The Proteome of Tetrasphaera elongata is adapted to Changing Conditions in Wastewater Treatment Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation and Sampling
2.2. Metabolomics & Proteomics
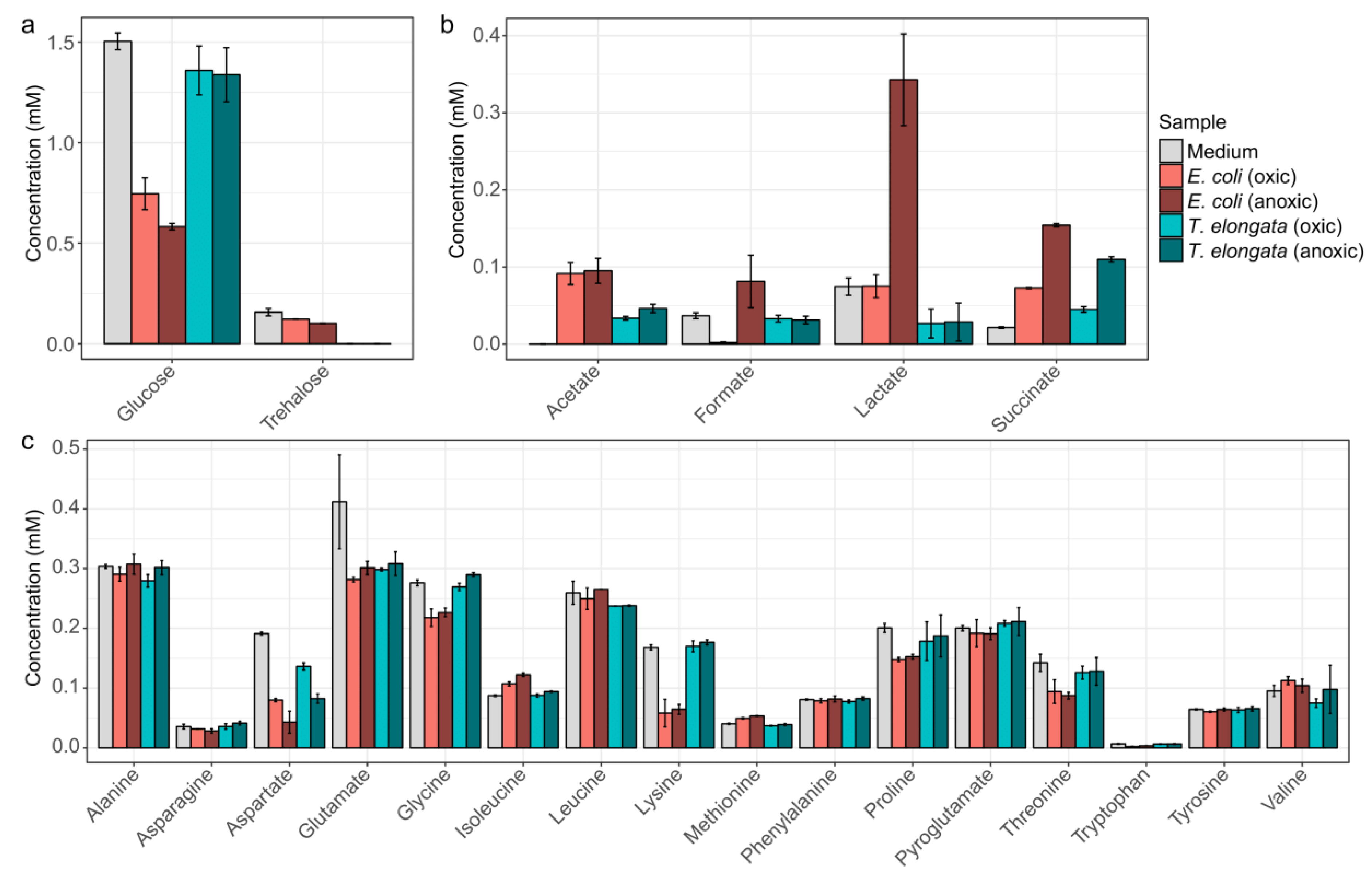
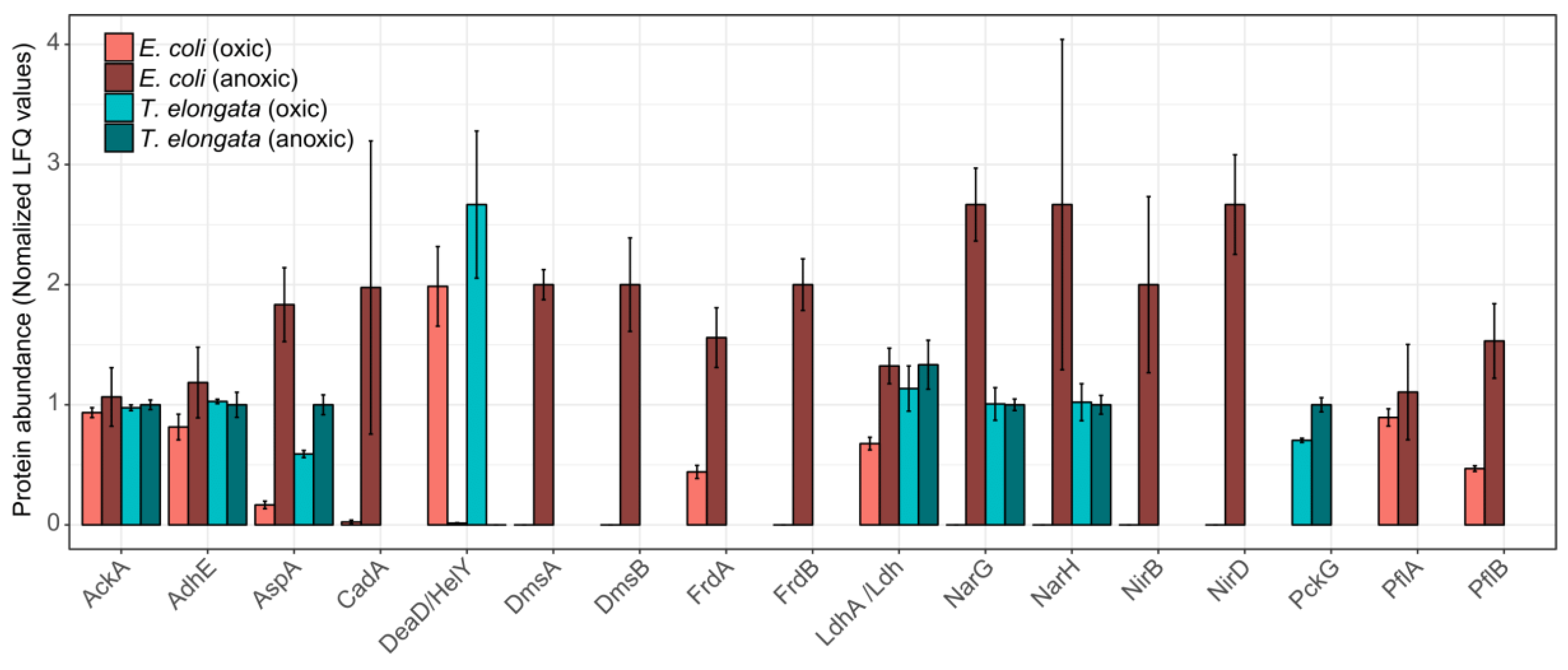
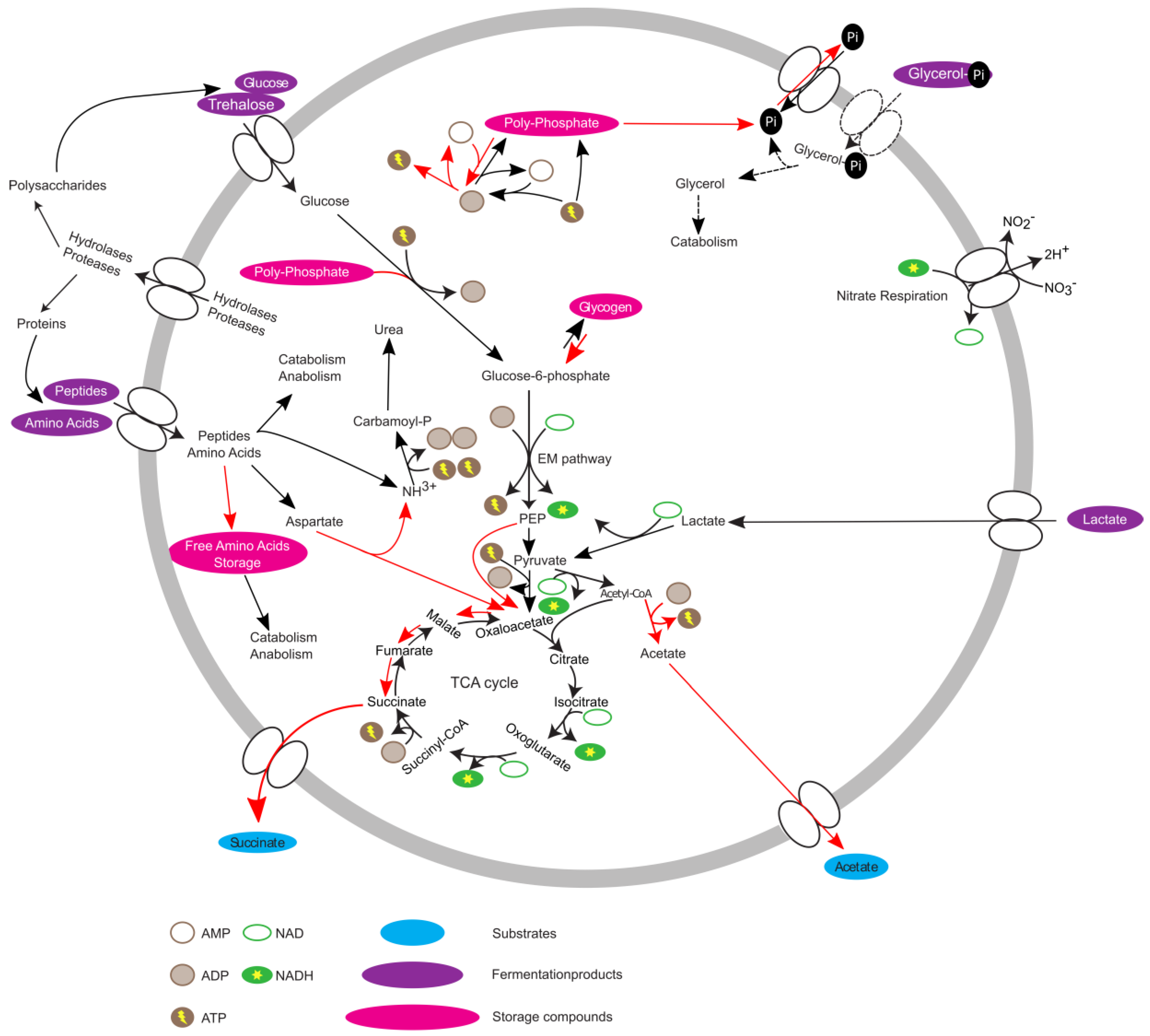
3. Results and Discussion
3.1. Efficient Growth under Anoxic Conditation
3.2. Cost-Effective Use of Substrates
3.3. Minor Changes in the Proteomes Upon Shift to Anoxic Conditions
3.4. PAO-Metabolism in T. elongata
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- van Loosdrecht, M.C.M.; Pot, M.A.; Heijnen, J.J. Importance of bacterial storage polymers in bioprocesses. Water Sci. Technol. 1997, 35. [Google Scholar] [CrossRef]
- Nielsen, P.H.; Saunders, A.M.; Hansen, A.A.; Larsen, P.; Nielsen, J.L. Microbial communities involved in enhanced biological phosphorus removal from wastewater--a model system in environmental biotechnology. Curr. Opin. Biotechnol. 2012, 23, 452–459. [Google Scholar] [CrossRef]
- Salman, V.; Bailey, J.V.; Teske, A. Phylogenetic and morphologic complexity of giant sulphur bacteria. Antonie Van Leeuwenhoek 2013, 104, 169–186. [Google Scholar] [CrossRef] [PubMed]
- López-Maury, L.; Marguerat, S.; Bähler, J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008, 9, 583–593. [Google Scholar] [CrossRef]
- Merchant, S.S.; Helmann, J.D. Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Physiol. 2012, 60, 91–210. [Google Scholar]
- Rosen, R.; Ron, E.Z. Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrom. Rev. 2002, 21, 244–265. [Google Scholar] [CrossRef] [PubMed]
- Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000, 3, 3–8. [Google Scholar] [PubMed]
- Unden, G.; Becker, S.; Bongaerts, J.; Schirawski, J.; Six, S. Oxygen regulated gene expression in facultatively anaerobic bacteria. Antonie Van Leeuwenhoek 1994, 66, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Seviour, R.J.; Nielsen, P.H. Microbial Ecology of Activated Sludge; IWA Publishing: London, UK, 2010; ISBN 9781843390329. [Google Scholar]
