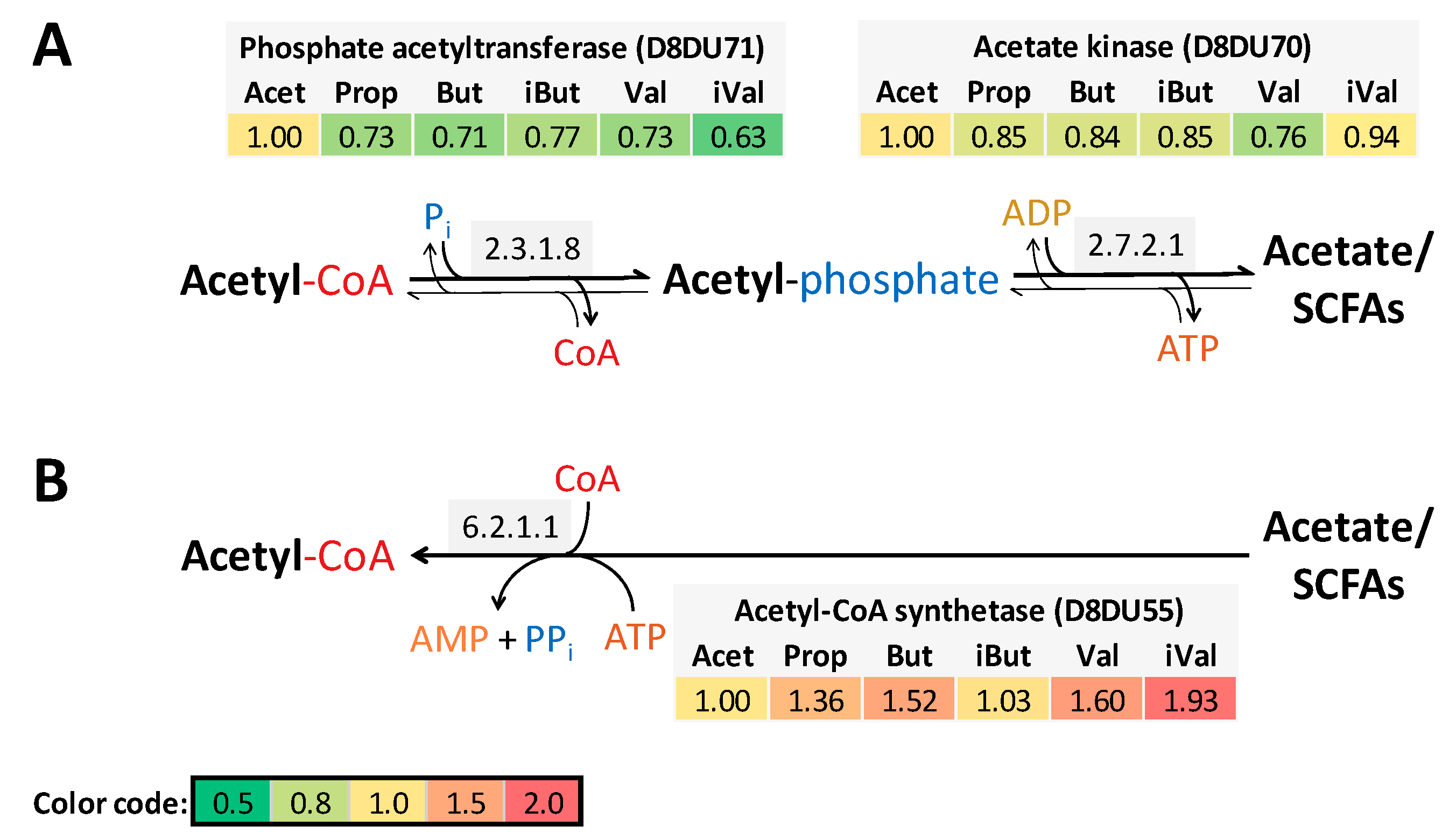
4.3. SCFAs are Incorporated into Long-Chain Fatty Acids of Lipid Membranes
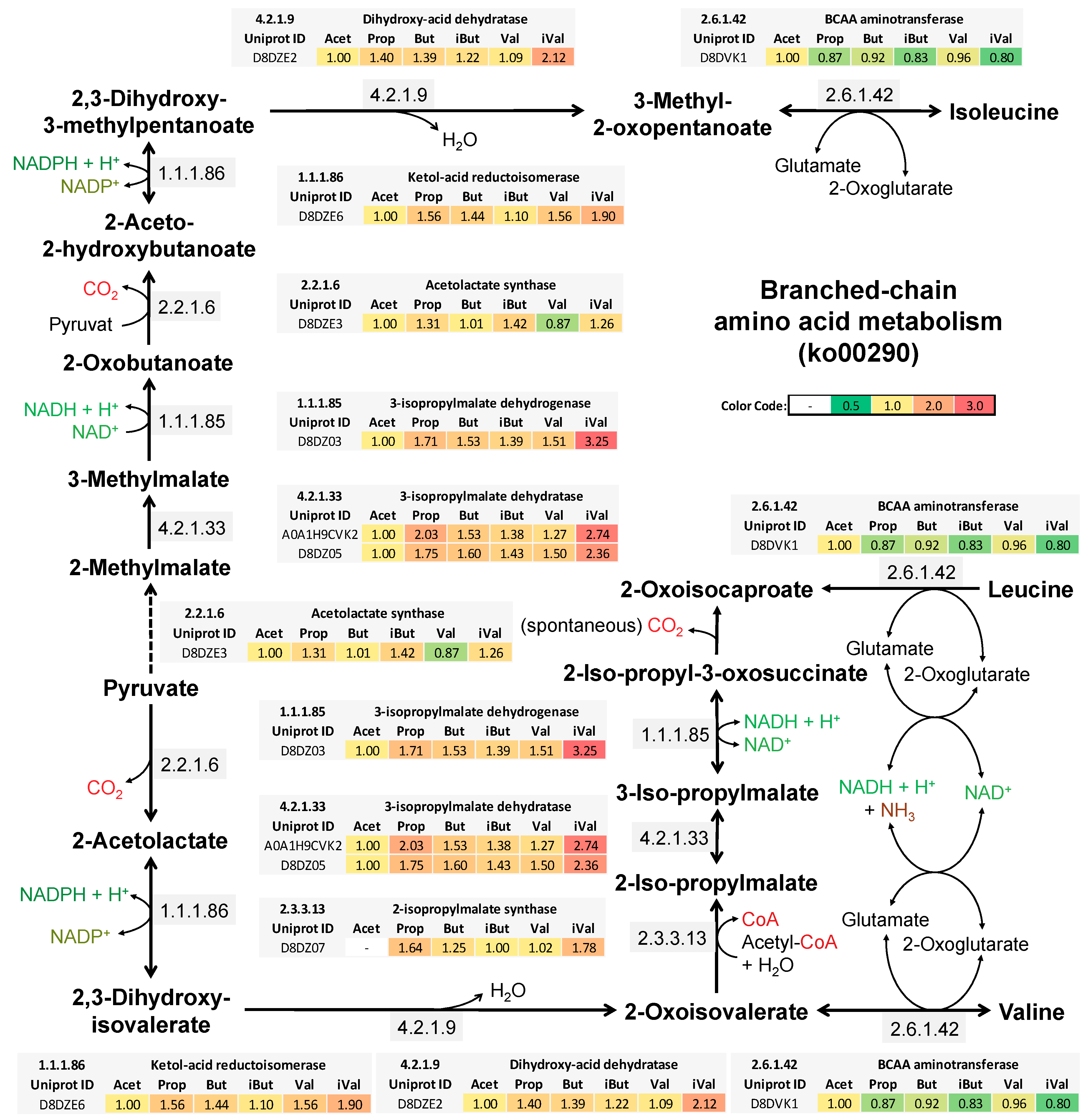
4.4. Isovaleric Acid Enhance Branched-Chain Amino Acid Synthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flint, H.J.; Bayer, E.A. Plant cell wall breakdown by anaerobic microorganisms from the mammalian digestive tract. Ann. N. Y. Acad. Sci. 2008, 1125, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Dehority, B. Characterization of several bovine rumen bacteria isolated with a xylan medium. J. Bacteriol. 1966, 91, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Hold, G.L.; Harmsen, H.J.; Stewart, C.S.; Flint, H.J. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 2141–2146. [Google Scholar] [PubMed]
- Dehority, B.; Scott, H.; Kowaluk, P. Volatile fatty acid requirements of cellulolytic rumen bacteria. J. Bacteriol. 1967, 94, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Torrungruang, K.; Jitpakdeebordin, S.; Charatkulangkun, O.; Gleebbua, Y. Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Treponema denticola/Prevotella intermedia co-infection are associated with severe periodontitis in a thai population. PLoS ONE 2015, 10, e0136646. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, G.R.; Wood, D.P.; Birchmeier, K.R. Detection and comparison of specific hemin binding by Porphyromonas gingivalis and Prevotella intermedia. J. Bacteriol. 1997, 179, 620–626. [Google Scholar] [CrossRef]
- Scott, H.; Dehority, B. Vitamin requirements of several cellulolytic rumen bacteria. J. Bacteriol. 1965, 89, 1169–1175. [Google Scholar] [CrossRef]
- Gibbons, R.J.; Macdonald, J.B. Hemin and vitamin k compounds as required factors for the cultivation of certain strains of Bacteroides melaninogenicus. J. Bacteriol. 1960, 80, 164–170. [Google Scholar] [CrossRef]
- Alvarez, H.; Santini, F.J.; Rearte, D.H.; Elizalde, J.C. Milk production and ruminal digestion in lactating dairy cows grazing temperate pastures and supplemented with dry cracked corn or high moisture corn. Anim. Feed Sci. Technol. 2001, 91, 183–195. [Google Scholar] [CrossRef]
- Byrne, C.; Chambers, E.; Morrison, D.; Frost, G. The role of short chain fatty acids in appetite regulation and energy homeostasis. Int. J. Obes. 2015, 39, 1331–1338. [Google Scholar] [CrossRef]
