Fluorescent Reporters and Biosensors for Probing the Dynamic Behavior of Protein Kinases
Abstract
:1. Introduction—Protein Kinases are Dynamic Enzymes
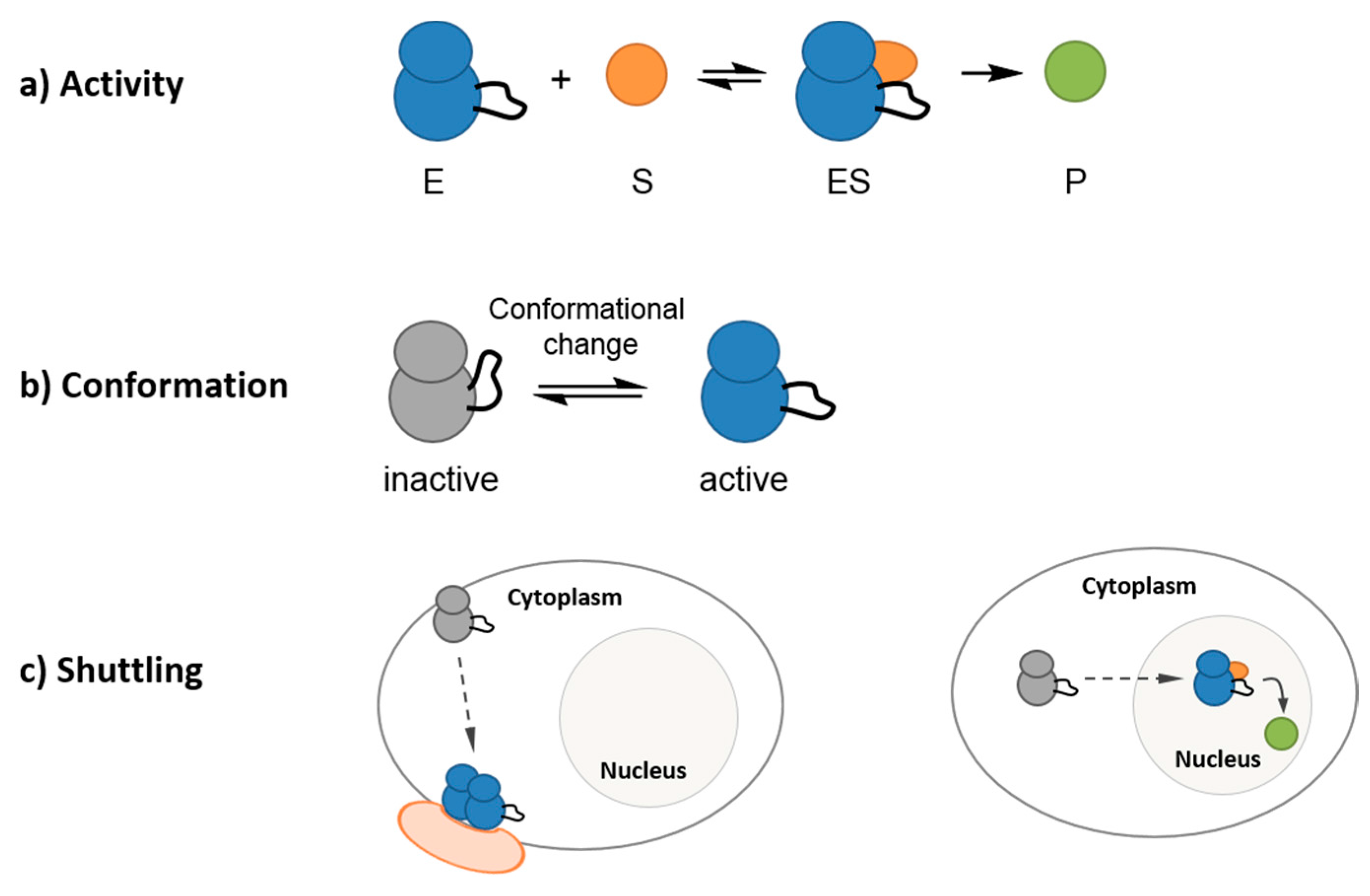

2. Fluorescent Reporters and Biosensors: Probing Kinase Dynamic Behavior in Space and in Time in a Continuous Fashion with High Temporal and Spatial Resolution
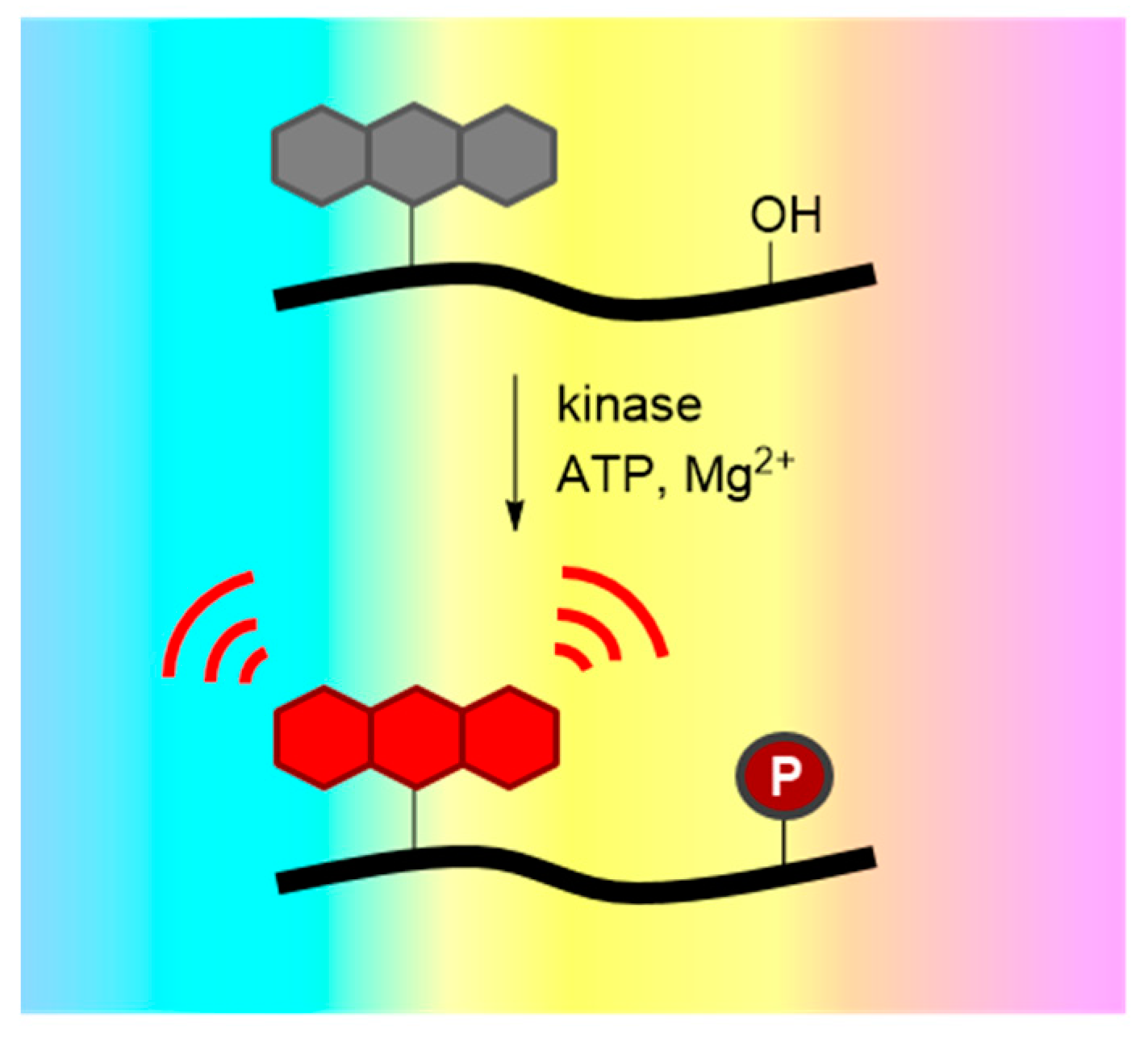
3. Probing Protein Kinase Activities in Vitro with Peptide and Protein Biosensors
3.1. Peptide and Polypeptide Kinase Biosensors
| Sensing Mechanism | Fluorophore | Protein Kinase | Assay a | Ref. |
|---|---|---|---|---|
| Environment-Sensitive Biosensors | ||||
| Probe proximal to phosphorylation site | NBD | PKC | In vitro (RE/CE); in cellulo to monitor the spatiotemporal dynamics of the PKC pathway | [36] |
| Phosphorylation-driven protein-protein interaction, based on an SH2 domain | NBD or Dapoxyl | Src | In vitro (RE) | [37] |
| CDKACT | Cy3 | CDK/Cyclin activity | In vitro (RE/CE); in cellulo probing dynamics and quantification of kinase activity | [38] |
| Merobody: fibronectin monobody conjugated to a probe | mero53 | Src | In vitro (CE); in cellulo quantification of Src activity at the edge of living cells, in correlation with protrusion and retraction activities | [39] |
| Quenching-Based Biosensors | ||||