- Nielsen, P.H.; Mielczarek, A.T.; Kragelund, C.; Nielsen, J.L.; Saunders, A.M.; Kong, Y.; Hansen, A.A.; Vollertsen, J. A conceptual ecosystem model of microbial communities in enhanced biological phosphorus removal plants. Water Res. 2010, 44, 5070–5088. [Google Scholar] [CrossRef]
- Takiguchi, N.; Kishino, M.; Kuroda, A.; Kato, J.; Ohtake, H. A laboratory-scale test of anaerobic digestion and methane production after phosphorus recovery from waste activated sludge. J. Biosci. Bioeng. 2004, 97, 365–368. [Google Scholar] [CrossRef]
- Nyberg, U.; Aspegren, H.; Andersson, B.; Jorgensen, P.E.; Jansen, J. la C. Circulation of phosphorus in a system with biological p-removal and sludge digestion. Water Sci. Technol. 1994, 30, 293–302. [Google Scholar] [CrossRef]
- García Martín, H.; Ivanova, N.; Kunin, V.; Warnecke, F.; Barry, K.W.; McHardy, A.C.; Yeates, C.; He, S.; Salamov, A.A.; Szeto, E.; et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006, 24, 1263–1269. [Google Scholar] [CrossRef]
- McMahon, K.D.; Read, E.K. Microbial contributions to phosphorus cycling in eutrophic lakes and wastewater. Annu. Rev. Microbiol. 2013, 67, 199–219. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, R.; Nguyen, H.T.T.; Saunders, A.M.; Nielsen, J.L.; Wimmer, R.; Le, V.Q.; McIlroy, S.J.; Petrovski, S.; Seviour, R.J.; Calteau, A.; et al. A metabolic model for members of the genus Tetrasphaera involved in enhanced biological phosphorus removal. ISME J. 2013, 7, 543–554. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Kristiansen, R.; Vestergaard, M.; Wimmer, R.; Nielsen, P.H. Intracellular accumulation of glycine in polyphosphate-accumulating organisms in activated sludge, a novel storage mechanism under dynamic anaerobic-aerobic conditions. Appl. Environ. Microbiol. 2015, 81, 4809–4818. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.T.; Le, V.Q.; Hansen, A.A.; Nielsen, J.L.; Nielsen, P.H. High diversity and abundance of putative polyphosphate-accumulating Tetrasphaera-related bacteria in activated sludge systems. FEMS Microbiol. Ecol. 2011, 76, 256–267. [Google Scholar] [CrossRef] [PubMed]
