Enzymes and Metabolites in Carbohydrate Metabolism of Desiccation Tolerant Plants
Abstract
:1. Introduction
2. Accumulation of Sugars in Response to Dehydration
3. Physiological Roles of Sugars Accumulating in Resurrection Plants
4. Studies on Enzymes Responsible for Sugar Accumulation in Resurrection Plants
5. Sugar Metabolism during Dehydration Studied by Proteomics
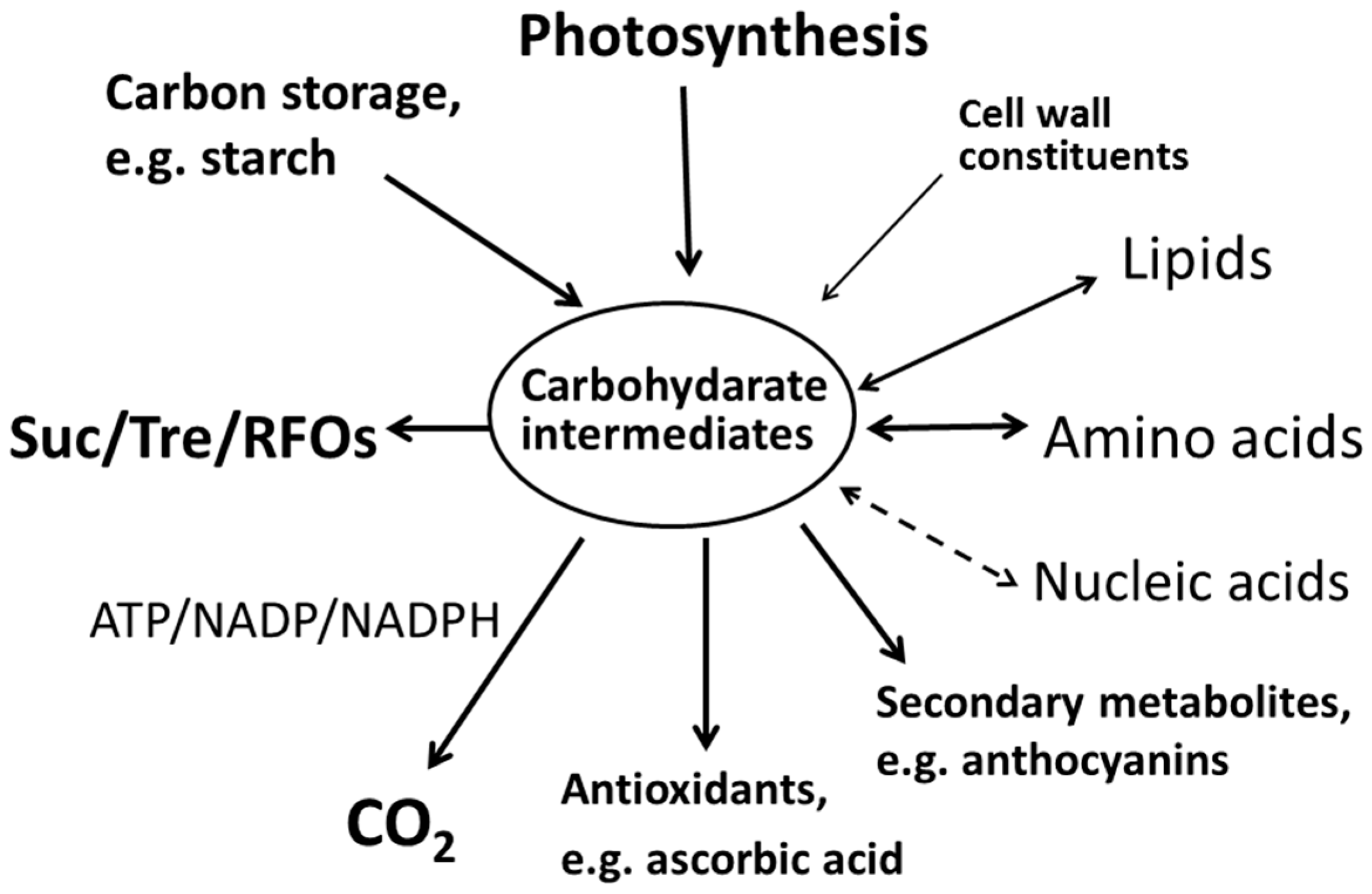
6. The Relationships among Sugar Accumulation, Energy Metabolism, and Other Metabolites
7. Unanswered Questions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Alpert, P. The Limits and Frontiers of Desiccation-tolerant Life. Integr. Comp. Biol. 2005, 45, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Alpert, P. Constraints of Tolerance: Why are Desiccation-tolerant Organisms so Small or Rare? J. Exp. Biol. 2006, 209, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D. Physiological Aspects of Desiccation Tolerance. Annu. Rev. Plant Physiol. 1979, 30, 195–238. [Google Scholar] [CrossRef]
- Tweddle, J.C.; Dickie, J.B.; Baskin, C.C.; Baskin, J.M. Ecological Aspects of Seed Desiccation Sensitivity. J. Ecol. 2003, 91, 294–304. [Google Scholar] [CrossRef]
- Alpert, P.; Oliver, M.J. Desiccation and Survival in Plants: Drying Without Dying; Black, M., Pritchard, H.W., Eds.; CABI: Wallingford, UK, 2002; pp. 3–43. [Google Scholar]
- Oliver, M.J.; Velten, J.; Mishler, B.D. Desiccation Tolerance in Bryophytes: A Reflection of the Primitive Strategy for Plant Survival in Dehydrating Habitats? Integr. Comp. Biol. 2005, 45, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Gaff, D.F.; Oliver, M. The Evolution of Desiccation Tolerance in Angiosperm Plants: A Rare yet Common Phenomenon. Funct. Plant Biol. 2013, 40, 315–328. [Google Scholar] [CrossRef]
- Oliver, M.J.; Tuba, Z.; Mishler, B.D. The Evolution of Vegetative Desiccation Tolerance in Land Plants. Plant Ecol. 2000, 151, 85–100. [Google Scholar] [CrossRef]
- Hoekstra, F.A.; Golovina, E.A.; Buitink, J. Mechanisms of Plant Desiccation Tolerance. Trends Plant Sci. 2001, 6, 431–438. [Google Scholar] [CrossRef]
- Moore, J.P.; Le, N.T.; Brandt, W.F.; Driouich, A.; Farrant, J.M. Towards a Systems-based Understanding of Plant Desiccation Tolerance. Trends Plant Sci. 2009, 14, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Farrant, J.M.; Cooper, K.; Hilgart, A.; Abdalla, K.O.; Bentley, J.; Thomson, J.A.; Dace, H.J.W.; Peton, N.; Mundree, S.G.; Rafudeen, M.S. A Molecular Physiological Review of Vegetative Desiccation Tolerance in the Resurrection Plant Xerophyta viscosa (Baker). Planta 2015, 242, 407–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeekens, S.; Ma, J.; Hanson, J.; Rolland, F. Sugar Signals and Molecular Networks Controlling Plant Growth. Curr. Opin. Plant Biol. 2010, 13, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Bolouri-Moghaddam, M.R.; Le Roy, K.; Xiang, L.; Rolland, F.; Van den Ende, W. Sugar Signalling and Antioxidant Network Connections in Plant Cells. FEBS J. 2010, 277, 2022–2037. [Google Scholar] [CrossRef] [PubMed]