- Von Engelhardt, W.; Rönnau, K.; Rechkemmer, G.; Sakata, T. Absorption of short-chain fatty acids and their role in the hindgut of monogastric animals. Anim. Feed Sci. Technol. 1989, 23, 43–53. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 2017, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Matsuoka, H.; Hirooka, K. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 2007, 66, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Yamada, T. Pathways for amino acid metabolism by Prevotella intermedia and Prevotella nigrescens. Oral Microbiol. Immunol. 2000, 15, 96–102. [Google Scholar] [CrossRef]
- Bhatia, S.K.; Yang, Y.-H. Microbial production of volatile fatty acids: Current status and future perspectives. Rev. Environ. Sci. Bio/Technol. 2017, 16, 327–345. [Google Scholar] [CrossRef]
- Hobson, P.N. Rumen bacteria. In Methods in Microbiology; Norris, J.R., Ribbons, D.W., Eds.; Elsevier Ltd.: London, UK, 1969; pp. 133–149. [Google Scholar]
- Cotta, M.A.; Russell, J.B. Effect of peptides and amino acids on efficiency of rumen bacterial protein synthesis in continuous culture. J. Dairy Sci. 1982, 65, 226–234. [Google Scholar] [CrossRef]
- Wischer, G.; Boguhn, J.; Steingaß, H.; Schollenberger, M.; Hartung, K.; Rodehutscord, M. Effect of monensin on in vitro fermentation of silages and microbial protein synthesis. Arch. Anim. Nutr. 2013, 67, 219–234. [Google Scholar] [CrossRef]
- Paczia, N.; Nilgen, A.; Lehmann, T.; Gätgens, J.; Wiechert, W.; Noack, S. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Fact. 2012, 11, 122. [Google Scholar] [CrossRef]
- Vlaeminck, B.; Fievez, V.; Cabrita, A.; Fonseca, A.; Dewhurst, R. Factors affecting odd-and branched-chain fatty acids in milk: A review. Anim. Feed Sci. Technol. 2006, 131, 389–417. [Google Scholar] [CrossRef]
- Allison, M.J.; Bryant, M.; Katz, I.; Keeney, M. Metabolic function of branched-chain volatile fatty acids, growth factors for Ruminococci ii.: Biosynthesis of higher branched-chain fatty acids and aldehydes. J. Bacteriol. 1962, 83, 1084–1093. [Google Scholar] [CrossRef]
- Deusch, S.; Camarinha-Silva, A.; Conrad, J.; Beifuss, U.; Rodehutscord, M.; Seifert, J. A structural and functional elucidation of the rumen microbiome influenced by various diets and microenvironments. Front. Microbiol. 2017, 8, 1605. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Jehmlich, N.; Schmidt, F.; Hartwich, M.; Bergen, M.V.; Richnow, H.-H.; Vogt, C. Incorporation of carbon and nitrogen atoms into proteins measured by protein-based stable isotope probing (protein-sip). Rapid Commun. Mass Spectrom. 2008, 22, 2889–2897. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using stagetips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Godoy, L.M.F.D.; Li, G.; Macek, B.; Mortensen, P.; Pesch, R.; Makarov, A.; Lange, O.; Horning, S.; Mann, M. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteom. 2005, 4, 2010–2021. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T. 2016 update of the pride database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by EggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Hungate, R.E. The Rumen and Its Microbes; Academic Press: London, UK, 1966. [Google Scholar]
- Hackmann, T.J.; Ngugi, D.K.; Firkins, J.L.; Tao, J. Genomes of rumen bacteria encode atypical pathways for fermenting hexoses to short-chain fatty acids. Environ. Microbiol. 2017, 19, 4670–4683. [Google Scholar] [CrossRef]
- Conner, R.L.; Reilly, A.E. The effects of isovalerate supplementation on growth and fatty acid composition of Tetrahymena pyriformis w. Biochim. Biophys. Acta 1975, 398, 209–216. [Google Scholar] [CrossRef]
- Grundy, F.J.; Waters, D.A.; Allen, S.; Henkin, T. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J. Bacteriol. 1993, 175, 7348–7355. [Google Scholar] [CrossRef] [PubMed]
- Valgepea, K.; Adamberg, K.; Nahku, R.; Lahtvee, P.-J.; Arike, L.; Vilu, R. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Sys. Biol. 2010, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.C.; Ebersole, J.L. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000 2005, 38, 72–122. [Google Scholar] [CrossRef]
- Yu, F.; Anaya, C.; Lewis, J.P. Outer membrane proteome of Prevotella intermedia 17: Identification of thioredoxin and iron-repressible hemin uptake loci. J. Proteom. 2007, 7, 403–412. [Google Scholar] [CrossRef]
- Lindt, F.; Blum, J.W. Occurrence of iron deficiency in growing cattle. Zent. Vet. A 1994, 41, 237–246. [Google Scholar] [CrossRef]
- Herold, S.; Paton, J.C.; Srimanote, P.; Paton, A.W. Differential effects of short-chain fatty acids and iron on expression of iha in shiga-toxigenic Escherichia coli. Microbiology 2009, 155, 3554–3563. [Google Scholar] [CrossRef]
- Caldwell, D.; White, D.; Bryant, M.; Doetsch, R. Specificity of the heme requirement for growth of Bacteroides ruminicola. J. Bacteriol. 1965, 90, 1645–1654. [Google Scholar] [CrossRef]
- Leung, K.-P.; Folk, S.P. Effects of porphyrins and inorganic iron on the growth of Prevotella intermedia. FEMS Microbiol. Lett. 2002, 209, 15–21. [Google Scholar] [CrossRef]
- Smalley, J.W.; Silver, J.; Birss, A.J.; Withnall, R.; Titler, P.J. The haem pigment of the oral anaerobes Prevotella nigrescens and Prevotella intermedia is composed of iron (iii) protoporphyrin ix in the monomeric form. Microbiology 2003, 149, 1711–1718. [Google Scholar] [CrossRef]
