Retrospective Proteomic Screening of 100 Breast Cancer Tissues
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Specimens
2.2. Sample Preparations
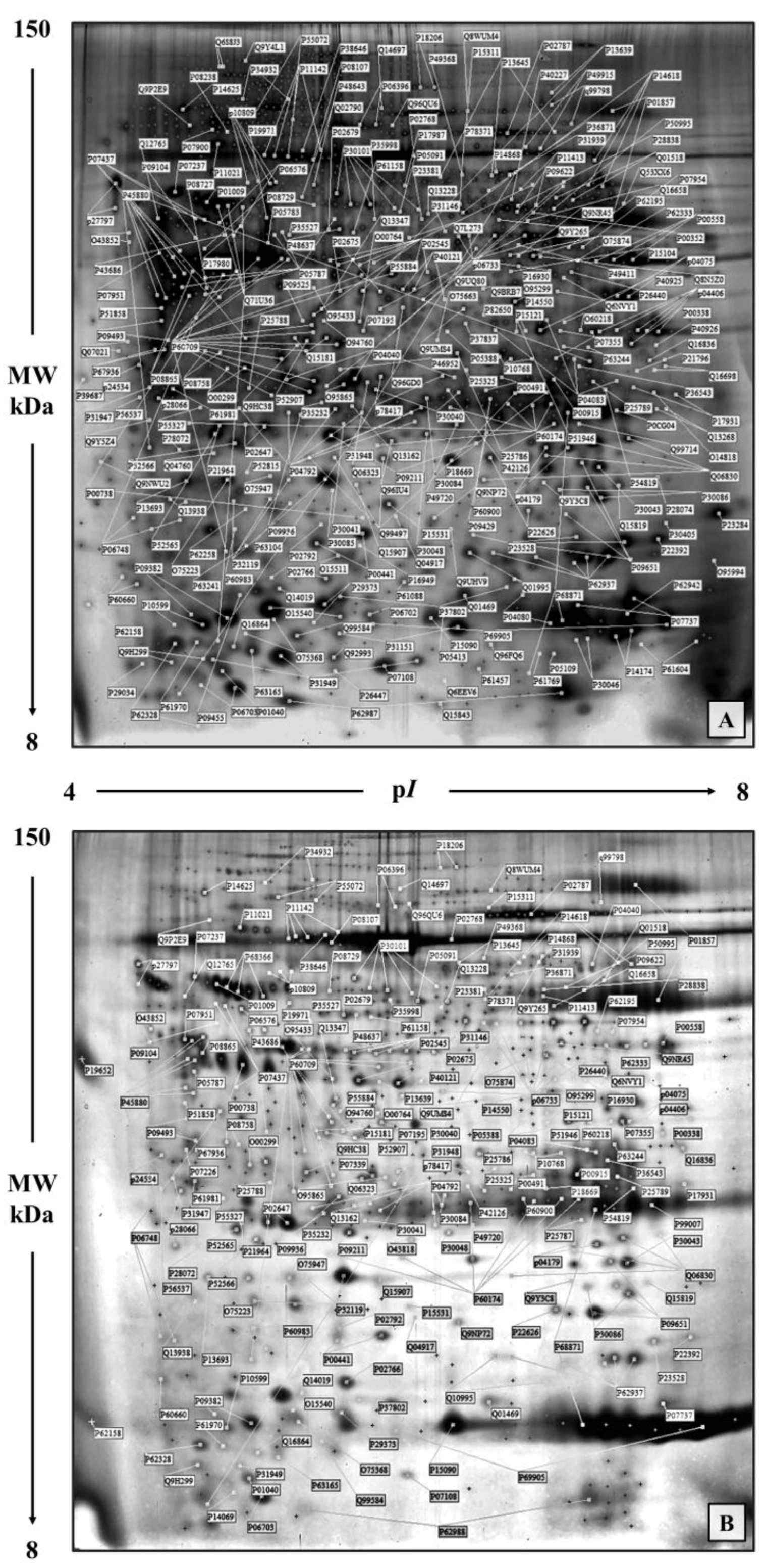
2.3. Two-Dimensional Gel Electrophoresis
2.4. Protein Identification
2.5. Western Blot
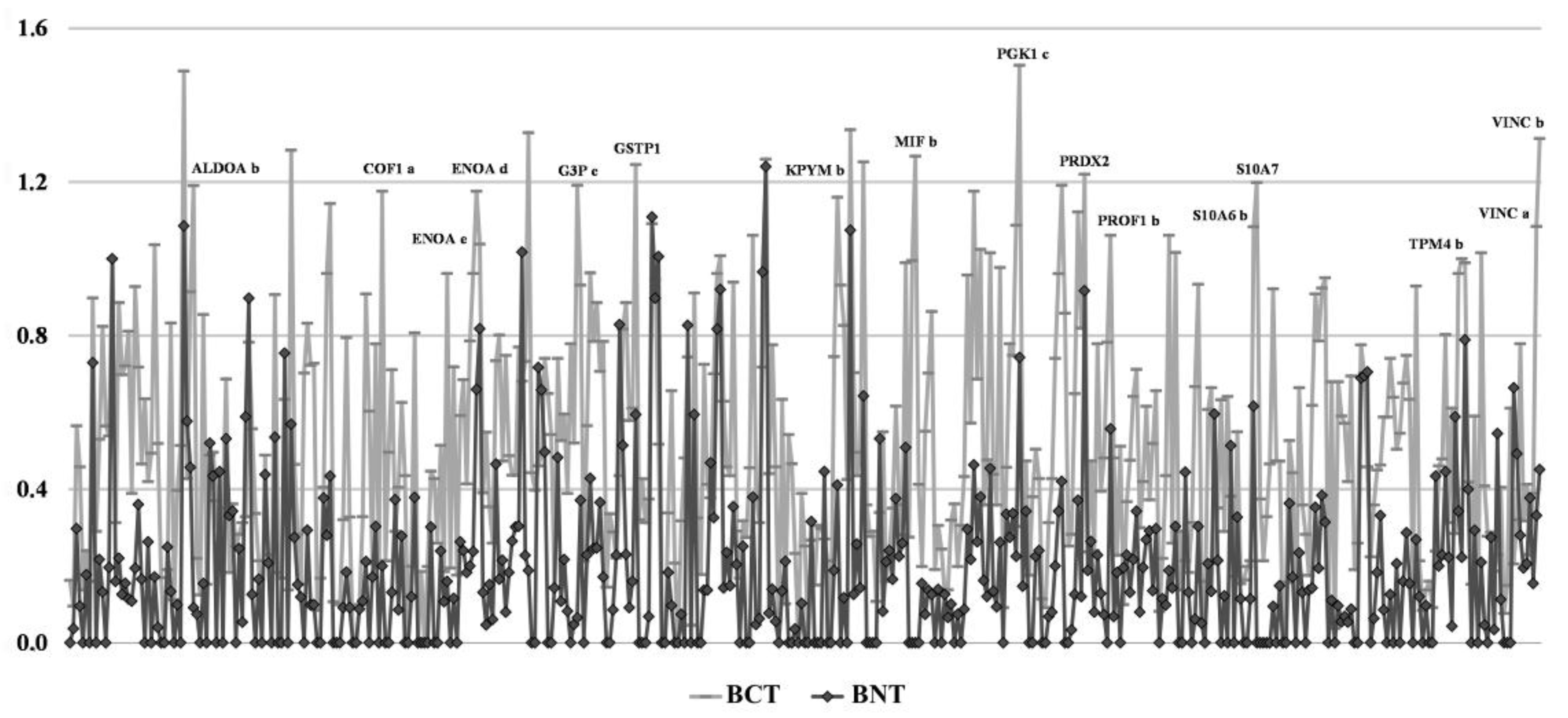
3. Results
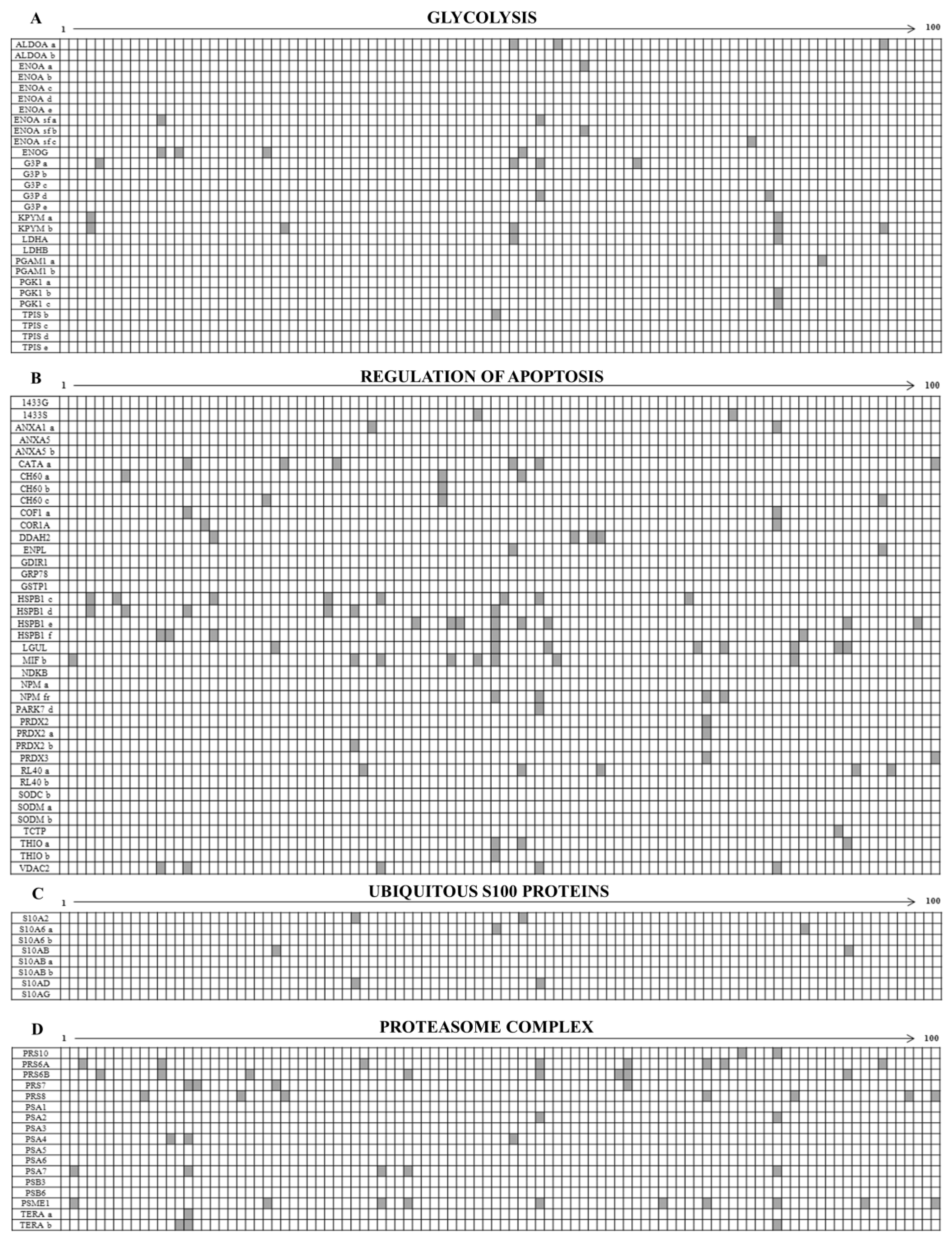
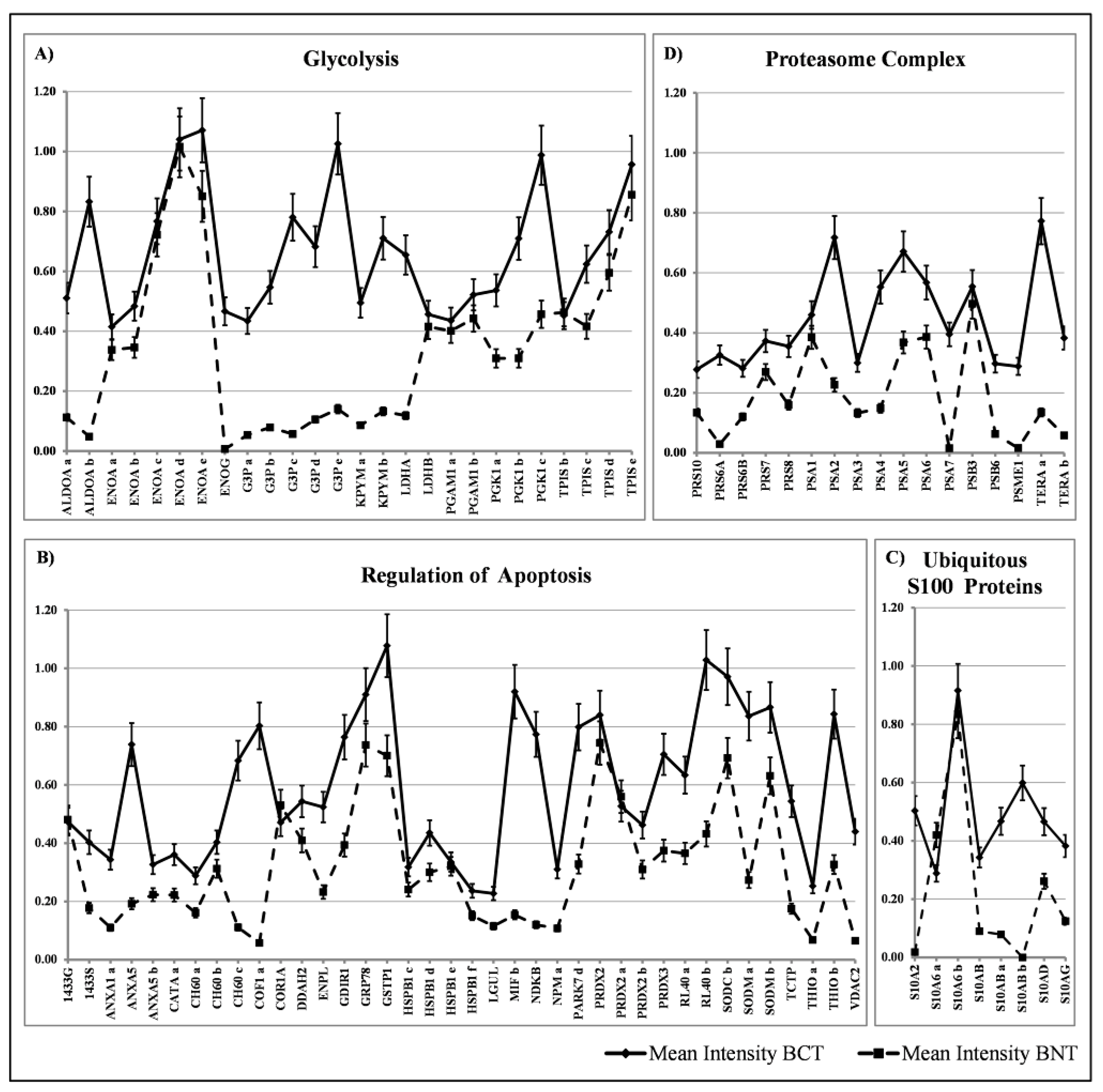
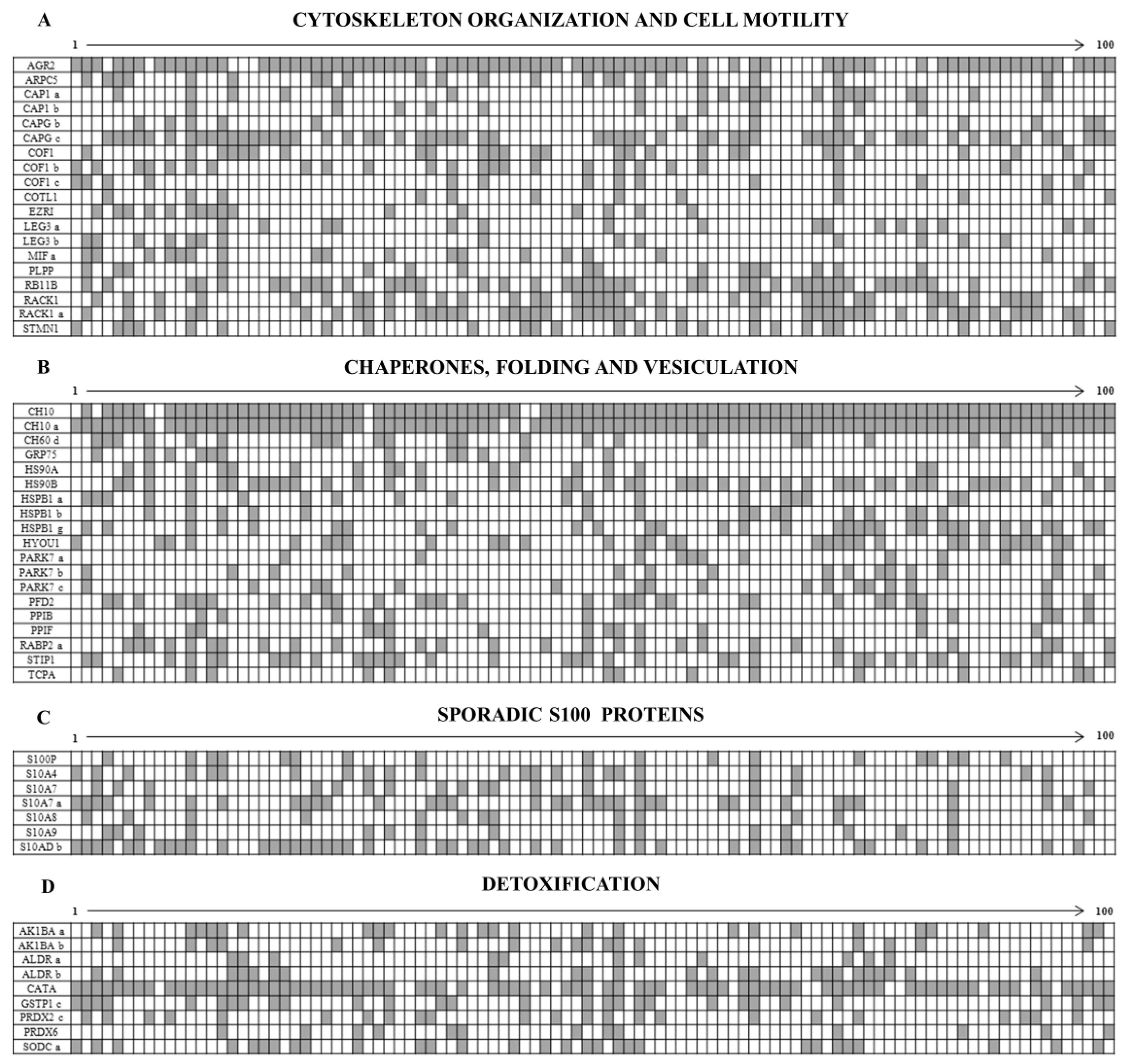
3.1. Ubiquitous Proteins