- Farrant, J.M.; Moore, J.P. Programming Desiccation-tolerance: From Plants to Seeds to Resurrection Plants. Curr. Opin. Plant Biol. 2011, 14, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, B.J.W.; Costa, M.C.D.; Maia, J.; Bentsink, L.; Ligterink, W.; Hilhorst, H.W.M. Acquisition and Loss of Desiccation Tolerance in Seeds: From Experimental Model to Biological Relevance. Planta 2015, 241, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Dinakar, C.; Bartels, D. Light Response, Oxidative Stress Management and Nucleic Acid Stability in Closely Related Linderniaceae Species Differing in Desiccation Tolerance. Planta 2012, 236, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Gechev, T.; Dinakar, C.; Benina, M.; Toneva, V.; Bartels, D. Molecular Mechanisms of Desiccation Tolerance in Resurrection Plants. Cell. Mol. Life Sci. 2012, 69, 3175–3186. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, C.A.; Gaff, D.F.; Neale, A.D. Drying Without Senescence in Resurrection Plants. Front. Plant Sci. 2014, 5, 36. [Google Scholar] [CrossRef] [PubMed]
- Marschall, M.; Proctor, M.C.F.; Smirnoff, N. Carbohydrate Composition and Invertase Activity of the Leafy Liverwort Porella platyphylla. New Phytol. 1998, 138, 343–353. [Google Scholar] [CrossRef]
- Adams, R.P.; Kendall, E.; Kartha, K.K. Comparison of Free Sugars in Growing and Desiccated Plants of Selaginella lepidophylla. Biochem. Syst. Ecol. 1990, 18, 107–110. [Google Scholar] [CrossRef]
- Ghasempour, H.R.; Gaff, D.F.; Williams, R.P.W.; Gianello, R.D. Contents of Sugars in Leaves of Drying Desiccation Tolerant Flowering Plants, Particularly Grasses. Plant Growth Regul. 1998, 24, 185–191. [Google Scholar] [CrossRef]
- Whittaker, A.; Bochicchio, A.; Vazzana, C.; Lindsey, G.; Farrant, J. Changes in Leaf Hexokinase Activity and Metabolite Levels in Response to Drying in the Desiccation-tolerant Species Sporobolus stapfianus and Xerophyta viscosa. J. Exp. Biol. 2001, 52, 961–969. [Google Scholar] [CrossRef]
- Peters, S.; Mundree, S.G.; Thomson, J.A.; Farrant, J.M.; Keller, F. Protection Mechanisms in the Resurrection Plant Xerophyta viscosa (Baker): Both Sucrose and Raffinose Family Oligosaccharides (RFOs) Accumulate in Leaves in Response to Water Deficit. J. Exp. Biol. 2007, 58, 1947–1956. [Google Scholar] [CrossRef] [PubMed]
- Albini, F.M.; Murelli, C.; Finzi, P.V.; Ferrarotti, M.; Cantoni, B.; Puliga, S.; Vazzana, C. Galactinol in the Leaves of the Resurrection Plant Boea hygroscopica. Phytochemistry 1999, 51, 499–505. [Google Scholar] [CrossRef]
- Bianchi, G.; Gamba, A.; Murelli, C.; Salamini, F.; Bartels, D. Novel Carbohydrate Metabolism in the Resurrection Plant Craterostigma plantagineum. Plant J. 1991, 1, 355–359. [Google Scholar] [CrossRef]
- Egert, A.; Eicher, B.; Keller, F.; Peters, S. Evidence for Water Deficit-induced Mass Increases of Raffinose Family Oligosaccharides (RFOs) in the Leaves of Three Craterostigma Resurrection Plant Species. Front. Physiol. 2015, 6, 206. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.P.; Hearshaw, M.; Ravenscroft, N.; Lindsey, G.G.; Farrant, J.M.; Brandt, W.F. Desiccation-Induced Ultrastructural and Biochemical Changes in the Leaves of the Resurrection Plant Myrothamnus flabellifolia. Aust. J. Bot. 2007, 55, 482–491. [Google Scholar] [CrossRef]
- Müller, J.; Sprenger, N.; Bortlik, K.; Boller, T.; Wiemken, A. Desiccation Increases Sucrose Levels in Ramonda and Haberlea, Two Genera of Resurrection Plants in the Gesneriaceae. Physiol. Plant. 1997, 100, 153–158. [Google Scholar] [CrossRef]