- Henry, C.A.; Judy, M.; Dyer, B.; Wagner, M.; Matthews, J.L. Sensitivity of Porphyromonas and Prevotella species in liquid media to argon laser. Photochem. Photobiol. 1995, 61, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Macy, J.; Probst, I.; Gottschalk, G. Evidence for cytochrome involvement in fumarate reduction and adenosine 5’-triphosphate synthesis by bacteroides fragilis grown in the presence of hemin. J. Bacteriol. 1975, 123, 436–442. [Google Scholar] [CrossRef]
- Deusch, S.; Bok, E.; Schleicher, L.; Seifert, J.; Steuber, J. Occurrence and function of the Na+-translocating NADH: Quinone oxidoreductase in Prevotella spp. Microorganisms 2019, 7, 117. [Google Scholar] [CrossRef]
- Sperry, J.; Appleman, M.; Wilkins, T.D. Requirement of heme for growth of Bacteroides fragilis. Appl. Environ. Microbiol. 1977, 34, 386–390. [Google Scholar] [CrossRef]
- Granick, S.; Beale, S.I. Hemes, chlorophylls, and related compounds: Biosynthesis and metabolic regulation. Adv. Enzymol. Relat. Areas Mol. Biol. 1978, 46, 33–203. [Google Scholar] [PubMed]
- Minato, H.; Ishibashi, S.; Hamaoka, T. Cellular fatty acid and sugar composition of representative strains of rumen bacteria. J. Gen. Appl. Microbiol. 1988, 34, 303–319. [Google Scholar] [CrossRef]
- Logar, R.M.; Zorec, M.; Kopečný, J. Reliable identification of Prevotella and Butyrivibrio spp. From rumen by fatty acid methyl ester profiles. Folia Microbiol. 2001, 46, 57–59. [Google Scholar] [CrossRef]
- Sun, Y.; Wilkinson, B.J.; Standiford, T.J.; Akinbi, H.T.; O’Riordan, M.X. Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J. Bacteriol. 2012, 194, 5274–5284. [Google Scholar] [CrossRef]
- Mitchell, N.J.; Seaton, P.; Pokorny, A. Branched phospholipids render lipid vesicles more susceptible to membrane-active peptides. Biochim. Biophys. Acta 2016, 1858, 988–994. [Google Scholar] [CrossRef]
- Singh, V.K.; Hattangady, D.S.; Giotis, E.S.; Singh, A.K.; Chamberlain, N.R.; Stuart, M.K.; Wilkinson, B.J. Insertional inactivation of branched-chain α-keto acid dehydrogenase in Staphylococcus aureus leads to decreased branched-chain membrane fatty acid content and increased susceptibility to certain stresses. Appl. Environ. Microbiol. 2008, 74, 5882–5890. [Google Scholar] [CrossRef]