- McIlroy, S.J.; Saunders, A.M.; Albertsen, M.; Nierychlo, M.; McIlroy, B.; Hansen, A.A.; Karst, S.M.; Nielsen, J.L.; Nielsen, P.H. MiDAS: the field guide to the microbes of activated sludge. Database 2015, 2015, bav062. [Google Scholar] [CrossRef] [PubMed]
- Stokholm-Bjerregaard, M.; McIlroy, S.J.; Nierychlo, M.; Karst, S.M.; Albertsen, M.; Nielsen, P.H. A critical assessment of the microorganisms proposed to be important to enhanced biological phosphorus removal in full-scale wastewater treatment systems. Front. Microbiol. 2017, 8, 718. [Google Scholar] [CrossRef]
- Goh, E.-B.; Bledsoe, P.J.; Chen, L.-L.; Gyaneshwar, P.; Stewart, V.; Igo, M.M. Hierarchical control of anaerobic gene expression in Escherichia coli K-12: the nitrate-responsive NarX-NarL regulatory system represses synthesis of the fumarate-responsive DcuS-DcuR regulatory system. J. Bacteriol. 2005, 187, 4890–4899. [Google Scholar] [CrossRef] [PubMed]
- Herbst, F.-A.; Danielsen, H.N.; Wimmer, R.; Nielsen, P.H.; Dueholm, M.S. Label-free quantification reveals major proteomic changes in Pseudomonas putida F1 during the exponential growth phase. Proteomics 2015, 15, 3244–3252. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.T.; Ahmed, S.; Muntel, J.; Cuevas Polo, N.; Bachur, R.; Kentsis, A.; Steen, J.; Steen, H. MStern blotting–Hhgh throughput polyvinylidene fluoride (PVDF) membrane-based proteomic sample preparation for 96-well plates. Mol. Cell. Proteomics 2015, 14, 2814–2823. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Uniprot Consortium Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014, 42, D191–D198. [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- Vizcaíno, J.A.; Côté, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 41, D1063–D1069. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Redder, P.; Hausmann, S.; Khemici, V.; Yasrebi, H.; Linder, P. Bacterial versatility requires DEAD-box RNA helicases. FEMS Microbiol. Rev. 2015, 39, 392–412. [Google Scholar] [CrossRef]