3.2. Sporadic Proteins
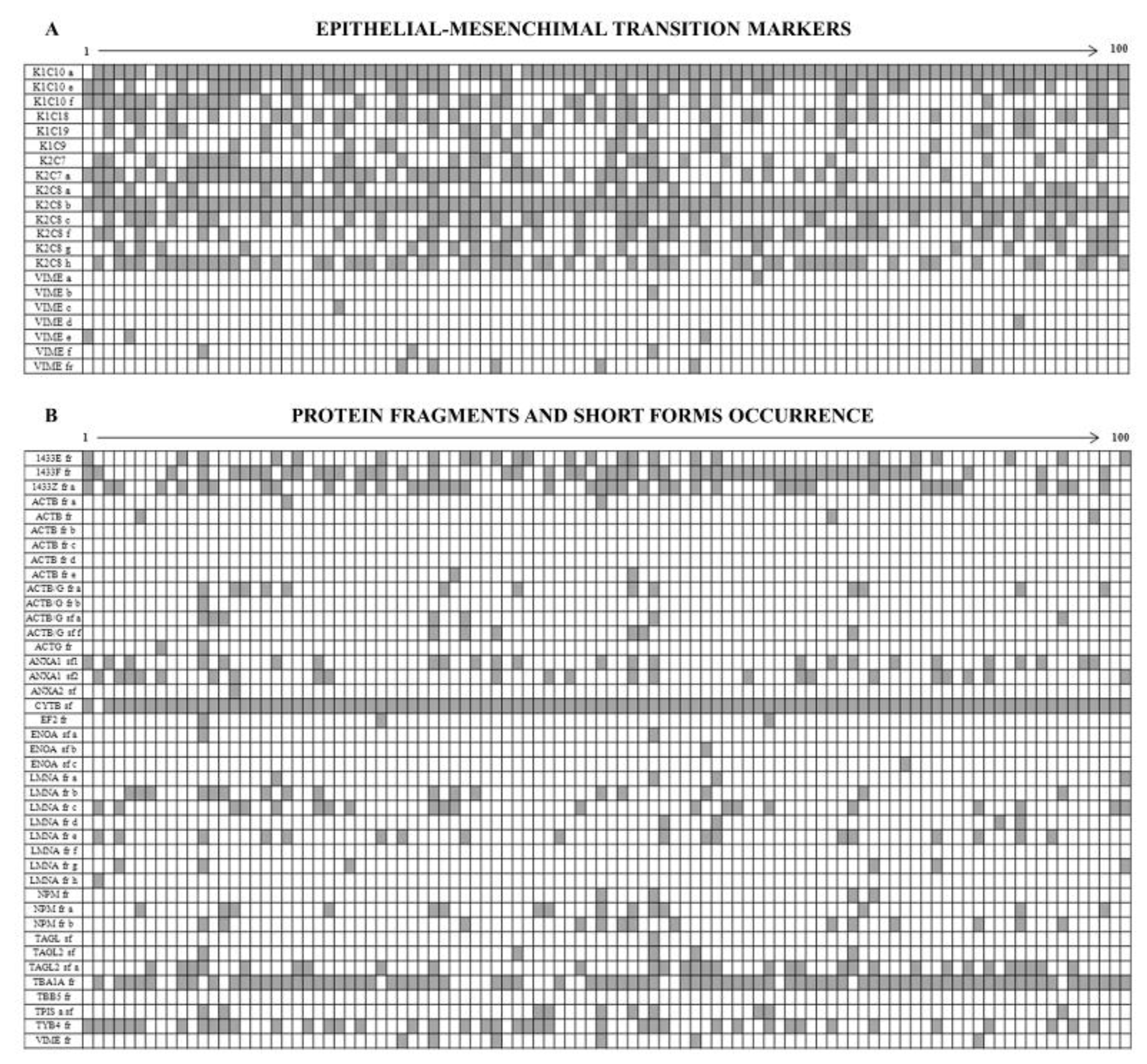
3.3. Intermediate Filament Proteins
3.4. The Enigma of Protein Fragments and Short Forms
4. Discussion
4.1. Ubiquitous Proteins
4.1.1. The Overexpressed Glycolytic Enzymes
4.1.2. Regulators of Apoptosis
4.1.3. Ubiquitous S100 Proteins
4.1.4. Proteasome Subunits
4.2. Epithelium-Mesenchymal Transition (EMT) Markers
4.3. Sporadic Proteins
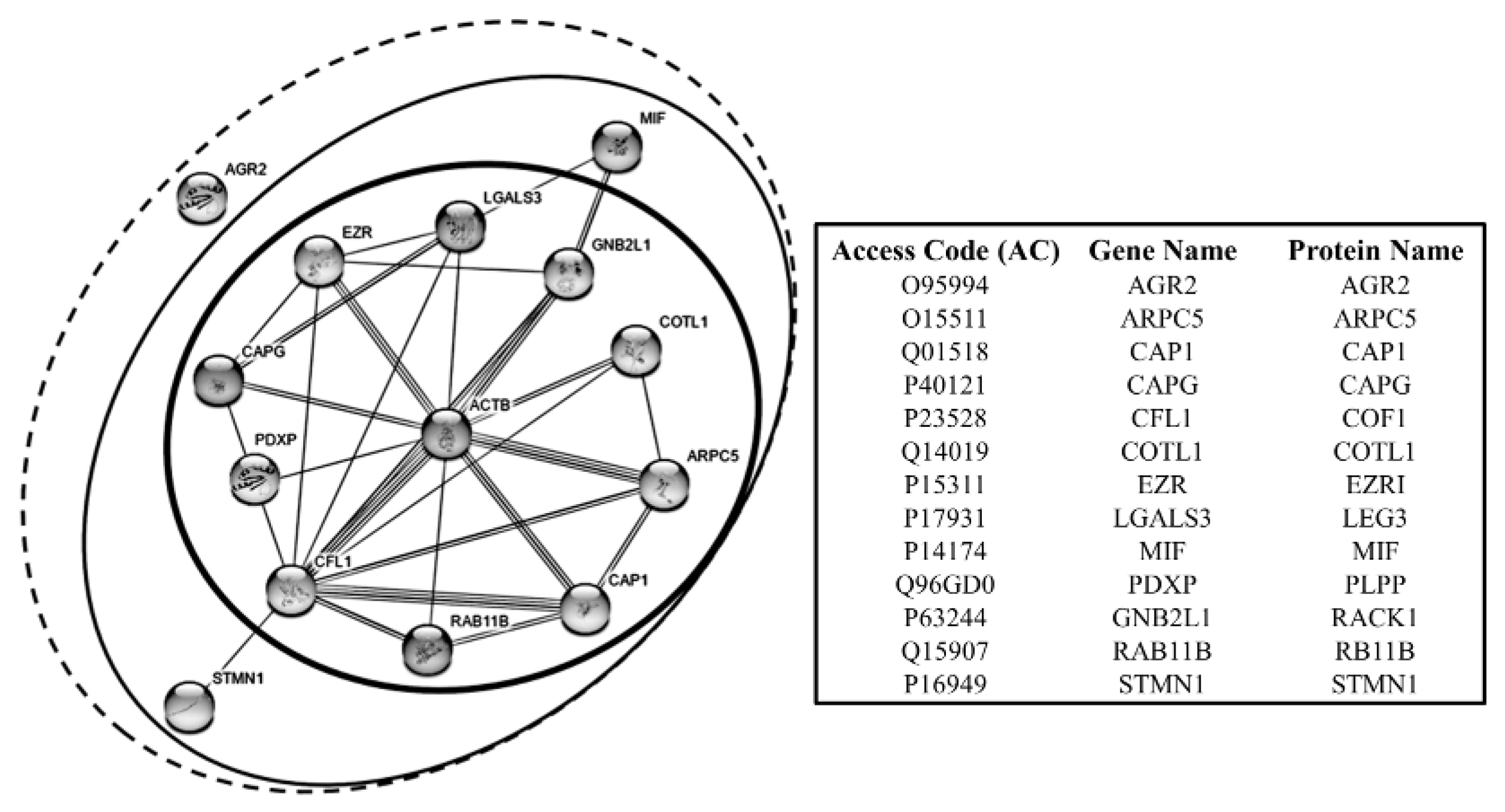
4.3.1. Cell Motility Proteins
4.3.2. Sporadic Heat Shock Proteins
4.3.3. Sporadic S100 Proteins
4.3.4. Detoxification Proteins
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hashim, D.; Boffetta, P.; La Vecchia, C.; Rota, M.; Bertuccio, P.; Malvezzi, M.; Negri, E. The global decrease in cancer mortality: Trends and disparities. Ann. Oncol. 2016, 27, 926–933. [Google Scholar] [CrossRef] [PubMed]
- Susan G. Komen. Available online: http://ww5.komen.org/BreastCancer/ChancesForSurvivalBasedOnCancerStage.html (accessed on 15 April, 2017).