- Illing, N.; Denby, K.J.; Collett, H.; Shen, A.; Farrant, J.M. The Signature of Seeds in Resurrection Plants: A Molecular and Physiological Comparison of Desiccation Tolerance in Seeds and Vegetative Tissues. Integr. Comp. Biol. 2005, 45, 771–787. [Google Scholar] [CrossRef] [PubMed]
- Matros, A.; Peshev, D.; Peukert, M.; Mock, H.-P.; Van den Ende, W. Sugars as Hydroxyl Radical Scavengers: Proof-of-concept by Studying the Fate of Sucralose in Arabidopsis. Plant J. 2015, 82, 822–839. [Google Scholar] [CrossRef] [PubMed]
- ElSayed, A.I.; Rafudeen, M.S.; Golldack, D. Physiological Aspects of Raffinose Family Oligosaccharides in Plants: Protection against Abiotic Stress. Plant Biol. 2014, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, R.; Wolf, S. Phloem Transport: Cellular Pathways and Molecular Trafficking. Annu. Rev. Plant Physiol. 2009, 60, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Chen, X.; Li, G. Involvement of Phospholipid Signaling in Plant Growth and Hormone Effects. Curr. Opin. Plant Biol. 2007, 10, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Thole, J.M.; Nielsen, E. Phosphoinositides in Plants: Novel Functions in Membrane Trafficking. Curr. Opin. Plant Biol. 2008, 11, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Ye, K. Nuclear Phosphoinositide Signaling Regulates Messenger RNA Export. RNA Biol. 2009, 6, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. Galactinol and Raffinose Constitute a Novel Function to Protect Plants from Oxidative Damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Peshev, D.; Vergauwen, R.; Moglia, A.; Hideg, E.; Van den Ende, W. Towards Understanding Vacuolar Antioxidant Mechanisms: A Role for Fructans? J. Exp. Biol. 2013, 64, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Drennan, P.M.; Smith, M.T.; Goldsworthy, D.; van Staden, J. The Occurrence of Trehalose in the Leaves of the Desiccation-tolerant Angiosperm Myrothamnus flabellifolius welw. J. Plant Physiol. 1993, 142, 493–496. [Google Scholar] [CrossRef]
- Liu, M.-S.; Chien, C.-T.; Lin, T.-P. Constitutive Components and Induced Gene Expression Are Involved in the Desiccation Tolerance of Selaginella tamariscina. Plant Cell Physiol. 2008, 49, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Yobi, A.; Wone, B.W.M.; Xu, W.; Alexander, D.C.; Guo, L.; Ryals, J.A.; Oliver, M.J.; Cushman, J.C. Comparative Metabolic Profiling between Desiccation-sensitive and Desiccation-tolerant Species of Selaginella Reveals Insights into the Resurrection Trait. Plant J. 2012, 72, 983–999. [Google Scholar] [CrossRef] [PubMed]
- Crowe, J.H. Trehalose as a “Chemical Chaperone”. In Molecular Aspects of the Stress Response: Chaperones, Membranes and Networks; Csermely, P., Vígh, L., Eds.; Springer Science & Business Media: New York, NY, USA, 2007; pp. 143–158. [Google Scholar]