| Self-reporting biosensor: tyrosine quencher | Pyrene | Src | In vitro (RE) | [40] |
| Cascade Yellow, Cascade Blue or Oregon Green | Src | In vitro (RE); in cellulo probing Src activation in response to stimulation | [41] | |
| Cascade Yellow or Oxazine | Abl, Lyn | In vitro (RE); in cellulo simultaneous visualization of Abl and Lyn kinases in chronic myelogenous leukemia drug-resistant cell lines | [42] | |
| Deep quench: probe/quencher/14-3-3 phosphoserine binding domain | Pyrene/Rose Bengal | PKA | In vitro (RE) | [43] |
| Coumarin/Acid Green | PKA | In vitro (RE) | [44] | |
| Quenching: probe/quencher | 5Fam, TAMRA, Atto620, Atto633 or Red681/Acid Blue or Evans Blue | PKA | In vitro (RE/CE); in cellulo to monitor endogenous cAMP-dependent protein kinase activity in erythrocytes | [45] |
| Metal-Ion Mediated Biosensors | ||||
| Ca2+-dependent | Fluorescein | PKCα | In vitro (RE) | [46] |
| Mg2+-dependent, BTF | Sox | PKA, PKC, Abl | In vitro (RE) | [47] |
| Mg2+-dependent, BTF, cell lysates | Sox | Akt, PKA, MK2 | In vitro (CE) | [48] |
| Mg2+-dependent, BTF, multiplexed assay in cell lysates | Sox | PKC, PKA, Akt1, MK2, CDK2, Pim2 | In vitro (CE) | [49] |
| Mg2+-dependent, RDF | Sox | PKC, Pim2, Akt1, MK2, PKA, Abl, Src, IRK | In vitro (RE) | [50] |
| Mg2+-dependent, RDF, protein-based docking domain (Sox-PNT) | Sox | ERK1/2 | In vitro (CE) | [51] |