- Laanbroek, H.J.; Lambers, J.T.; de Vos, W.M.; Veldkamp, H. L-Aspartate fermentation by a free-living Campylobacter species. Arch. Microbiol. 1978, 117, 109–114. [Google Scholar] [CrossRef]
- Crow, V.L. Metabolism of Aspartate by Propionibacterium freudenreichii subsp. shermanii: Effect on Lactate Fermentation. Appl. Environ. Microbiol. 1986, 52, 359–365. [Google Scholar]
- Mendz, G.L.; Hazell, S.L. Aminoacid utilization by Helicobacter pylori. Int. J. Biochem. Cell Biol. 1995, 27, 1085–1093. [Google Scholar] [CrossRef]
- Camarasa, C.; Grivet, J.-P.; Dequin, S. Investigation by 13C-NMR and tricarboxylic acid (TCA) deletion mutant analysis of pathways for succinate formation in Saccharomyces cerevisiae during anaerobic fermentation. Microbiology 2003, 149, 2669–2678. [Google Scholar] [CrossRef]
- Cecchini, G.; Schröder, I.; Gunsalus, R.P.; Maklashina, E. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta - Bioenerg. 2002, 1553, 140–157. [Google Scholar] [CrossRef]
- Schnorpfeil, M.; Janausch, I.G.; Biel, S.; Kröger, A.; Unden, G. Generation of a proton potential by succinate dehydrogenase of Bacillus subtilis functioning as a fumarate reductase. Eur. J. Biochem. 2001, 268, 3069–3074. [Google Scholar] [CrossRef]
- Harden, M.M.; He, A.; Creamer, K.; Clark, M.W.; Hamdallah, I.; Martinez, K.A.; Kresslein, R.L.; Bush, S.P.; Slonczewski, J.L. Acid-adapted strains of Escherichia coli K-12 obtained by experimental evolution. Appl. Environ. Microbiol. 2015, 81, 1932–1941. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Lin, E.C.C. Adaptation of Escherichia coli to redox environments by gene expression. Mol. Microbiol. 1993, 9, 9–15. [Google Scholar] [CrossRef] [PubMed]
- McIlroy, S.J.; Albertsen, M.; Andresen, E.K.; Saunders, A.M.; Kristiansen, R.; Stokholm-Bjerregaard, M.; Nielsen, K.L.; Nielsen, P.H. ‘Candidatus Competibacter’-lineage genomes retrieved from metagenomes reveal functional metabolic diversity. ISME J. 2014, 8, 613–624. [Google Scholar] [PubMed]
- Saunders, A.M.; Mabbett, A.N.; McEwan, A.G.; Blackall, L.L. Proton motive force generation from stored polymers for the uptake of acetate under anaerobic conditions. FEMS Microbiol. Lett. 2007, 274, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ishige, K.; Kornberg, A. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. 2002, 99, 16678–16683. [Google Scholar] [CrossRef]