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Bishop, K.; Altekruse, S.F.; Kosary, C.L.; Yu, M.; Ruhl, J.; Tatalovich, Z.; et al. SEER Cancer Statistics Review 1975–2013; National Cancer Institute: Bethesda, MD, USA, 2016.
- Weigel, M.T.; Dowsett, M. Current and emerging biomarkers in breast cancer: Prognosis and prediction. Endocr. Relat. Cancer. 2010, 17, R245–R262. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y. Development of cancer diagnostics from biomarkers to clinical tests. Transl. Cancer Res. 2015, 4, 270–279. [Google Scholar]
- Breast Cancer Fund. Available online: http://www.breastcancerfund.org/clear-science/biology-of-breast-cancer/breast-cancer-subtypes (accessed on 10 April 2017).
- Pucci-Minafra, I. Extracellular matrix in breast cancer: Permissive and restrictive influences emanating from the stroma. In Extracellular Matrix: Pathobiology and Signalling, 1st ed.; Karamanos, N.K., Ed.; De Gruyter: Boston, MA, USA, 2012; pp. 610–625. [Google Scholar]
- Palazzolo, G.; Albanese, N.N.; Di Cara, G.; Gygax, D.; Vittorelli, M.L.; Pucci-Minafra, I. Proteomic analysis of exosome-like vesicles derived from breast cancer cells. Anticancer Res. 2012, 32, 847–860. [Google Scholar] [PubMed]
- Minafra, L.; Bravatà, V.; Forte, G.I.; Cammarata, F.P.; Gilardi, M.C.; Messa, C. Gene expression profiling of epithelial-mesenchymal transition in primary breast cancer cell culture. Anticancer Res. 2014, 34, 2173–2183. [Google Scholar] [PubMed]
- Minafra, L.; Norata, R.; Bravatà, V.; Viola, M.; Lupo, C.; Gelfi, C.; Messa, C. Unmasking epithelial-mesenchymal transition in a breast cancer primary culture: A study report. BMC Res. Notes 2012, 5, 343. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Waldemarson, S.; Kurbasic, E.; Krogh, M.; Cifani, P.; Berggård, T.; Borg, Å.; James, P. Proteomic analysis of breast tumors confirms the mRNA intrinsic molecular subtypes using different classifiers: A large-scale analysis of fresh frozen tissue samples. Breast Cancer Res. 2016, 18, 69. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Di Cara, G.; Marengo, G.; Albanese, N.N.; Marabeti, M.R.; Musso, R.; Cancemi, P.; Pucci-Minafra, I. Proteomic Profiling of Trastuzumab (herceptin®)-sensitive and resistant SKBR-3 Breast Cancer Cells. Anticancer Res. 2013, 33, 489–503. [Google Scholar] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pucci-Minafra, I.; Fontana, S.; Cancemi, P.; Basiricò, L.; Caricato, S.; Minafra, S. A contribution to breast cancer cell proteomics: detection of new sequences. Proteomics 2002, 2, 919–927. [Google Scholar] [CrossRef]
- Pucci Minafra, I.; Di Cara, G.; Musso, R.; Peri, G.; Valentino, B.; D’Arienzo, M.; Martini, D.; Raspanti, M.; Minafra, S. Proteomic profiling of In Vitro bone-conditioned skbr3 breast cancer cells. J. Proteom. Bioinf. 2016, 9, 75–83. [Google Scholar]
- DAVID Bioinformatics Resources 6.7. Available online: https://david-d.ncifcrf.gov/home.jsp (accessed on 15 March 2017).
- STRING: Functional Protein Association Networks. Available online: http://string-db.org/ (accessed on 15 March 2017).
- Cancemi, P.; Di Cara, G.; Albanese, N.N.; Costantini, F.; Marabeti, M.R.; Musso, R.; Lupo, C.; Roz, E.; Pucci-Minafra, I. Large-scale proteomic identification of S100 proteins in breast cancer tissues. BMC Cancer 2010, 10, 476. [Google Scholar] [CrossRef] [PubMed]
- Pucci-Minafra, I.; Cancemi, P.; Marabeti, M.R.; Albanese, N.N.; Di Cara, G.; Taormina, P.; Marrazzo, A. Proteomic profiling of 13 paired ductal infiltrating breast carcinomas and non-tumoral adjacent counterparts. Proteom. Clin. Appl. 2007, 1, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Pucci-Minafra, I.; Cancemi, P.; Albanese, N.N.; Di Cara, G.; Marabeti, M.R.; Marrazzo, A.; Minafra, S. New protein clustering of breast cancer tissue proteomics using actin content as a cellularity indicator. J. Proteom. Res. 2008, 7, 1412–1418. [Google Scholar] [CrossRef] [PubMed]
- Pucci-Minafra, I. Breast cancer proteomics. In Omics Approaches in Breast Cancer, 1st ed.; Barh, D., Ed.; Springer: New Delhi, India, 2014; Volume 9, pp. 183–209. [Google Scholar]
- Warburg, O. Über den stoffwechsel der carcinomzelle. Biochemistry 1924, 152, 309–344. [Google Scholar] [CrossRef]
- Pucci-Minafra, I.; Fontana, S.; Cancemi, P.; Alaimo, G.; Minafra, S. Proteomic patterns of cultured breast cancer cells and epithelial mammary cells. Ann. N. Y. Acad. Sci. 2002, 963, 122–139. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhao, F.; Cui, Y. Proteomics using mammospheres as a model system to identify proteins deregulated in breast cancer stem cells. Curr. Mol. Med. 2013, 13, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Altenberg, B.; Greulich, K.O. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 2004, 84, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Chaerkady, R.; Harsha, H.C.; Nalli, A.; Gucek, M.; Vivekanandan, P.; Akhtar, J.; Cole, R.N.; Simmers, J.; Schulick, R.D.; Singh, S.; et al. A quantitative proteomic approach for identification of potential biomarkers in hepatocellular carcinoma. J. Proteom. Res. 2008, 7, 4289–4298. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Guan, Z.; Hao, L.; Song, Y.; Wang, L.; Gong, L.; Liu, L.; Qi, X.; Hou, Z.; Shao, S. Fructose-bisphosphate aldolase a is a potential metastasis-associated marker of lung squamous cell carcinoma and promotes lung cell tumorigenesis and migration. PLoS ONE 2014, 9, e85804. [Google Scholar] [CrossRef] [PubMed]
- Poschmann, G.; Sitek, B.; Sipos, B.; Ulrich, A.; Wiese, S.; Stephan, C.; Warscheid, B.; Klöppel, G.; Borght, A.V.; Ramaekers, F.C.S.; et al. Identification of proteomic differences between squamous cell carcinoma of the lung and bronchial epithelium. Mol. Cell. Proteom. 2009, 8, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, T.T.; Zhou, Y.; Wang, W.; Qiu, X.C.; Gao, J.; Li, C.X.; Long, H.; Ma, B.A.; Ma, Q.; et al. Proteomic profiling of osteosarcoma cells identifies ALDOA and SULT1A3 as negative survival markers of human osteosarcoma. Mol. Carcinog. 2014, 53, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, X.; Wu, M.; Yang, J.; Liu, M.; Zhang, W.; Xiang, B.; Wang, X.; Li, X.; Li, G.; et al. New prognosis biomarkers identified by dynamic proteomic analysis of colorectal cancer. Mol. Biosyst. 2012, 8, 3077–3088. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, S.; Kim, J.H.; Lee, T.G.; Hirata, M.; Suh, P.G.; Ryu, S.H. Phospholipase D2 directly interacts with aldolase via Its PH domain. Biochemistry 2002, 41, 3414–3421. [Google Scholar] [CrossRef] [PubMed]
- Kao, A.W.; Noda, Y.; Johnson, J.H.; Pessin, J.E.; Saltiel, A.R. Aldolase mediates the association of F-actin with the insulin-responsive glucose transporter GLUT4. J. Biol. Chem. 1999, 274, 17742–17747. [Google Scholar] [CrossRef] [PubMed]
- Buscaglia, C.A.; Penesetti, D.; Tao, M.; Nussenzweig, V. Characterization of an aldolase-binding site in the Wiskott-Aldrich syndrome protein. J. Biol. Chem. 2006, 281, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Lincet, H.; Icard, P. How do glycolytic enzymes favour cancer cell proliferationby nonmetabolic functions? Oncogene 2015, 34, 3751–3759. [Google Scholar] [CrossRef] [PubMed]