| Mg2+-dependent, RDF, protein-based docking domain | Sox | p38α | In vitro (CE) | [52] |
| Sox | ERK1/2, p38α/β and JNK1/2/3 | In vitro (CE) | [53] | |
| Lanthanide-based biosensor | Tb3+/Eu3+ (Carbostyril 123) | Src, Abl | In vitro (RE) | [54] |
| Photoactivatable Biosensors | ||||
| Probe proximal to phosphorylation site, caged serine | NBD | PKC | In cellulo to monitor PKC activity in HeLa cells following microinjection and selective photoactivation | [55] |
| Probe proximal to phosphorylation site, caged serine | NBD | PKCβ | In cellulo to monitor PKCβ activity throughout mitosis in PtK2 cells | [56] |
| Self-reporting biosensor, caged tyrosine | Cascade Yellow | Src | In cellulo to follow the timing of kinase activity following microinjection and photoactivation in A549 cells | [41] |
| Quenching: probe/quencher, caged serine | Atto633/Evans Blue | PKA | In cellulo to monitor endogenous cAMP-dependent protein kinase activity in erythrocytes following microinjection and photoactivation | [45] |
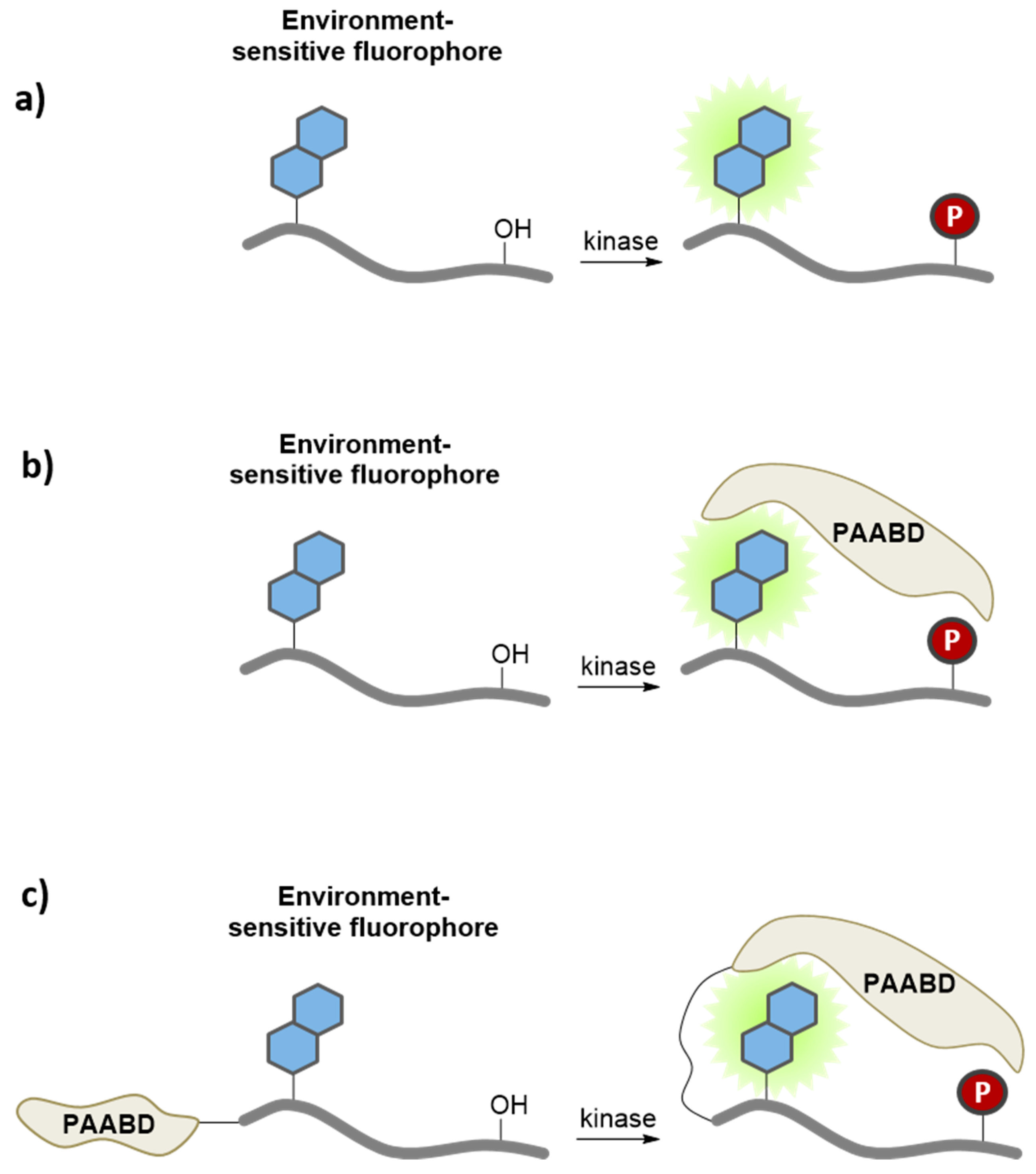
3.2. Environment-Sensitive Kinase Biosensors
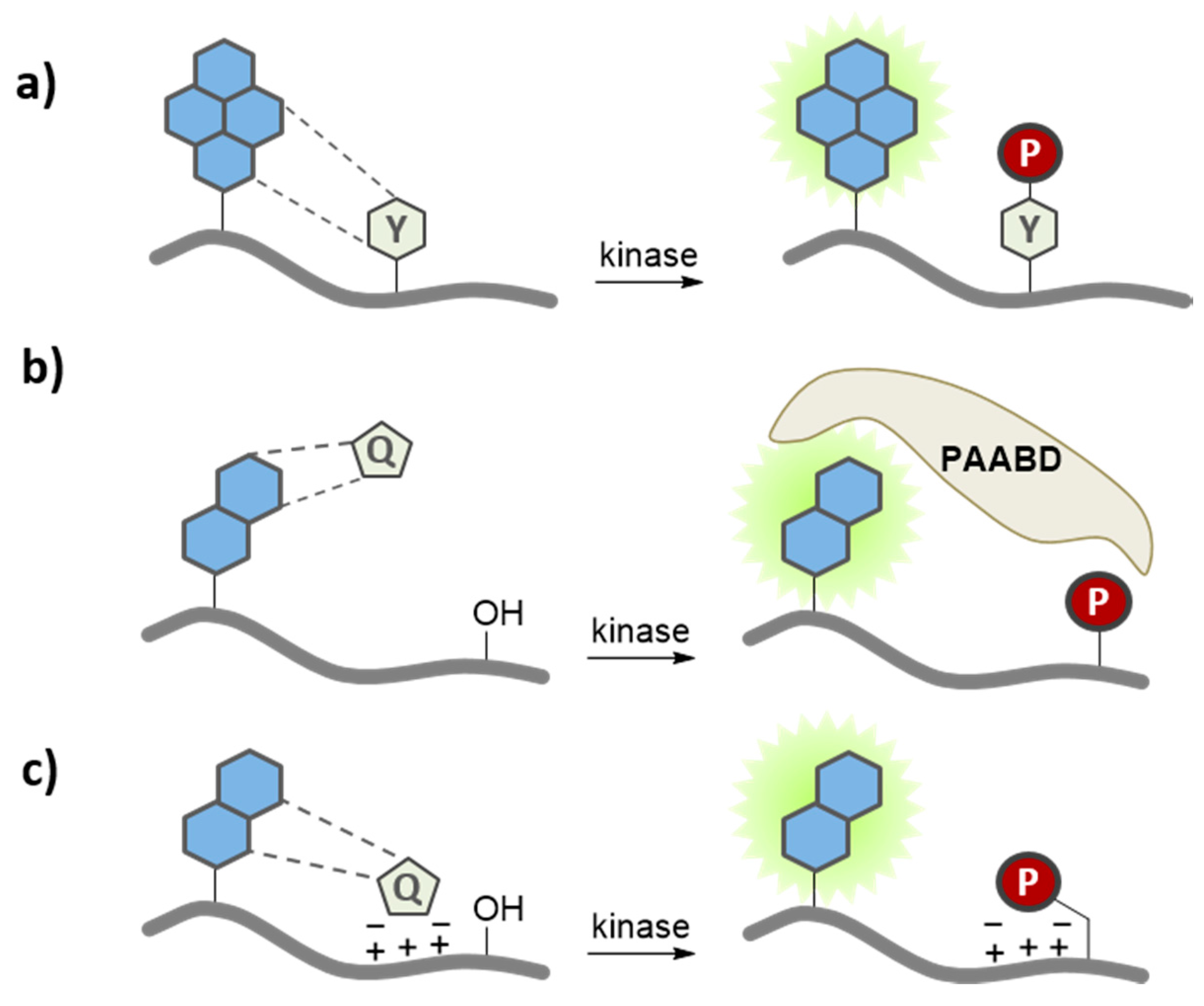
3.3. Quenching Based Kinase Biosensors
3.4. Metal-Ion Mediated Kinase Biosensors
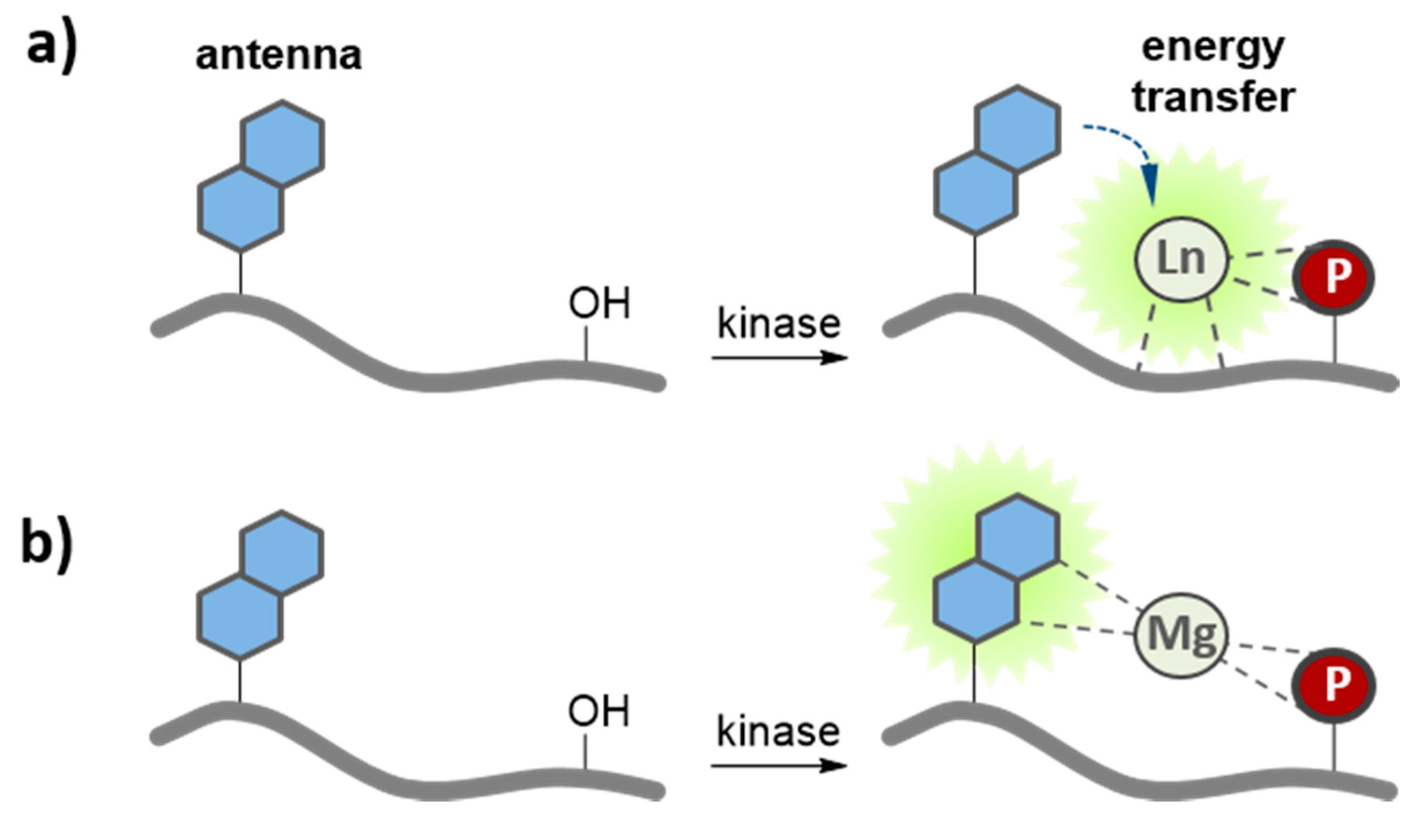
3.5. Application of Peptide and Protein Biosensors of PKs in Living Cells