- Kaneda, T. Iso-and anteiso-fatty acids in bacteria: Biosynthesis, function, and taxonomic significance. Microbiol. Rev. 1991, 55, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Sirobhushanam, S. Alternative Pathway for Provision of Acyl CoA Precursors for Fatty Acid Biosynthesis: Purification and Kinetic Characterization of Phosphotransbutyrylase and Butyrate Kinase from Listeria Monocytogenes. Ph.D. Thesis, Illinois State University, Normal, IL, USA, 2016. [Google Scholar]
- Reiser, K.; Davis, M.A.; Hynes, M.J. Aspergillus nidulans contains six possible fatty acyl-CoA synthetases with FaaB being the major synthetase for fatty acid degradation. Arch. Microbiol. 2010, 192, 373–382. [Google Scholar] [CrossRef] [PubMed]
- DiRusso, C.C.; Black, P.N. Bacterial long chain fatty acid transport: Gateway to a fatty acid-responsive signaling system. J. Biol. Chem. 2004, 279, 49563–49566. [Google Scholar] [CrossRef]
- El-Shazly, K. Degradation of protein in the rumen of the sheep. 2. The action of rumen microorganisms on amino acids. Biochem. J. 1952, 51, 647–653. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pasolli, E.; Tett, A.; Tarallo, S.; Naccarati, A.; De Angelis, M.; Neviani, E.; Cocolin, L.; Gobbetti, M.; Segata, N. Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe 2019, 25, 444.e3–453.e3. [Google Scholar] [CrossRef] [PubMed]
- Glasemacher, J.; Bock, A.K.; Schmid, R.; Schönheit, P. Purification and properties of acetyl-CoA synthetase (ADP-forming), an archaeal enzyme of acetate formation and ATP synthesis, from the hyperthermophile Pyrococcus furiosus. Eur. J. Biochem. 1997, 244, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.A.; Joshi, M.; Jeoung, N.H. Mechanisms responsible for regulation of branched-chain amino acid catabolism. Biochem. Biophys. Res. Commun. 2004, 313, 391–396. [Google Scholar] [CrossRef]
- Allison, M.J.; Baetz, A.L.; Wiegel, J. Alternative pathways for biosynthesis of leucine and other amino acids in Bacteroides ruminicola and Bacteroides fragilis. Appl. Environ. Microbiol. 1984, 48, 1111–1117. [Google Scholar] [CrossRef]
- Commichau, F.M.; Forchhammer, K.; Stülke, J. Regulatory links between carbon and nitrogen metabolism. Curr. Opin. Microbiol. 2006, 9, 167–172. [Google Scholar] [CrossRef]
- Den Hengst, C.D.; Groeneveld, M.; Kuipers, O.P.; Kok, J. Identification and functional characterization of the Lactococcus lactis CodY-regulated branched-chain amino acid permease BcaP (CtrA). J. Bacteriol. 2006, 188, 3280–3289. [Google Scholar] [CrossRef]