- Sarkar, S.; Davies, J.E.; Huang, Z.; Tunnacliffe, A.; Rubinsztein, D.C. Trehalose, a Novel mTOR-independent Autophagy Enhancer, Accelerates the Clearance of Mutant Huntingtin and α-Synuclein. J. Biol. Chem. 2007, 282, 5641–5652. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Njaci, I.; Moghaddam, L.; Long, H.; Dickman, M.B.; Zhang, X.; Mundree, S. Trehalose Accumulation Triggers Autophagy during Plant Desiccation. PLoS Genet. 2015, 11, e1005705. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.R.; Fischer, E.; Baron, M.; Van Den Dries, N.; Facchinelli, F.; Kutzer, M.; Rahmanzadeh, R.; Remus, D.; Bartels, D. Lindernia brevidens: A Novel Desiccation-tolerant Vascular Plant, Endemic to Ancient Tropical Rainforests. Plant J. 2008, 54, 938–948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bartels, D. Physiological Factors Determine the Accumulation of D-glycero-D-ido-octulose (D-g-D-i-oct) in the Desiccation Tolerant Resurrection Plant Craterostigma plantagineum. Funct. Plant Biol. 2016, 43, 684–694. [Google Scholar] [CrossRef]
- Huber, S.C.; Huber, J.L. Role and Regulation of Sucrose-phosphate Synthase in Higher Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Ingram, J.; Chandler, J.W.; Gallagher, L.; Salamini, F.; Bartels, D. Analysis of cDNA Clones Encoding Sucrose-phosphate Synthase in Relation to Sugar Interconversions Associated with Dehydration in the Resurrection Plant Craterostigma plantagineum Hochst. Plant Physiol. 1997, 115, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Salerno, G.L.; Curatti, L. Origin of Sucrose Metabolism in Higher Plants: When, How and Why? Trends Plant Sci. 2003, 8, 63–69. [Google Scholar] [CrossRef]
- Winter, H.; Huber, S.C. Regulation of Sucrose Metabolism in Higher Plants: Localization and regulation of Activity of Key Enzymes. Crit. Rev. Plant Sci. 2000, 19, 31–67. [Google Scholar] [CrossRef]
- Kleines, M.; Elster, R.-C.; Rodrigo, M.-J.; Blervacq, A.-S.; Salamini, F.; Bartels, D. Isolation and Expression Analysis of Two Stress-responsive Sucrose-synthase Genes from the Resurrection Plant Craterostigma plantagineum (Hochst.). Planta 1999, 209, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Velasco, R.; Salamini, F.; Bartels, D. Dehydration and ABA Increase mRNA Levels and Enzyme Activity of Cytosolic GAPDH in the Resurrection Plant Craterostigma plantagineum. Plant Mol. Biol. 1994, 26, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose Metabolism in Plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Zentella, R.; Mascorro-Gallardo, J.O.; Van Dijck, P.; Folch-Mallol, J.; Bonini, B.; Van Vaeck, C.; Gaxiola, R.; Covarrubias, A.A.; Nieto-Sotelo, J.; Thevelein, J.M.; et al. A Selaginellalepidophylla Trehalose-6-Phosphate Synthase Complements Growth and Stress-tolerance Defects in a Yeast tps1 Mutant. Plant Physiol. 1999, 119, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Pampurova, S.; Van Dijck, P. The Desiccation Tolerant Secrets of Selaginella lepidophylla: What We Have Learned so far? Plant Physiol. Biochem. 2014, 80, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Márquez-Escalante, J.A.; Figueroa-Soto, C.G.; Valenzuela-Soto, E.M. Isolation and Partial Characterization of Trehalose 6-phosphate Synthase Aggregates from Selaginella lepidophylla plants. Biochimie 2006, 88, 1505–1510. [Google Scholar] [CrossRef] [PubMed]
- Loewus, F.A.; Murthy, P.P.N. Myo-Inositol Metabolism in Plants. Plant Sci. 2000, 150, 1–19. [Google Scholar] [CrossRef]
- Majee, M.; Patra, B.; Mundree, S.G.; Majumder, A.L. Molecular Cloning, Bacterial Overexpression and Characterization of l-myo-inositol 1-Phosphate Synthase from a Monocotyledonous Resurrection Plant, Xerophyta viscosa Baker. J. Plant Biochem. Biotechnol. 2005, 14, 95–99. [Google Scholar] [CrossRef]