4. Probing Protein Kinase Activities in Living Cells with Genetically-Encoded FRET Biosensors
| Protein Kinase | Biosensor Name | AFP FRET Pairs | Cellular Process | Ref. |
|---|---|---|---|---|
| Abl/EGFR | CrkII-based reporter | CFP/YFP | Rapid, dynamic and transient phosphorylation by CrkII upon epidermal growth factor stimulation. | [88] |
| c-Abl | Picchu | CFP/YFP | Specific phosphorylation by c-Abl. | [89] |
| Bcr-Abl | Pickles | ECFP/Venus | Clinical diagnosis of Bcr-Abl activity in CML patient cells: monitoring disease status, response to therapy, and the onset of drug-resistance within a heterogeneous population. Comparative assessment of inhibitor efficacy: evaluation of second generation inhibitors or novel compounds to treat drug-resistant mutants. | [24] |
| Aurora B | Aurora B sensor | CFP/YFP | Dynamics of Aurora B activity during anaphase. | [90] |
| AKT | AktAR | Cerulean/cpVenus | PKB/Akt signaling and dynamics in living cells. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. | [91] |
| AKT | AktUS | CFP/YFP | PKB/Akt dynamics in living cells, in the Golgi and mitochondria | [92] |
| AKT | BKAR | ECFP/Citrine | Spatio-temporal dynamics of PKB/Akt activity in real time in living cells, in the nucleus, cytosol, and plasma membrane. | [93] |