- Mamczur, P.; Gamian, A.; Kolodziej, J.; Dziegiel, P.; Rakus, D. Nuclear localization of aldolase A correlates with cell proliferation. Biochim. Biophys. Acta 2013, 1833, 2812–2822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lin, J.D.; Zuo, X.Y.; Zhuang, Y.X.; Hong, C.Q.; Zhang, G.J.; Cui, X.J.; Cui, Y.K. Elevated transcriptional levels of aldolase A (ALDOA) associates with cell cycle-related genes in patients with NSCLC and several solid tumors. BioData Min. 2017, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Liu, S.; Sun, M.Z. Novel insight into the role of GAPDH playing in tumor. Clin. Transl. Oncol. 2013, 15, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Demarse, N.A.; Ponnusamy, S.; Spicer, E.K.; Apohan, E.; Baatz, J.E.; Ogretmen, B.; Davies, C. Direct binding of glyceraldehyde 3-phosphate dehydrogenase to telomeric DNA protects telomeres against chemotherapy-induced rapid degradation. J. Mol. Biol. 2009, 394, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Carujo, S.; Estanyol, J.M.; Ejarque, A.; Agell, N.; Bachs, O.; Pujol, M.J. Glyceraldehyde 3-phosphate dehydrogenase is a SET-binding protein and regulates cyclin B-cdk1 activity. Oncogene 2006, 25, 4033–4042. [Google Scholar] [CrossRef] [PubMed]
- Inoki, K.; Li, Y.; Xu, T.; Guan, K.L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003, 17, 1829–1834. [Google Scholar] [CrossRef] [PubMed]
- Tristan, C.; Shahani, N.; Sedlak, T.W.; Sawa, A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell. Signal. 2011, 23, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Sirover, M.A. Subcellular dynamics of multifunctional protein regulation: Mechanisms of GAPDH intracellular translocation. J. Cell. Biochem. 2012, 113, 2193–2200. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wang, Z.; Lei, Q.Y. Acetylation control of metabolic enzymes in cancer: An updated version. Acta Biochim. Biophys. Sin. 2014, 46, 204–213. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sirover, M.A. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: Biochemical mechanisms and regulatory control. Biochim. Biophys. Acta 2011, 1810, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, G.S.; Dmitriev, A.A.; Snezhkina, A.V.; Kudryavtseva, A.V. Deregulation of glycolysis in cancer: Glyceraldehyde-3-phosphate dehydrogenase as a therapeutic target. Expert Opin. Ther. Targets 2013, 17, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, H.K.; Han, Y.M.; Kim, J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int. J. Biochem. Cell Biol. 2008, 40, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.J.; Knight, J.R.P.; Granchi, C.; Rani, R.; Minutolo, F.; Milner, J.; Phillips, R.M. Identification of LDH-A as a therapeutic target for cancer cell killing via (I) p53/NAD(H)-dependent and (II) p53-independent pathways. Oncogenesis 2014, 3, e102. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Chun, J.; Pan, C.; Alesi, G.N.; Li, D.; Magliocca, K.R.; Kang, Y.; Chen, Z.G.; Shin, D.M.; Khuri, F.R.; et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene 2017. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Zhang, C.; Zhang, Q.; Sahin, O.; Wang, H.; Xu, J.; Xiao, Y.; Zhang, J.; Rehman, S.K.; Li, P.; et al. Upregulation of lactate dehydrogenase a by 14–3-3ζ leads to increased glycolysis critical for breast cancer initiation and progression. Oncotarget 2016, 7, 35270–35283. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Liang, X.; Zhang, X.; Liu, T.; Shi, Q.; Song, Y.; Jiang, Y.; Wu, H.; Jiang, Y.; Lu, X.; et al. Phosphoglycerate kinase-1 is a predictor of poor survival and a novel prognostic biomarker of chemoresistance to paclitaxel treatment in breast cancer. Br. J. Cancer 2015, 112, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.S.; Glatzle, J.; Bajaeifer, K.; Bühler, S.; Lehmann, T.; Königsrainer, I.; Vollmer, J.P.; Sipos, B.; Ahmad, S.S.; Northoff, H.; et al. Phosphoglycerate kinase 1 as a promoter of metastasis in colon cancer. Int. J. Oncol. 2013, 43, 586–590. [Google Scholar] [PubMed]
- Hu, H.; Zhu, W.; Qin, J.; Chen, M.; Gong, L.; Li, L.; Liu, X.; Tao, Y.; Yin, H.; Zhou, H.; et al. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology 2017, 65, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Dai, J.; Jung, Y.; Wei, C.L.; Wang, Y.; Havens, A.M.; Hogg, P.J.; Keller, E.T.; Pienta, K.J.; et al. A glycolytic mechanism regulating an angiogenic switch in prostate cancer. Cancer Res. 2007, 67, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Daly, E.B.; Wind, T.; Jiang, X.M.; Sun, L.; Hogg, P.J. Secretion of phosphoglycerate kinase from tumour cells is controlled by oxygen-sensing hydroxylases. Biochim. Biophys. Acta 2004, 1691, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Vizin, T.; Kos, J. Gamma-enolase: A well-known tumour marker, with a less-known role in cancer. Radiol. Oncol. 2015, 49, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Portt, L.; Norman, G.; Clapp, C.; Greenwood, M.; Greenwood, M.T. Anti-apoptosis and cell survival: A review. Biochim. Biophys. Acta 2011, 1813, 238–259. [Google Scholar] [CrossRef] [PubMed]
- Ichim, G.; Tait, S.W. A fate worse than death: Apoptosis as an oncogenic process. Nat. Rev. Cancer 2016, 16, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Available online: http://www.geneontology.org/ (accessed on 17 March 2017).
- Wilker, E.W.; Grant, R.A.; Artim, S.C.; Yaffe, M.B. A structural basis for 14-3-3sigma functional specificity. J. Biol. Chem. 2005, 280, 18891–18898. [Google Scholar] [CrossRef] [PubMed]
- Aghazadeh, Y.; Papadopoulos, V. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov. Today 2016, 21, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Zuo, D.; Xue, B.; Muthuswamy, S.; Muller, W.J. A novel role for 14-3-3 sigma in regulating epithelial cell polarity. Genes Dev. 2010, 24, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Freeman, A.K.; Morrison, D.K. 14-3-3 Proteins: Diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011, 22, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Masters, S.C.; Fu, H. 14-3-3 proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 2001, 276, 45193–45200. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, M. 14-3-3 proteins in apoptosis. Braz. J. Med. Biol. Res. 2003, 36, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lu, H. 14-3-3 Gamma inhibition of MDMX-mediated p21 turnover independent of p53. J. Biol. Chem. 2011, 286, 5136–5142. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Creutz, C.E.; Moss, S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Mussunoor, S.; Murray, G.I. The role of annexins in tumour development and progression. J. Pathol. 2008, 216, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Wehder, L.; Arndt, S.; Murzik, U.; Bosserhoff, A.K.; Kob, R.; von Eggeling, F.; Melle, C. Annexin A5 is involved in migration and invasion of oral carcinoma. Cell Cycle 2009, 8, 1552–1558. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.C.; Huang, C.Y.; Pan, T.L.; Chen, W.Y.; Ho, C.T.; Liu, T.Z.; Chang, Y.J. Proteomic characterization of annexin l (ANX1) and heat shock protein 27 (HSP27) as biomarkers for invasive hepatocellular carcinoma cells. PLoS ONE 2015, 10, e0139232. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Guo, C.; Guan, H.; Liu, S.; Sun, M.Z. Annexin A5 as a potential marker in tumors. Clin. Chim. Acta 2014, 427, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Rezvani, H.R.; Mazurier, F.; Cario-André, M.; Pain, C.; Ged, C.; Taïeb, A.; de Verneuil, H. Protective effects of catalase overexpression on UVB-induced apoptosis in normal human keratinocytes. J. Biol. Chem. 2006, 281, 17999–18007. [Google Scholar] [CrossRef] [PubMed]
- Kanellos, G.; Frame, M.C. Cellular functions of the ADF/cofilin family at a glance. J. Cell Sci. 2016, 129, 3211–3218. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhou, G.; Vedantam, S.; Li, P.; Field, J. Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J. Cell Sci. 2008, 121, 2913–2920. [Google Scholar] [CrossRef] [PubMed]
- Bruneel, A.; Labas, V.; Mailloux, A.; Sharma, S.; Royer, N.; Vinh, J.; Pernet, P.; Vaubourdolle, M.; Baudin, B. Proteomics of human umbilical vein endothelial cells applied to etoposide-induced apoptosis. Proteomics 2005, 5, 3876–3884. [Google Scholar] [CrossRef] [PubMed]