- Marte, B. Proteomics. Nature 2003, 422, 191. [Google Scholar] [CrossRef]
- Harten, J.B.; Eickmeier, W.G. Enzyme Dynamics of the Resurrection Plant Selaginella lepidophylla (Hook. & Grev.) Spring during Rehydration. Plant Physiol. 1986, 82, 61–64. [Google Scholar] [PubMed]
- Wang, X.; Chen, S.; Zhang, H.; Shi, L.; Cao, F.; Guo, L.; Xie, Y.; Wang, T.; Yan, X.; Dai, S. Desiccation Tolerance Mechanism in Resurrection Fern-Ally Selaginella tamariscina Revealed by Physiological and Proteomic Analysis. J. Proteome Res. 2010, 9, 6561–6577. [Google Scholar] [CrossRef] [PubMed]
- Oliver, M.J.; Jain, R.; Balbuena, T.S.; Agrawal, G.; Gasulla, F.; Thelen, J.J. Proteome Analysis of Leaves of the Desiccation-tolerant Grass, Sporobolus stapfianus, in Response to Dehydration. Phytochemistry 2011, 72, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Hu, J.; Guo, S.; Wang, J.; Cheng, Y.; Dang, X.; Wu, L.; He, Y. Proteome Analysis of Physcomitrella Patens Exposed to Progressive Dehydration and Rehydration. J. Exp. Biol. 2011, 63, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Wang, Z.; Shang, H.; Yang, W.; Hu, Z.; Phillips, J.; Deng, X. Proteome Analysis of Leaves from the Resurrection Plant Boea hygrometrica in Response to Dehydration and Rehydration. Planta 2007, 225, 1405–1420. [Google Scholar] [CrossRef] [PubMed]
- Ingle, R.A.; Schmidt, U.G.; Farrant, J.M.; Thomson, J.A.; Mundree, S.G. Proteomic Analysis of Leaf Proteins during Dehydration of the Resurrection Plant Xerophyta viscosa. Plant Cell Environ. 2007, 30, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, K.O.; Rafudeen, M.S. Analysis of the Nuclear Proteome of the Resurrection Plant Xerophyta viscosa in Response to Dehydration Stress using iTRAQ with 2DLC and Tandem Mass Spectrometry. J. Proteom. 2012, 75, 2361–2374. [Google Scholar] [CrossRef] [PubMed]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.-M.; Krüger, A.; TauqeerAlam, M.; et al. The return of Metabolism: Biochemistry and Physiology of the Pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef] [PubMed]
- Bernacchia, G.; Schwall, G.; Lottspeich, F.; Salamini, F.; Bartels, D. The Transketolase Gene Family of the Resurrection Plant Craterostigma Plantagineum: Differential Expression during the Rehydration Phase. EMBO J. 1995, 14, 610–618. [Google Scholar] [PubMed]
- Willige, B.C.; Kutzer, M.; Tebartz, F.; Bartels, D. Subcellular Localization and Enzymatic Properties of Differentially Expressed Transketolase Genes Isolated from the Desiccation Tolerant Resurrection Plant Craterostigma plantagineum. Planta 2009, 229, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Linnemann, T.V.; Schreiber, L.; Bartels, D. The Role of Transketolase and Octulose in the Resurrection Plant Craterostigma plantagineum. J. Exp. Bot. 2016, 67, 3551–3559. [Google Scholar] [CrossRef] [PubMed]
- Norwood, M.; Toldi, O.; Richter, A.; Scott, P. Investigation into the Ability of Roots of the Poikilohydric Plant Craterostigma plantagineum to Survive Dehydration Stress. J. Exp. Biol. 2003, 54, 2313–2321. [Google Scholar] [CrossRef] [PubMed]