| AMPK | AMPKAR | ECFP/cpVenus | Probing AMPK activity upon cellular stress. | [94] |
| ATM | ATOMIC | CFP/YFP | Monitoring ATM kinase activity in living cells and in response to double strand breaks. | [95] |
| CAMKII | Camui | CFP/YFP | Activation of calcium/calmodulin-dependent protein kinase II in living neurons and in cardiomyocytes | [96] |
| CDK1/CyclinB activity | CDK1 sensor | mCerulean/YPet | Progressive activation of CyclinB1-Cdk1 at the G2/M transition in living cells, just before nuclear envelope breakdown, contributing to initiate prophase. | [97] |
| ERK | EKAR | EGFP/mRFP1 | Spatiotemporal signaling dynamics of ERK kinase in HEK293 cells after epidermal growth factor stimulation, in neurons from intact brain tissue by fluorescence lifetime imaging, in the dendrites and nucleus of hippocampal pyramidal neurons in brain slices after theta-burst stimuli or trains of back-propagating action potentials. | [98] |
| ERK1 | Erkus | CFP/YFP | Spatiotemporal dynamics of cytosolic and nuclear activity of ERK in living cells | [99] |
| IR | Phocus | CFP/YFP | Phosphorylation by the insulin receptor in living cells. | [100] |
| JNK Kinase | JNKAR | EGFP/citrine | Spatiotemporal dynamics of JNK activity-signaling properties and behavior of the JNK cascade in living cells. | [101] |