- Ishige, K.; Zhang, H.; Kornberg, A. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. 2002, 99, 16684–16688. [Google Scholar] [CrossRef] [PubMed]
- Nocek, B.; Kochinyan, S.; Proudfoot, M.; Brown, G.; Evdokimova, E.; Osipiuk, J.; Edwards, A.M.; Savchenko, A.; Joachimiak, A.; Yakunin, A.F. Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria. Proc. Natl. Acad. Sci. 2008, 105, 17730–17735. [Google Scholar] [CrossRef]
- Szymona, M.; Ostrowski, W. Inorganic polyphosphate glucokinase of Mycobacterium phlei. Biochim. Biophys. Acta 1964, 85, 283–295. [Google Scholar] [CrossRef]
- Marques, R.; Santos, J.; Nguyen, H.; Carvalho, G.; Noronha, J.P.; Nielsen, P.H.; Reis, M.A.M.; Oehmen, A. Metabolism and ecological niche of Tetrasphaera and Ca. Accumulibacter in enhanced biological phosphorus removal. Water Res. 2017, 122, 159–171. [Google Scholar] [CrossRef]
- Fernando, E.Y.; McIlroy, S.J.; Nierychlo, M.; Herbst, F.-A.; Petriglieri, F.; Schmid, M.C.; Wagner, M.; Nielsen, J.L.; Nielsen, P.H. Resolving the individual contribution of key microbial populations to enhanced biological phosphorus removal with Raman–FISH. ISME J. 2019, 1. [Google Scholar] [CrossRef] [PubMed]
- Félix, M.-A.; Barkoulas, M. Pervasive robustness in biological systems. Nat. Rev. Genet. 2015, 16, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Andersson, A.F.; Lefsrud, M.G.; Wexler, M.; Shah, M.; Zhang, B.; Hettich, R.L.; Bond, P.L.; VerBerkmoes, N.C.; Banfield, J.F. Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J. 2008, 2, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Wexler, M.; Richardson, D.J.; Bond, P.L. Radiolabelled proteomics to determine differential functioning of Accumulibacter during the anaerobic and aerobic phases of a bioreactor operating for enhanced biological phosphorus removal. Environ. Microbiol. 2009, 11, 3029–3044. [Google Scholar] [PubMed]