- Dinakar, C.; Djilianov, D.; Bartels, D. Photosynthesis in Desiccation Tolerant Plants: Energy Metabolism and Antioxidative Stress Defense. Plant Sci. 2012, 182, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Farrant, J.M.; Brandt, W.; Lindsey, G.G. An Overview of Mechanisms of Desiccation Tolerance in Selected Angiosperm Resurrection Plants. Plant Stress 2007, 1, 72–84. [Google Scholar]
- Norwood, M.; Truesdale, M.R.; Richter, A.; Scott, P. Photosynthetic Carbohydrate Metabolism in the Resurrection Plant Craterostigma plantagineum. J. Exp. Biol. 2000, 51, 159–165. [Google Scholar] [CrossRef]
- Whittaker, A.; Martinelli, T.; Farrant, J.M.; Bochicchio, A.; Vazzana, C. Sucrose Phosphate Synthase Activity and the Co-ordination of Carbon Partitioning during Sucrose and Amino Acid Accumulation in Desiccation-tolerant Leaf Material of the C4 Resurrection Plant Sporobolus stapfianus during Dehydration. J. Exp. Biol. 2007, 58, 3775–3787. [Google Scholar] [CrossRef] [PubMed]
- Vicré, M.; Lerouxel, O.; Farrant, J.; Lerouge, P.; Driouich, A. Composition and Desiccation-induced Alterations of the Cell Wall in the Resurrection Plant Craterostigma wilmsii. Physiol. Plant. 2004, 120, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Gaff, D.; McGregor, G. The Effect of Dehydration and Rehydration on the Nitrogen Content of Various Fractions from Resurrection Plants. Biol. Plant. 1979, 21, 92–99. [Google Scholar] [CrossRef]
- Tymms, M.J.; Gaff, D.F. Proline Accumulation during Water Stress in Resurrection Plants. J. Exp. Biol. 1979, 30, 165–168. [Google Scholar] [CrossRef]
- Oliver, M.J.; Guo, L.; Alexander, D.C.; Ryals, J.A.; Wone, B.W.M.; Cushman, J.C. A Sister Group Contrast Using Untargeted Global Metabolomic Analysis Delineates the Biochemical Regulation Underlying Desiccation Tolerance in Sporobolus stapfianus. Plant Cell 2011, 23, 1231–1248. [Google Scholar] [CrossRef] [PubMed]
- Quartacci, M.F.; Forli, M.; Rascio, N.; Vecchia, F.D.; Bochicchio, A.; Navari-Izzo, F. Desiccation-tolerant Sporobolus stapfianus: Lipid Composition and Cellular Ultrastructure during Dehydration and Rehydration. J. Exp. Biol. 1997, 48, 1269–1279. [Google Scholar]
- Quartacci, M.F.; Glišić, O.; Stevanović, B.; Navari-Izzo, F. Plasma Membrane Lipids in the Resurrection Plant Ramonda serbica Following Dehydration and Rehydration. J. Exp. Biol. 2002, 53, 2159–2166. [Google Scholar] [CrossRef]
- Gasulla, F.; vomDorp, K.; Dombrink, I.; Zähringer, U.; Gisch, N.; Dörmann, P.; Bartels, D. The Role of Lipid Metabolism in the Acquisition of Desiccation Tolerance in Craterostigma plantagineum: A Comparative Approach. Plant J. 2013, 75, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Oliver, M.J.; O’Mahony, P.; Wood, A.J. “To Dryness and beyond”—Preparation for the Dried State and Rehydration in Vegetative Desiccation-tolerant Plants. Plant Growth Regul. 1998, 24, 193–201. [Google Scholar] [CrossRef]
- Steyn, W.J.; Wand, S.J.E.; Holcroft, D.M.; Jacobs, G. Anthocyanins in Vegetative Tissues: A Proposed Unified Function in Photoprotection. New Phytol. 2002, 155, 349–361. [Google Scholar] [CrossRef]
- Sgherri, C.L.M.; Loggini, B.; Bochicchio, A.; Navari-Izzo, F. Antioxidant System in Boea hygroscopica: Changes in Response to Desiccation and Rehydration. Phytochemistry 1994, 37, 377–381. [Google Scholar] [CrossRef]