| FAK Kinase | FAK sensor | ECFP/YPet | Focal adhesion kinase activity and activation at membrane microdomains. | [102] |
| Histone Phosphorylation | CFP/YFP | Histone phosphorylation in living cells. | [103] | |
| MLCK | MLCK-FIP (Ca2+/calmodulin) | CFP/YFP | Transient and regional myosin light chain kinase activation in lamella and cleavage furrows. Spatial and temporal pattern of MLCK activation, revealing enrichment at the spindle equator during late metaphase and maximal activation just before cleavage furrow constriction. | [104] |
| PKA | ART | BGFP/RGFP | cAMP-induced dynamics of PKA activation in COS-7 transfected cells. | [105] |
| PKA | AKAR1 | ECFP/YFP | PKA activity following substrate tethering. | [106] |
| PKA | AKAR2 | ECFP/Citrine | Insulin disrupts β-adrenergic signaling to protein kinase A in adipocytes. | [107] |
| PKA | AKAR | EGFP/cpVenus | Subcellular dynamics of PKA activity. | [108] |
| PKA | AKAR3 | CFP/YFP | Detection of dynamic PKA activity in the sarcoplasmic reticulum of cardiomyocytes. | [109] |
| PKC | CKAR | ECFP/Citrine | Oscillatory activity of PKC at the plasma membrane in response to histamine, associated with calcium oscillation. | [110] |
| PKC-delta | deltaCKAR | CFP/YFP | Monitoring PKCdelta activity. | [111] |
| PKC | KPC-1 (pleckstrin based) | GFP/EYFP | PKC activation through phorbol ester stimulation or upon activation of physiologically relevant pathways | [112] |