- Atasoglu, C.; Valdés, C.; Walker, N.D.; Newbold, C.J.; Wallace, R.J. De novo synthesis of amino acids by the ruminal bacteria Prevotella bryantii B14, Selenomonas ruminantium HD4, and Streptococcus bovis ES1. Appl. Environ. Microbiol. 1998, 64, 2836–2843. [Google Scholar] [CrossRef] [PubMed]


| Cultivation Condition | Mean ± SD of OD600 | ∆pH | Production/Consumption (mM) of ____ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C2 | C3 | iC4 | C4 | iC5 | C5 | |||||
| 8 h incubation | Acet | 1.44 | ±0.19 | 1.34 | 9.55 | 0.00 | 0.00 | 0.00 | 0.06 | 0.00 |
| Prop | 1.50 | ±0.17 | 1.32 | 10.03 | 0.70 | 0.00 | −0.04 | 0.08 | 0.07 | |
| But | 1.43 | ±0.16 | 1.33 | 10.14 | −0.05 | 0.00 | −0.10 | 0.09 | 0.00 | |
| iBut | 1.23 | ±0.07 | 0.85 | 6.06 | 0.00 | −0.49 | 0.00 | 0.08 | 0.00 | |
| Val | 1.73 | ±0.04 | 1.47 | 11.77 | 0.00 | 0.00 | −0.03 | 0.16 | −0.69 | |
| iVal | 1.16 | ±0.09 | 0.56 | 6.21 | 0.00 | 0.00 | 0.00 | −0.24 | 0.00 | |
| Cultivation Condition | Mean ± SD of OD600 | ∆pH | Production/Consumption (mM) of ____ | |||||||
| C2 | C3 | iC4 | C4 | iC5 | C5 | |||||
| 24 h incubation | Acet | 1.67 | ±0.04 | 1.38 | 13.94 | 0.00 | 0.00 | 0.00 | 0.08 | 0.00 |
| Prop | 1.64 | ±0.26 | 1.57 | 16.05 | 0.27 | 0.00 | −0.04 | 0.07 | 0.06 | |
| But | 1.66 | ±0.15 | 1.61 | 16.79 | −0.10 | 0.00 | −0.43 | 0.10 | 0.00 | |
| iBut | 1.64 | ±0.06 | 1.49 | 14.02 | 0.00 | −0.87 | 0.00 | 0.09 | 0.00 | |
| Val | 1.77 | ±0.13 | 1.62 | 15.59 | 0.00 | 0.00 | −0.01 | 0.17 | −0.89 | |
| iVal | 1.54 | ±0.34 | 1.05 | 10.82 | 0.00 | 0.00 | 0.00 | −0.46 | 0.00 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trautmann, A.; Schleicher, L.; Deusch, S.; Gätgens, J.; Steuber, J.; Seifert, J. Short-Chain Fatty Acids Modulate Metabolic Pathways and Membrane Lipids in Prevotella bryantii B14. Proteomes 2020, 8, 28. https://doi.org/10.3390/proteomes8040028
Trautmann A, Schleicher L, Deusch S, Gätgens J, Steuber J, Seifert J. Short-Chain Fatty Acids Modulate Metabolic Pathways and Membrane Lipids in Prevotella bryantii B14. Proteomes. 2020; 8(4):28. https://doi.org/10.3390/proteomes8040028
Chicago/Turabian StyleTrautmann, Andrej, Lena Schleicher, Simon Deusch, Jochem Gätgens, Julia Steuber, and Jana Seifert. 2020. "Short-Chain Fatty Acids Modulate Metabolic Pathways and Membrane Lipids in Prevotella bryantii B14" Proteomes 8, no. 4: 28. https://doi.org/10.3390/proteomes8040028
APA StyleTrautmann, A., Schleicher, L., Deusch, S., Gätgens, J., Steuber, J., & Seifert, J. (2020). Short-Chain Fatty Acids Modulate Metabolic Pathways and Membrane Lipids in Prevotella bryantii B14. Proteomes, 8(4), 28. https://doi.org/10.3390/proteomes8040028