- Wolucka, B.A.; Persiau, G.; Van Doorsselaere, J.; Davey, M.W.; Demol, H.; Vandekerckhove, J.; Van Montagu, M.; Zabeau, M.; Boerjan, W. Partial Purification and Identification of GDP-mannose 3′′,5′′-epimerase of Arabidopsis thaliana, a Key Enzyme of the Plant Vitamin C Pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 14843–14848. [Google Scholar] [CrossRef] [PubMed]
- Sgherri, C.; Stevanovic, B.; Navari-Izzo, F. Role of Phenolics in the Antioxidative Status of the Resurrection Plant Ramonda serbica during Dehydration and Rehydration. Physiol. Plant. 2004, 122, 478–485. [Google Scholar] [CrossRef]
- Tapia, H.; Young, L.; Fox, D.; Bertozzi, C.R.; Koshland, D. Increasing Intracellular Trehalose is Sufficient to Confer Desiccation Tolerance to Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2015, 112, 6122–6127. [Google Scholar] [CrossRef] [PubMed]
- Petitjean, M.; Teste, M.A.; François, J.M.; Parrou, J. Yeast Tolerance to Various Stresses Relies on the Trehalose-6P Synthase (Tps1) Protein, not on Trehalose. J. Biol. Chem. 2015, 290, 16177–16190. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Fujita, M.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Engineering Drought Tolerance in Plants: Discovering and Tailoring Genes to Unlock the Future. Curr. Opin. Biotechnol. 2006, 17, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N.J.; Urwin, P.E. The Interaction of Plant Biotic and Abiotic Stresses: From Genes to the Field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [PubMed]
- Beranova-Giorgianni, S. Proteome Analysis by Two-dimensional Gel Electrophoresis and Mass Spectrometry: Strengths and Limitations. TrAC Trends Anal. Chem. 2003, 22, 273–281. [Google Scholar] [CrossRef]
- Dinakar, C.; Bartels, D. Desiccation Tolerance in Resurrection Plants: New Insights from Transcriptome, Proteome and Metabolome Analysis. Front. Plant Sci. 2013, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perround, P.F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella Genome Reveals Evolutionary Insights into the Conquest of Land by Plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Yang, G.; Zhang, L.; Yang, X.; Zhao, S.; Ji, Z.; Zhou, Q.; Hu, M.; Wang, Y.; Chen, M.; et al. The resurrection genome of Boea hygrometrica: A blueprint for survival of dehydration. Proc. Natl. Acad. Sci. USA 2015, 112, 5833–5837. [Google Scholar] [CrossRef] [PubMed]
- VanBuren, R.; Bryant, D.; Edger, P.P.; Tang, H.; Burgess, D.; Challabathula, D.; Spittle, K.; Hall, R.; Gu, J.; Lyons, E.; et al. Single-molecule Sequencing of the Desiccation-tolerant Grass Oropetium thomaeum. Nature 2015, 527, 508–511. [Google Scholar] [CrossRef] [PubMed]
| Species * | Sugar Composition in Percentage | References | |
|---|---|---|---|
| Hydrated | Desiccated | ||
| Porella platyphylla A | Glucose 0.9% | Glucose 2.6% | (Marschall et al., 1998) [19] |
| Fructose 0.7% | Fructose 2.3% | ||
| Sucrose 39.4% | Sucrose 44.6% | ||
| Fructan 59.0% | Fructan 50.6% | ||
| Selaginella lepidophylla B | Fructose 0.1% | Fructose 0% | (Adams et al., 1990) [20] |
| Glucose 3.1% | Glucose 0.2% | ||
| Mannitol 0% | Mannitol 0.1% | ||
| myo-Inostitol 0.1% | myo-Inostitol 0.03% | ||
| Sucrose 6.9% | Sucrose 23.1% | ||
| Trehalose 89.8% | Trehalose 75.6% | ||
| Trisaccharides 0.1% | Trisaccharides 0.2% | ||
| Eragrostis nindensis C | Sucrose 77.2% | Sucrose 68.6% | (Ghasempour et al., 1998) [21] |