| PKA and PKC | KPAC-1 (pleckstrin based) | Monitoring PKA and PKC activities independently in living cells. | [113] | |
| PKD | DKAR | CFP/YFP | Monitoring protein kinase D dynamics and its dependence on calcium through positive feedback regulation of diacylglycerol production. | [114] |
| Plk1 | Plk sensor | CFP/YFP | Mitotic Plk1 kinase activity in human cells in a physiological context and upon checkpoint recovery. | [115] |
| SAP3K | SAP3K activity reporter | Venus/SECFP | Stimulus-specific distinctions in spatial and temporal dynamics of SAP3K activity towards MKK6 SAP2K in living cells: response to epidermal growth factor and osmostress at the plasma membrane, anisomycin and UV in the cytoplasm, etoposide in the nucleus. | [116] |
| Src | Src sensor | CFP/YFP | Dynamics of Src activation following mechanical stimuli. | [117] |
| c-Src | Srcus | CFP/YFP | Src activation by steroids in the cytosol and at the plasma membrane. Epidermal growth factor directs sex-specific steroid signaling through Src activation. | [118] |
| Syk | Syk sensor | ECFP/Ypet | Imaging and quantifying real-time activation of Syk upon immunoreceptor activation and following stimulation by the platelet-derived growth factor. | [119] |