- Rehklau, K.; Gurniak, C.B.; Conrad, M.; Friauf, E.; Ott, M.; Rust, M.B. ADF/cofilin proteins translocate to mitochondria during apoptosis but are not generally required for cell death signaling. Cell Death Differ. 2012, 19, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Maimaiti, Y.; Liu, Z.; Tan, J.; Abudureyimu, K.; Huang, B.; Liu, C.; Guo, Y.; Wang, C.; Nie, X.; Zhou, J.; Huang, T. Dephosphorylated cofilin expression is associated with poor prognosis in cases of human breast cancer: A tissue microarray analysis. OncoTargets Ther. 2016, 9, 6461–6466. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jolly, C.; Morimoto, R.I. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 2000, 92, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Bakthisaran, R.; Tangirala, R.; Rao, C.M. Small heat shock proteins: Role in cellular functions and pathology. Biochim. Biophys. Acta 2015, 1854, 291–319. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat shock proteins and cancer. Trends Pharmacol. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef] [PubMed]
- Ciocca, D.R.; Calderwood, S.K. Heat shock proteins in cancer: Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005, 10, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Ağababaoğlu, İ.; Önen, A.; Demir, A.B.; Aktaş, S.; Altun, Z.; Ersöz, H.; Şanl, A.; Özdemir, N.; Akkoçlu, A. Chaperonin (HSP60) and annexin-2 are candidate biomarkers for non-small cell lung carcinoma. Medicine 2017, 96, e5903. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhang, Y.; Fu, Y.; Chan, L.; Lee, A.S. Novel mechanism of anti-apoptotic function of 78-kDa glucose-regulated protein (GRP78): Endocrine resistance factor in breast cancer, through release of B-cell lymphoma 2 (BCL-2) from BCL-2-interacting killer (BIK). J. Biol. Chem. 2011, 286, 25687–25696. [Google Scholar] [CrossRef] [PubMed]
- Spike, B.T.; Kelber, J.A.; Booker, E.; Kalathur, M.; Rodewald, R.; Lipianskaya, J.; La, J.; He, M.; Wright, T.; Klemke, R.; et al. CRIPTO/GRP78 signaling maintains fetal and adult mammary stem cells Ex Vivo. Stem Cell Rep. 2014, 2, 427–439. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gray, P.C.; Vale, W. Cripto/GRP78 modulation of the TGF-β pathway in development and oncogenesis. FEBS Lett. 2012, 586, 1836–1845. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hong, F.; Gewirth, D.; Guo, B.; Liu, B.; Li, Z. The molecular chaperone gp96/GRP94 interacts with Toll-like Receptors and Integrins via its C-terminal hydrophobic domain. J. Biol. Chem. 2012, 287, 6735–6742. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.X.; Hong, F.; Zhang, Y.; Ansa-Addo, E.; Li, Z. GRP94/gp96 in cancer: Biology, structure, immunology, and drug development. Adv. Cancer Res. 2016, 129, 165–190. [Google Scholar] [PubMed]
- Kawatani, M.; Okumura, H.; Honda, K.; Kanoh, N.; Muroi, M.; Dohmae, N.; Takami, M.; Kitagawa, M.; Futamura, Y.; Imoto, M.; et al. The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I. Proc. Natl. Acad. Sci. USA 2008, 105, 11691–11696. [Google Scholar] [CrossRef] [PubMed]
- Conroy, H.; Mawhinney, L.; Donnelly, S.C. Inflammation and cancer: Macrophage migration inhibitory factor (MIF)—The potential missing link. Q. J. Med. 2010, 103, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Nobre, C.; de Araújo, J.M.; Fernandes, T.A.; Cobucci, R.N.; Lanza, D.C.; Andrade, V.S.; Fernandes, J.V. Macrophage migration inhibitory factor (MIF): Biological activities and relation with cancer. Pathol. Oncol. Res. 2016, 23, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Richard, V.; Kindt, N.; Saussez, S. Macrophage migration inhibitory factor involvement in breast cancer (Review). Int. J. Oncol. 2015, 47, 1627–1633. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S. Breast cancer advocacy and basic research: A scientist’s perspective. Breast Dis. 1998, 10, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, G.; Sobel, M.E.; Liotta, L.A.; Steeg, P.S. Association of low nm23 RNA levels in human primary infiltrating ductal breast carcinomas with lymph node involvement and other histopathological indicators of high metastatic potential. Cancer Res. 1989, 49, 5185–5190. [Google Scholar] [PubMed]
- Yan, J.; Yang, Q.; Huang, Q. Metastasis suppressor genes. Histol. Histopathol. 2013, 28, 285–292. [Google Scholar] [PubMed]
- Box, J.K.; Paquet, N.; Adams, M.N.; Boucher, D.; Bolderson, E.; O’Byrne, K.J.; Richard, D.J. Nucleophosmin: From structure and function to disease development. BMC Mol. Biol. 2016, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Pang, Q.; Christianson, T.A.; Koretsky, T.; Carlson, H.; David, L.; Keeble, W.; Faulkner, G.R.; Speckhart, A.; Bagby, G.C. Nucleophosmin interacts with and inhibits the catalytic function of eukaryotic initiation factor 2 kinase PKR. J. Biol. Chem. 2003, 278, 41709–41717. [Google Scholar] [CrossRef] [PubMed]
- Gasser, T.; Müller-Myhsok, B.; Wszolek, Z.K.; Dürr, A.; Vaughan, J.R.; Bonifati, V.; Meco, G.; Bereznai, B.; Oehlmann, R.; Agid, Y.; et al. Genetic complexity and Parkinson’s disease. Science 1997, 277, 388–390. [Google Scholar] [PubMed]
- Vasseur, S.; Afzal, S.; Tardivel-Lacombe, J.; Park, D.S.; Iovanna, J.L.; Mak, T.W. DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses. Proc. Natl. Acad. Sci. USA 2009, 106, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.H.; Peters, M.; Jang, Y.; Shi, W.; Pintilie, M.; Fletcher, G.C.; De Luca, C.; Liepa, J.; Zhou, L.; Snow, B.; et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell 2005, 7, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.; Hadar, R.; Schlossberg, A.; Sternlicht, T.; Slipicevic, A.; Skrede, M.; Risberg, B.; Flørenes, V.A.; Kopolovic, J.; Reich, R. Expression and clinical role of DJ-1, a negative regulator of PTEN, in ovarian carcinoma. Hum. Pathol. 2008, 39, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mukherjee, S.; Fan, X.; Salameh, A.; Mujoo, K.; Huang, Z.; Li, L.; To’a Salazar, G.; Zhang, N.; An, Z. Novel association of DJ-1 with HER3 potentiates HER3 activation and signaling in cancer. Oncotarget 2016, 7, 65758–65769. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, J.; Subbaram, S.; Connor, K.M.; Rodriguez, A.M.; Tirosh, O.; Beckman, J.S.; Jourd’Heuil, D.; Melendez, J.A. Manganese superoxide dismutase protects from TNF-alpha-induced apoptosis by increasing the steady-state production of H2O2. Antioxid. Redox Signal. 2006, 8, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.Y.; Kim, H.J.; Lee, K.J.; Lee, K. Translationally controlled tumor protein induces epithelial to mesenchymal transition and promotes cell migration, invasion and metastasis. Sci. Rep. 2015, 5, 8061. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.H.; Chen, L.; Guan, X.Y. Role of translationally controlled tumor protein in cancer progression. Biochem. Res. Int. 2012, 2012, 369384. [Google Scholar] [CrossRef] [PubMed]
- Karlenius, T.C.; Tonissen, K.F. Thioredoxin and cancer: A Role for thioredoxin in all states of tumor oxygenation. Cancers 2010, 2, 209–232. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Yu, H.; Lu, Y.; Yin, L. Diagnostic and prognostic value of serum thioredoxin and DJ-1 in non-small cell lung carcinoma patients. Tumour Biol. 2016, 37, 1949–1958. [Google Scholar] [CrossRef] [PubMed]
- Raninga, P.V.; Trapani, G.D.; Tonissen, K.F. Cross talk between two antioxidant systems, thioredoxin and DJ-1: Consequences for cancer. Oncoscience 2014, 1, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.H.; Sheiko, T.V.; Fisher, J.K.; Craigen, W.J.; Korsmeyer, S.J. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science 2003, 301, 513–517. [Google Scholar] [CrossRef] [PubMed]
- HGNC (HUGO Gene Nomenclature Committee). Available online: http://www.genenames.org/cgi-bin/genefamilies/set/459 (accessed on 17 March 2017).