- Oyserman, B.O.; Noguera, D.R.; del Rio, T.G.; Tringe, S.G.; McMahon, K.D. Metatranscriptomic insights on gene expression and regulatory controls in Candidatus Accumulibacter phosphatis. ISME J. 2016, 10, 810–822. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Choi, P.J.; Li, G.-W.; Chen, H.; Babu, M.; Hearn, J.; Emili, A.; Xie, X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010, 329, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Gedeon, T.; Bokes, P. Delayed protein synthesis reduces the correlation between mRNA and protein fluctuations. Biophys. J. 2012, 103, 377–385. [Google Scholar] [CrossRef]

| OD600nm | Protein Concentration | |||
|---|---|---|---|---|
| E. coli | T. elongata | E. coli | T. elongata | |
| Aerobic phase (t = 0 h) | 0.183 ± 0.003 | 0.174 ± 0.026 | 107.8 ± 0.8 µg/mL | 78.4 ± 3.1 µg/mL |
| Anoxic phase (t = 3 h) | 0.214 ± 0.009 | 0.209 ± 0.032 | 114.0 ± 5.1 µg/mL | 88.7 ± 1.8 µg/mL |
| Absolute increase | 0.031 ± 0.009 | 0.036 ± 0.015 | 6.2 ± 5.3 µg/µL | 10.3 ± 3.8 µg/µL |
| Relative increase | 16.8 ± 4.8% | 20.7 ± 9.2% | 5.8 ± 5.0% | 13.2 ± 5.3% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herbst, F.-A.; Dueholm, M.S.; Wimmer, R.; Nielsen, P.H. The Proteome of Tetrasphaera elongata is adapted to Changing Conditions in Wastewater Treatment Plants. Proteomes 2019, 7, 16. https://doi.org/10.3390/proteomes7020016
Herbst F-A, Dueholm MS, Wimmer R, Nielsen PH. The Proteome of Tetrasphaera elongata is adapted to Changing Conditions in Wastewater Treatment Plants. Proteomes. 2019; 7(2):16. https://doi.org/10.3390/proteomes7020016
Chicago/Turabian StyleHerbst, Florian-Alexander, Morten S. Dueholm, Reinhard Wimmer, and Per Halkjær Nielsen. 2019. "The Proteome of Tetrasphaera elongata is adapted to Changing Conditions in Wastewater Treatment Plants" Proteomes 7, no. 2: 16. https://doi.org/10.3390/proteomes7020016
APA StyleHerbst, F.-A., Dueholm, M. S., Wimmer, R., & Nielsen, P. H. (2019). The Proteome of Tetrasphaera elongata is adapted to Changing Conditions in Wastewater Treatment Plants. Proteomes, 7(2), 16. https://doi.org/10.3390/proteomes7020016