| Glucose 13.0% | Glucose 6.5% | ||
| Fructose 4.5% | Fructose 9.0% | ||
| Trehalose 5.4 | Trehalose 2.2% | ||
| Raffinose 0.0% | Raffinose 8.0 % | ||
| Stachyose 0.0% | Stachyose 5.7% | ||
| Sporobolus stapfianus C | Sucrose 83.6% | Sucrose 99.7% | (Whittaker et al., 2001) [22] |
| Glucose 8.2% | Glucose 0.1% | ||
| Fructose 8.2% | Fructose 0.1% | ||
| Xerophyta viscosa C | Fructose 11.1% | Fructose 2.2% | (Peters et al., 2007) [23] |
| Glucose 16.6% | Glucose 2.8% | ||
| Sucrose 13.8% | Sucrose 39.8% | ||
| Galactinol 31.0% | Galactinol 0.0% | ||
| myo-Inositol 4.4% | myo-Inositol 0.5% | ||
| Raffinose 15.2% | Raffinose 29.3% | ||
| Stachyose 7.5% | Stachyose 18.1% | ||
| Verbascose 0.4% | Verbascose 7.4% | ||
| Boea hygroscopica D | Fructose 7.0% | Fructose 1.9% | (Albini et al., 1999) [24] |
| Glucose 9.2% | Glucose 3.0% | ||
| Alditols 1.8% | Alditols 0 % | ||
| myo-Inositol 2.3% | myo-Inositol 0% | ||
| Sucrose 13.4% | Sucrose 91.0% | ||
| Cellobiose 0.9% | Cellobiose 0% | ||
| Gentiobiose 0.9% | Gentiobiose 0% | ||
| Galactinol 21.1% | Galactinol 1.0% | ||
| Raffinose l8.l % | Raffinose 2.5% | ||
| Melezitose 1.4% | Melezitose 0% | ||
| Maltotriose 0.9% | Maltotriose 0% | ||
| Stachyose 10.0% | Stachyose (traces) | ||
| Pentasaccharide 10.1% | Pentasaccharide (traces) | ||
| Hexasaccharide 2.8% | Hexasaccharide 0% | ||
| Craterostigma plantagineum D | Glucose 1% | Glucose 3% | (Bianchi et al., 1991) [25] |
| Fructose 2% | Fructose 2% | ||
| Sucrose 5% | Sucrose 90% | ||
| Octulose 89% | Octulose 4% | ||
| myo-Inositol 1% | myo-Inositol 1% | ||
| Craterostigma pumilum D | Octulose 94.2% | Octulose 10.3% | (Egert et al., 2015) [26] |
| Sucrose 2.4% | Sucrose 80.3% | ||
| Galactinol 0.3% | Galactinol 2.1% | ||
| Raffinose 0.8% | Raffinose 1.5% | ||
| Stachyose 1.1% | Stachyose 2.6% | ||
| Verbascose 1.3% | Verbascose 3.1% | ||
| Myrothamnus flabellifolia D | Trehalose 30.7% | Trehalose 38.1% | (Moore et al., 2007) [27] |
| Fructose 25.9% | Fructose 7.0% | ||
| Sucrose 20.6% | Sucrose 38.9% | ||
| Glucose 17.0% | Glucose 12.1% | ||
| Stachyose 5.6% | Stachyose 1.7% | ||
| Raffinose 0.2% | Raffinose 2.2% | ||
| Ramonda nathaliae D | Glucose 3.6% | Glucose 2.1% | (Müller et al., 1997) [28] |
| Fructose 4.6% | Fructose 2.1% | ||
| myo-Inositol 1.0% | myo-Inositol 0.8% | ||
| Sucrose 40.3% | Sucrose 82.6% | ||
| Raffinose 34.3% | Raffinose 9.4% | ||
| Galactinol 16.1% | Galactinol 2.9% | ||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Song, X.; Bartels, D. Enzymes and Metabolites in Carbohydrate Metabolism of Desiccation Tolerant Plants. Proteomes 2016, 4, 40. https://doi.org/10.3390/proteomes4040040
Zhang Q, Song X, Bartels D. Enzymes and Metabolites in Carbohydrate Metabolism of Desiccation Tolerant Plants. Proteomes. 2016; 4(4):40. https://doi.org/10.3390/proteomes4040040
Chicago/Turabian StyleZhang, Qingwei, Xiaomin Song, and Dorothea Bartels. 2016. "Enzymes and Metabolites in Carbohydrate Metabolism of Desiccation Tolerant Plants" Proteomes 4, no. 4: 40. https://doi.org/10.3390/proteomes4040040
APA StyleZhang, Q., Song, X., & Bartels, D. (2016). Enzymes and Metabolites in Carbohydrate Metabolism of Desiccation Tolerant Plants. Proteomes, 4(4), 40. https://doi.org/10.3390/proteomes4040040