- Salama, I.; Malone, P.S.; Mihaimeed, F.; Jones, J.L. A review of the S100 proteins in cancer. Eur. J. Surg. Oncol. 2008, 34, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, E.; Fritz, G.; Vetter, S.W.; Heizmann, C.W. Binding of S100 proteins to RAGE: An update. Biochim. Biophys. Acta 2009, 1793, 993–1007. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.; Passey, R.J.; Endoh, Y.; Rahimi, F.; Youssef, P.; Yen, T.; Geczy, C.L. Regulation of S100A8 by glucocorticoids. J. Immunol. 2005, 174, 2318–2326. [Google Scholar] [CrossRef] [PubMed]
- Gläser, R.; Harder, J.; Lange, H.; Bartels, J.; Christophers, E.; Schröder, J.M. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat. Immunol. 2005, 6, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.C.; Tsai, S.T.; Jin, Y.T.; Wu, L.W. Cyclooxygenase-2 is involved in S100A2-mediated tumor suppression in squamous cell carcinoma. Mol. Cancer Res. 2006, 4, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Bulk, E.; Sargin, B.; Krug, U.; Hascher, A.; Jun, Y.; Knop, M.; Kerkhoff, C.; Gerke, V.; Liersch, R.; Mesters, R.M.; et al. S100A2 induces metastasis in non-small cell lung cancer. Clin. Cancer Res. 2009, 15, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, F.; Umeda, Y.; Shimamoto, S.; Tsuchiya, M.; Tokumitsu, H.; Tokuda, M.; Kobayashi, R. S100 proteins modulate protein phosphatase 5 function: A link between CA2+ signal transduction and protein dephosphorylation. J. Biol. Chem. 2012, 287, 13787–13798. [Google Scholar] [CrossRef] [PubMed]
- Anania, M.C.; Miranda, C.; Vizioli, M.G.; Mazzoni, M.; Cleris, L.; Pagliardini, S.; Manenti, G.; Borrello, M.G.; Pierotti, M.A.; Greco, A. S100A11 overexpression contributes to the malignant phenotype of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, E1591–E1600. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Wang, K.; Yue, Y.; Tian, T.; Xu, A.; Hao, J.; Xiao, X.; He, D. Selective expression of S100A11 in lung cancer and its role in regulating proliferation of adenocarcinomas cells. Mol. Cell. Biochem. 2012, 359, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Pierce, A.; Barron, N.; Linehan, R.; Ryan, E.; O’Driscoll, L.; Daly, C.; Clynes, M. Identification of a novel, functional role for S100A13 in invasive lung cancer cell lines. Eur. J. Cancer 2008, 44, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Massi, D.; Landriscina, M.; Piscazzi, A.; Cosci, E.; Kirov, A.; Paglierani, M.; Di Serio, C.; Mourmouras, C.; Santucci, M.; Marchionni, N.; et al. S100A13 is a new angiogenic marker in human melanoma. Mod. Pathol. 2010, 23, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, D.; Bruland, O.; Parajuli, H.; Osman, T.A.; Teh, M.; Johannessen, A.C.; Costea, D.E. S100A16 promotes differentiation and contributes to a less aggressive tumor phenotype in oral squamous cell carcinoma. BMC Cancer 2015, 15, 631. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Xue, Y.; Liang, C.; Zhang, R.; Zhang, Z.; Li, H.; Su, D.; Liang, X.; Zhang, Y.; Huang, Q.; et al. S100A16 promotes cell proliferation and metastasis via AKT and ERK cell signaling pathways in human prostate cancer. Tumour. Biol. 2016, 37, 12241–12250. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Ichikawa-Tomikawa, N.; Shishito, N.; Nishiura, K.; Miura, T.; Hozumi, A.; Chiba, H.; Yoshida, S.; Ohtake, T.; Sugino, T. Co-expression of S100A14 and S100A16 correlates with a poor prognosis in human breast cancer and promotes cancer cell invasion. BMC Cancer 2015, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Brewer, M.D.; Guo, L.; Wang, R.; Jiang, P.; Yang, X. Enhanced degradation of misfolded proteins promotes tumorigenesis. Cell Rep. 2017, 18, 3143–3154. [Google Scholar] [CrossRef] [PubMed]
- Barrière, G.; Tartary, M.; Rigaud, M. Epithelial mesenchymal transition: A new insight into the detection of circulating tumor cells. ISRN Oncol. 2012, 2012, 382010. [Google Scholar] [CrossRef] [PubMed]
- Pucci-Minafra, I.; Minafra, S.; La Rocca, G.; Barranca, M.; Fontana, S.; Alaimo, G.; Okada, Y. Zymographic analysis of circulating and tissue forms of colon carcinoma gelatinase A (MMP-2) and B (MMP-9) separated by mono- and two-dimensional electrophoresis. Matrix Biol. 2001, 20, 419–427. [Google Scholar] [CrossRef]
- Pollard, T.D.; Borisy, G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003, 112, 453–465. [Google Scholar] [CrossRef]
- Smith, B.A.; Daugherty-Clarke, K.; Goode, B.L.; Gelles, J. Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc. Natl. Acad. Sci. USA 2013, 110, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, D.; Kurisu, S.; Takenawa, T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 2005, 96, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Nohata, N.; Watanabe-Takano, H.; Yoshino, H.; Hidaka, H.; Fujimura, L.; Fuse, M.; Yamasaki, T.; Enokida, H.; Nakagawa, M.; et al. Actin-related protein 2/3 complex subunit 5 (ARPC5) contributes to cell migration and invasion and is directly regulated by tumor-suppressive microRNA-133a in head and neck squamous cell carcinoma. Int. J. Oncol. 2012, 40, 1770–1778. [Google Scholar] [PubMed]
- Zhang, H.; Zhou, G.L. CAP1 (cyclase-associated protein 1) exerts distinct functions in the proliferation and metastatic potential of breast cancer cells mediated by ERK. Sci. Rep. 2016, 6, 25933. [Google Scholar] [CrossRef] [PubMed]
- Westbrook, J.A.; Cairns, D.A.; Peng, J.; Speirs, V.; Hanby, A.M.; Holen, I.; Wood, S.L.; Ottewell, P.D.; Marshall, H.; Banks, R.E.; et al. CAPG and GIPC1: Breast cancer biomarkers for bone metastasis development and treatment. J. Natl. Cancer Inst. 2016, 108, djv360. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.; Beltzner, C.C.; Pollard, T.D. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 2009, 19, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.L.; Zhang, H.; Field, J. Mammalian CAP (Cyclase-associated protein) in the world of cell migration: Roles in actin filament dynamics and beyond. Cell Adh. Migr. 2014, 8, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Cordero, J.J.; Magalhaes, M.A.; Eddy, R.J.; Hodgson, L.; Condeelis, J. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 2013, 14, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Katahira, T.; Ohashi, K.; Mizuno, K.; Sugiyama, S.; Nakamura, H. Coactosin accelerates cell dynamism by promoting actin polymerization. Dev. Biol. 2013, 379, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Guo, C.; Meng, X.; Yu, Y.; Jin, Y.; Tong, D.; Geng, J.; Huang, Q.; Qi, J.; Liu, A.; et al. Differential expression of PAI-RBP1, C1orf142, and COTL1 in non-small cell lung cancer cell lines with different tumor metastatic potential. J. Investig. Med. 2012, 60, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Hoskin, V.; Szeto, A.; Ghaffari, A.; Greer, P.A.; Côté, G.P.; Elliott, B.E. Ezrin regulates focal adhesion and invadopodia dynamics by altering calpain activity to promote breast cancer cell invasion. Mol. Biol. Cell 2015, 26, 3464–3479. [Google Scholar] [CrossRef] [PubMed]
- Asp, N.; Kvalvaag, A.; Sandvig, K.; Pust, S. Regulation of ErbB2 localization and function in breast cancer cells by ERM proteins. Oncotarget 2016, 7, 25443–25460. [Google Scholar] [CrossRef] [PubMed]
- Shetty, P.; Bargale, A.; Patil, B.R.; Mohan, R.; Dinesh, U.S.; Vishwanatha, J.K.; Gai, P.B.; Patil, V.S.; Amsavardani, T.S. Cell surface interaction of annexin A2 and galectin-3 modulates epidermal growth factor receptor signaling in Her-2 negative breast cancer cells. Mol. Cell. Biochem. 2016, 411, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Fukumori, T.; Takenaka, Y.; Yoshii, T.; Kim, H.R.; Hogan, V.; Inohara, H.; Kagawa, S.; Raz, A. CD29 and CD7 mediate galectin-3-induced type II T-cell apoptosis. Cancer Res. 2003, 63, 8302–8311. [Google Scholar] [PubMed]
- De Oliveira, F.L.; Gatto, M.; Bassi, N.; Luisetto, R.; Ghirardello, A.; Punzi, L.; Doria, A. Galectin-3 in autoimmunity and autoimmune diseases. Exp. Biol. Med. 2015, 240, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Luo, M.; Liang, X.; Wang, D.; Gu, X.; Duan, C.; Gu, H.; Chen, G.; Zhao, X.; Zhao, Z.; et al. Galectin-3 as a marker and potential therapeutic target in breast cancer. PLoS ONE 2014, 9, e103482. [Google Scholar] [CrossRef] [PubMed]
- Gohla, A.; Birkenfeld, J.; Bokoch, G.M. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 2005, 7, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.X.; Xu, J.D.; Xu, J.W.; Liu, X.L.; Cheng, Y.Y.; Li, Q.Q.; Xu, Z.D.; Liu, X.P. RACK1 promotes breast carcinoma migration/metastasis via activation of the RhoA/Rho kinase pathway. Breast Cancer Res. Treat. 2011, 126, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Emery, G.; Ramel, D. Cell coordination of collective migration by Rab11 and Moesin. Commun. Integr. Biol. 2013, 6, e24587. [Google Scholar] [CrossRef] [PubMed]
- Kuang, X.Y.; Jiang, H.S.; Li, K.; Zheng, Y.Z.; Liu, Y.R.; Qiao, F.; Li, S.; Hu, X.; Shao, Z.M. The phosphorylation-specific association of STMN1 with GRP78 promotes breast cancer metastasis. Cancer Lett. 2016, 377, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Fessart, D.; Domblides, C.; Avril, T.; Eriksson, L.A.; Begueret, H.; Pineau, R.; Malrieux, C.; Dugot-Senant, N.; Lucchesi, C.; Chevet, E.; et al. Secretion of protein disulphide isomerase AGR2 confers tumorigenic properties. eLife 2016, 5, e13887. [Google Scholar] [CrossRef] [PubMed]
- Ondrouskova, E.; Sommerova, L.; Nenutil, R.; Coufal, O.; Bouchal, P.; Vojtesek, B.; Hrstka, R. AGR2 associates with HER2 expression predicting poor outcome in subset of estrogen receptor negative breast cancer patients. Exp. Mol. Pathol. 2017, 102, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Dumartin, L.; Whiteman, H.J.; Weeks, M.E.; Hariharan, D.; Dmitrovic, B.; Iacobuzio-Donahue, C.A.; Brentnall, T.A.; Bronner, M.P.; Feakins, R.M.; Timms, J.F.; et al. AGR2 is a novel surface antigen that promotes the dissemination of pancreatic cancer cells through regulation of cathepsins B and D. Cancer Res. 2011, 71, 7091–7102. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M.; Mah, V.; Maresh, E.L.; Bagryanova, L.; Horvath, S.; Chia, D.; Goodglick, L.; Liu, A.Y. High expression of AGR2 in lung cancer is predictive of poor survival. BMC Cancer 2015, 15, 655. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Mallik, M.; Lakhotia, S.C. Heat shock genes-integrating cell survival and death. J. Biosci. 2007, 32, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, R.; Takano, S.; Kaur, K.; Deocaris, C.C.; Pereira-Smith, O.M.; Reddel, R.R.; Kaul, S.C. Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis. Int. J. Cancer 2006, 118, 2973–2980. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Ji, M.; Chen, L.; Liu, Q.; Che, S.; Xu, M.; Lin, Z. The clinicopathological significance of Mortalin overexpression in invasive ductal carcinoma of breast. J. Exp. Clin. Cancer Res. 2016, 35, 42. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liao, Q.; Li, X.; Wang, H.; Wei, F.; Chen, J.; Yang, J.; Zeng, Z.; Guo, X.; Chen, P.; et al. HYOU1, regulated by LPLUNC1, is up-regulated in nasopharyngeal carcinoma and associated with poor prognosis. J. Cancer 2016, 7, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Jarosz, D. Hsp90: A global regulator of the genotype-to-phenotype map in cancers. Adv. Cancer Res. 2016, 129, 225–247. [Google Scholar] [PubMed]
- Calderwood, S.K.; Neckers, L. Hsp90 in Cancer: Transcriptional roles in the nucleus. Adv. Cancer Res. 2016, 129, 89–106. [Google Scholar] [PubMed]
- Fang, F.; Flegler, A.J.; Du, P.; Lin, S.; Clevenger, C.V. Expression of cyclophilin B is associated with malignant progression and regulation of genes implicated in the pathogenesis of breast cancer. Am. J. Pathol. 2009, 174, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Zheng, J.; Galbaugh, T.L.; Fiorillo, A.A.; Hjort, E.E.; Zeng, X.; Clevenger, C.V. Cyclophilin B as a co-regulator of prolactin-induced gene expression and function in breast cancer cells. J. Mol. Endocrinol. 2010, 44, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Javadov, S.; Kuznetsov, A. Mitochondrial permeability transition and cell death: The role of cyclophilin d. Front. Physiol. 2013, 4, 76. [Google Scholar] [CrossRef] [PubMed]
- Bigi, A.; Beltrami, E.; Trinei, M.; Stendardo, M.; Pelicci, P.G.; Giorgio, M. Cyclophilin D counteracts P53-mediated growth arrest and promotes Ras tumorigenesis. Oncogene 2016, 35, 5132–5143. [Google Scholar] [CrossRef] [PubMed]
- Millán-Zambrano, G.; Chávez, S. Nuclear functions of prefoldin. Open Biol. 2014, 4, 140085. [Google Scholar] [CrossRef] [PubMed]
- López, V.; González-Peramato, P.; Suela, J.; Serrano, A.; Algaba, F.; Cigudosa, J.C.; Vidal, A.; Bellmunt, J.; Heredero, O.; Sánchez-Carbayo, M. Identification of prefoldin amplification (1q23.3-q24.1) in bladder cancer using comparative genomic hybridization (CGH) arrays of urinary DNA. J. Transl. Med. 2013, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Vreeland, A.C.; Levi, L.; Zhang, W.; Berry, D.C.; Noy, N. Cellular retinoic acid-binding protein 2 inhibits tumor growth by two distinct mechanisms. J. Biol. Chem. 2014, 289, 34065–34073. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Parameswaran, N.; Li, M.; Wang, Y.; Jackson, M.W.; Liu, H.; Xin, W.; Zhou, L. CRABP-II enhances pancreatic cancer cell migration and invasion by stabilizing interleukin 8 expression. Oncotarge 2016. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.L.; Chao, A.; Jung, S.M.; Tsai, C.N.; Lin, C.Y.; Chen, S.H.; Sue, S.C.; Wang, T.H.; Wang, H.S.; Lai, C.H. Stress-induced phosphoprotein-1 maintains the stability of JAK2 in cancer cells. Oncotarget 2016, 7, 50548–50563. [Google Scholar] [CrossRef] [PubMed]
- Guest, S.T.; Kratche, Z.R.; Bollig-Fischer, A.; Haddad, R.; Ethier, S.P. Two members of the TRiC chaperonin complex, CCT2 and TCP1 are essential for survival of breast cancer cells and are linked to driving oncogenes. Exp. Cell Res. 2015, 332, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Boudiaf-Benmammar, C.; Cresteil, T.; Melki, R. The cytosolic chaperonin CCT/TRiC and cancer cell proliferation. PLoS ONE 2013, 8, e60895. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, M.K.; Nigavekar, S.S.; Arumugam, T.; Logsdon, C.D.; Schmidt, A.M.; Park, J.C.; Huang, EH. RAGE activation by S100P in colon cancer stimulates growth, migration, and cell signaling pathways. Dis. Colon Rectum 2007, 50, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, T.; Simeone, D.M.; Van Golen, K.; Logsdon, C.D. S100P promotes pancreatic cancer growth, survival, and invasion. Clin. Cancer Res. 2005, 11, 5356–5364. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.L.; Padilla, L.; Dakhel, S.; Coll, T.; Hervas, R.; Adan, J.; Masa, M.; Mitjans, F.; Martinez, J.M.; Coma, S.; et al. Therapeutic targeting of tumor growth and angiogenesis with a novel anti-S100A4 monoclonal antibody. PLoS ONE 2013, 8, e72480. [Google Scholar] [CrossRef] [PubMed]
- Cancemi, P.; Di Cara, G.; Albanese, N.N.; Costantini, F.; Marabeti, M.R.; Musso, R.; Riili, I.; Lupo, C.; Roz, E.; Pucci-Minafra, I. Differential occurrence of S100A7 in breast cancer tissues: A proteomic-based investigation. Proteom. Clin. Appl. 2012, 6, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Curtis, L.; Pind, M.; Murphy, L.C.; Watson, P.H. S100A7 (psoriasin) influences immune response genes in human breast cancer. Exp. Cell Res. 2007, 313, 3016–3025. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; Watson, P.H. S100A7 (psoriasin) oncostatin-M and interleukin-6 in human breast cancer. Oncogene 2010, 29, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- Nasser, M.W.; Qamri, Z.; Deol, Y.S.; Ravi, J.; Powell, C.A.; Trikha, P.; Schwendener, R.A.; Bai, X.F.; Shilo, K.; Zou, X.; et al. S100A7 enhances mammary tumorigenesis through upregulation of inflammatory pathways. Cancer Res. 2012, 72, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xu, C.; Jin, Q.; Liu, Z. S100 protein family in human cancer. Am. J. Cancer Res. 2014, 4, 89–115. [Google Scholar] [PubMed]
- Oberley, T.D. Oxidative damage and cancer. Am. J. Pathol. 2002, 160, 403–408. [Google Scholar] [CrossRef]
- Matkowskyj, K.A.; Bai, H.; Liao, J.; Zhang, W.; Li, H.; Rao, S.; Omary, R.; Yang, G.Y. Aldoketoreductase family 1B10 (AKR1B10) as a biomarker to distinguish hepatocellular carcinoma from benign liver lesions. Hum. Pathol. 2014, 45, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Shen, Y.; Peng, X.; Zhang, S.; Wang, M.; Xu, G.; Zheng, X.; Wang, J.; Lu, C. Aberrant promoter methylation of cancer-related genes in human breast cancer. Oncol. Lett. 2016, 12, 5145–5155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Zhang, F.; Hong, C.Q.; Giuliano, A.E.; Cui, X.J.; Zhou, G.J.; Zhang, G.J.; Cui, Y.K. Critical protein GAPDH and its regulatory mechanisms in cancer cells. Cancer Biol. Med. 2015, 12, 10–22. [Google Scholar] [PubMed]
- Chung, Y.T.; Matkowskyj, K.A.; Li, H.; Bai, H.; Zhang, W.; Tsao, M.S.; Liao, J.; Yang, G.Y. Overexpression and oncogenic function of aldo-keto reductase family 1B10 (AKR1B10) in pancreatic carcinoma. Mod. Pathol. 2012, 25, 758–766. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pucci-Minafra, I.; Di Cara, G.; Musso, R.; Cancemi, P.; Albanese, N.N.; Roz, E.; Minafra, S. Retrospective Proteomic Screening of 100 Breast Cancer Tissues. Proteomes 2017, 5, 15. https://doi.org/10.3390/proteomes5030015
Pucci-Minafra I, Di Cara G, Musso R, Cancemi P, Albanese NN, Roz E, Minafra S. Retrospective Proteomic Screening of 100 Breast Cancer Tissues. Proteomes. 2017; 5(3):15. https://doi.org/10.3390/proteomes5030015
Chicago/Turabian StylePucci-Minafra, Ida, Gianluca Di Cara, Rosa Musso, Patrizia Cancemi, Nadia Ninfa Albanese, Elena Roz, and Salvatore Minafra. 2017. "Retrospective Proteomic Screening of 100 Breast Cancer Tissues" Proteomes 5, no. 3: 15. https://doi.org/10.3390/proteomes5030015
APA StylePucci-Minafra, I., Di Cara, G., Musso, R., Cancemi, P., Albanese, N. N., Roz, E., & Minafra, S. (2017). Retrospective Proteomic Screening of 100 Breast Cancer Tissues. Proteomes, 5(3), 15. https://doi.org/10.3390/proteomes5030015