| ZAP-70 | ROZA | CFP/YFP | Dynamics of the ZAP-70 tyrosine kinase activity in T-cell lines and primary human lymphocytes with subcellular resolution during the formation of an immunological synapse. | [120] |
| MARK | MARK sensor | ECFP/Citrine | Evaluation of microtubule affinity regulating kinase activity in living neurons. | [121] |
| RSK | Eevee-RSK | ECFP/Ypet | Probing RSK activity and quantitative evaluation of kinase inhibitors in living cells. | [122] |
| S6K | Eevee-S6K | Turquosie-GL/Ypet | Probing S6K activity and quantitative evaluation of kinase inhibitors in living cells. | [122] |
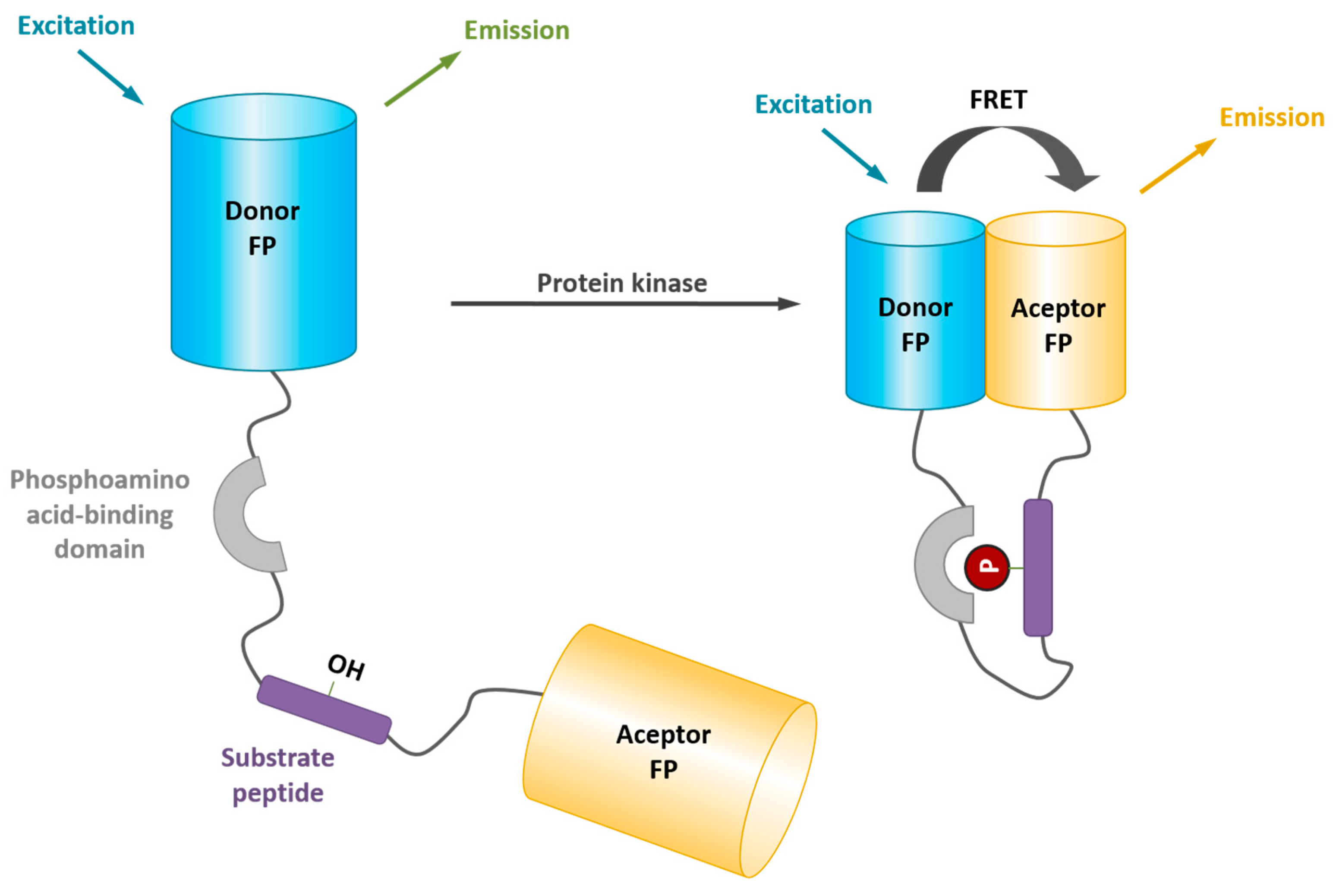
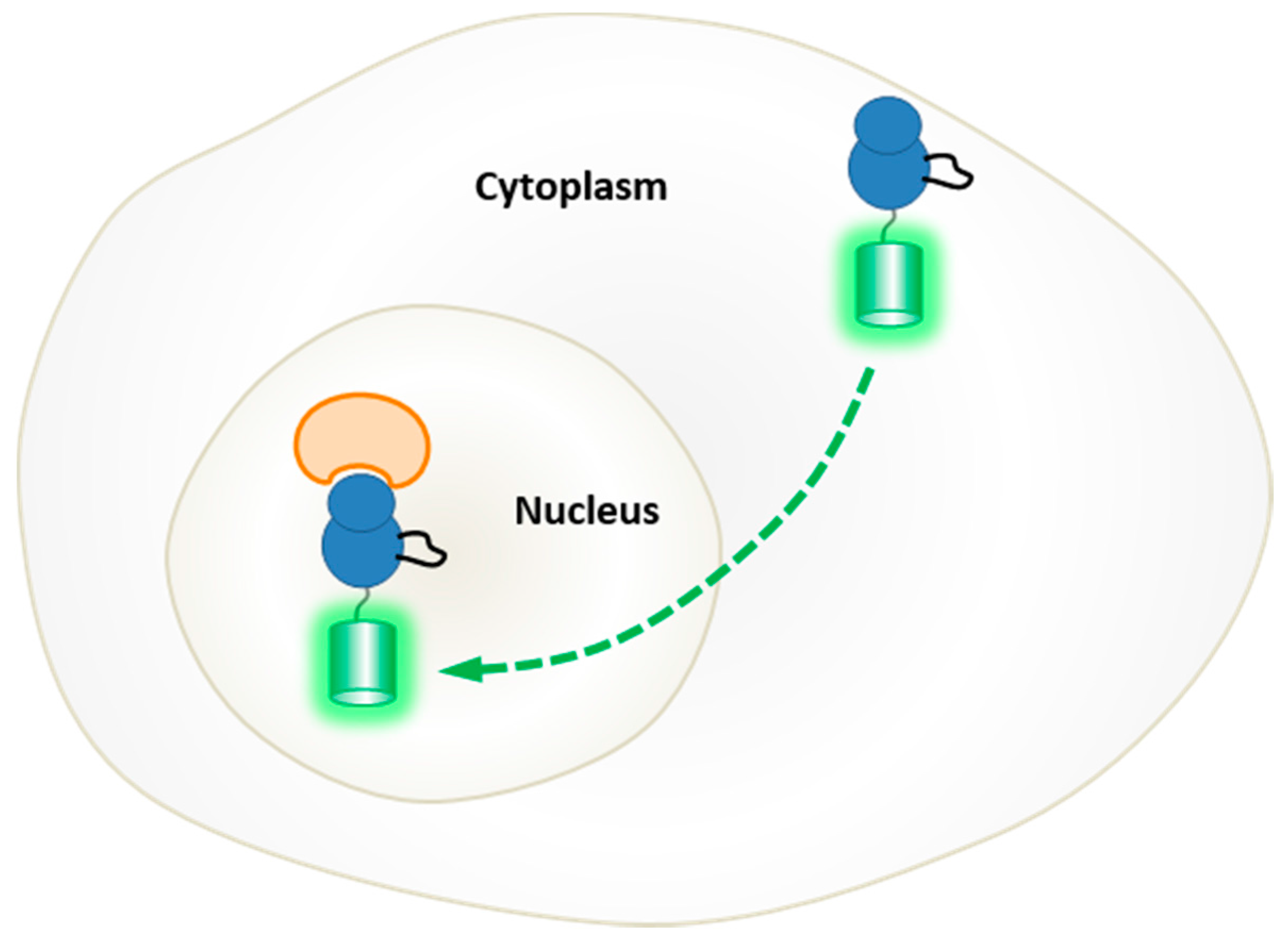
5. Probing the Spatial and Temporal Dynamics of Protein Kinases in Living Cells
5.1. Genetically-Encoded Reporters of Protein Kinases
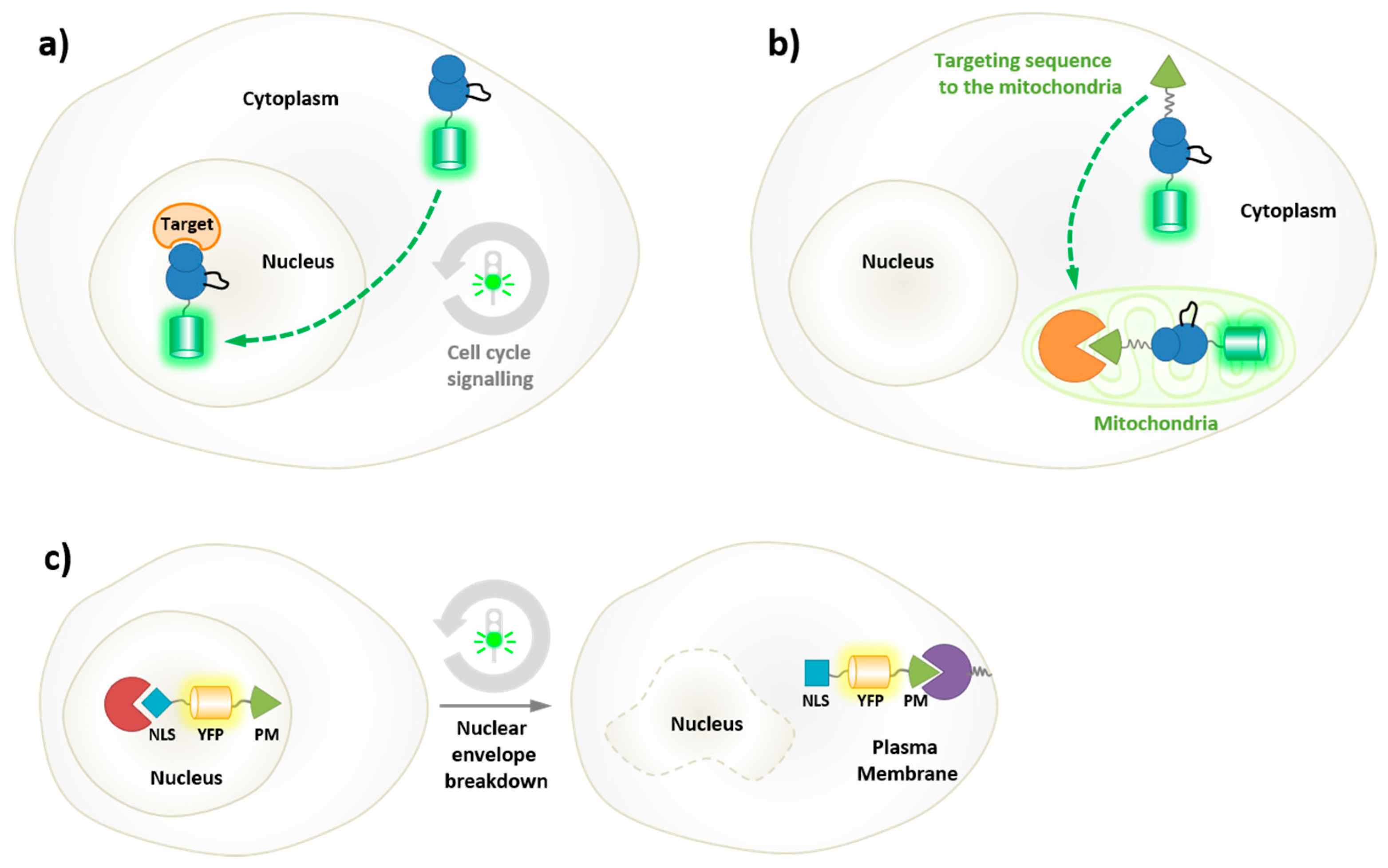
5.2. Biorthogonal Labeling and Intracellular Labeling Strategies

6. Probing the Conformational Dynamics of Protein Kinases
6.1. Conformational Dynamics of Protein Kinases
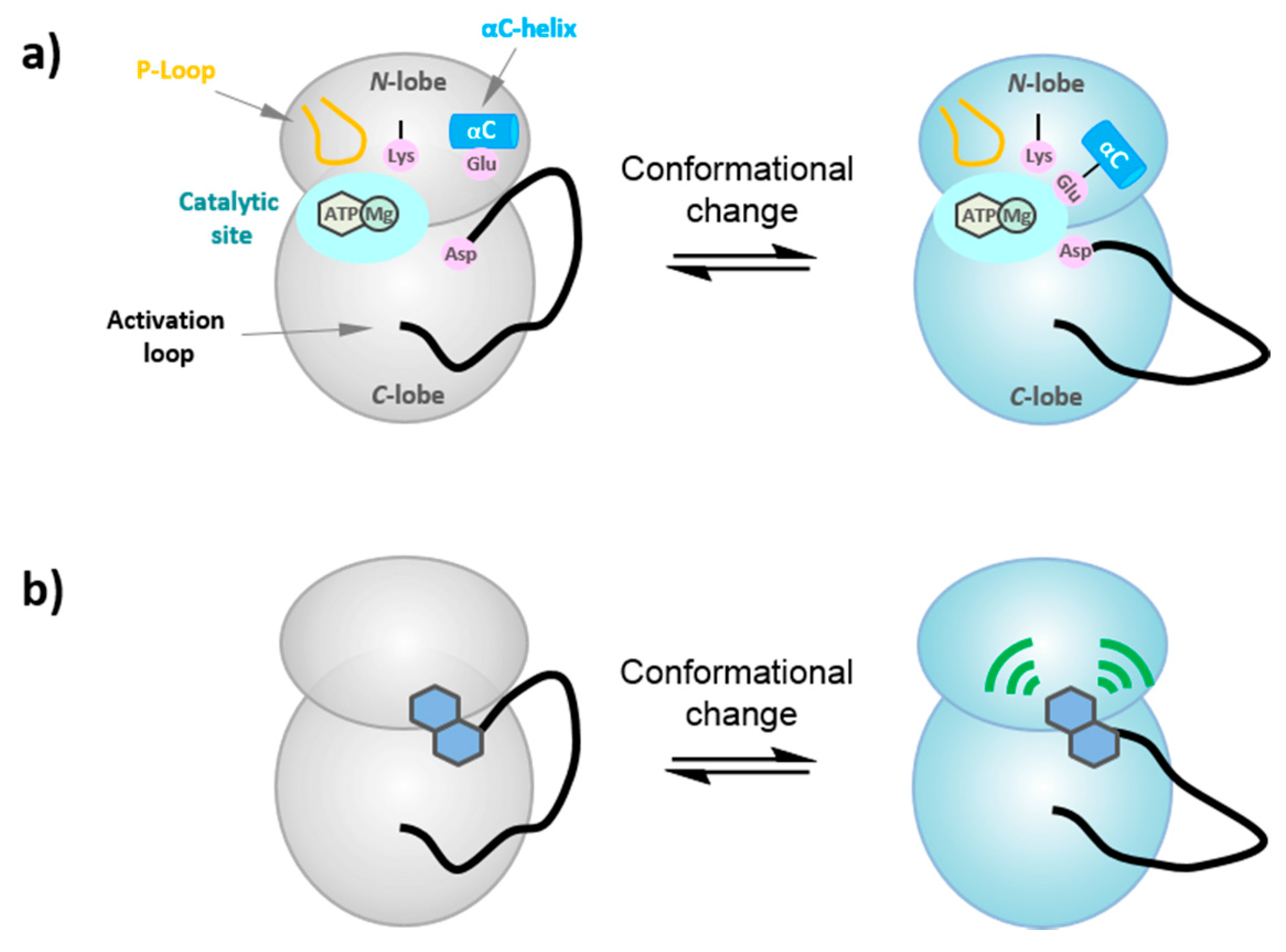
6.2. FLIK Technology
7. Recent Developments and Cutting-Edge Approaches—What Is in the Pipeline?
7.1. Near Infrared and Infrared Probes
7.2. Photoactivation Strategies
7.2.1. Caged Compounds
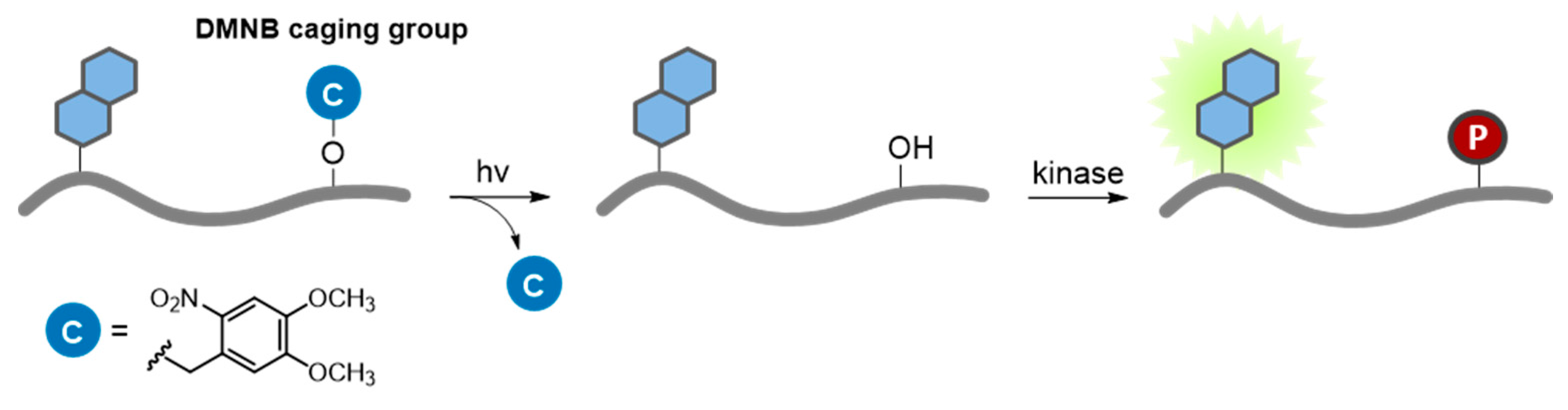
7.2.2. Photocontrollable Fluorescent Proteins—Photoswitching and Optogenetics
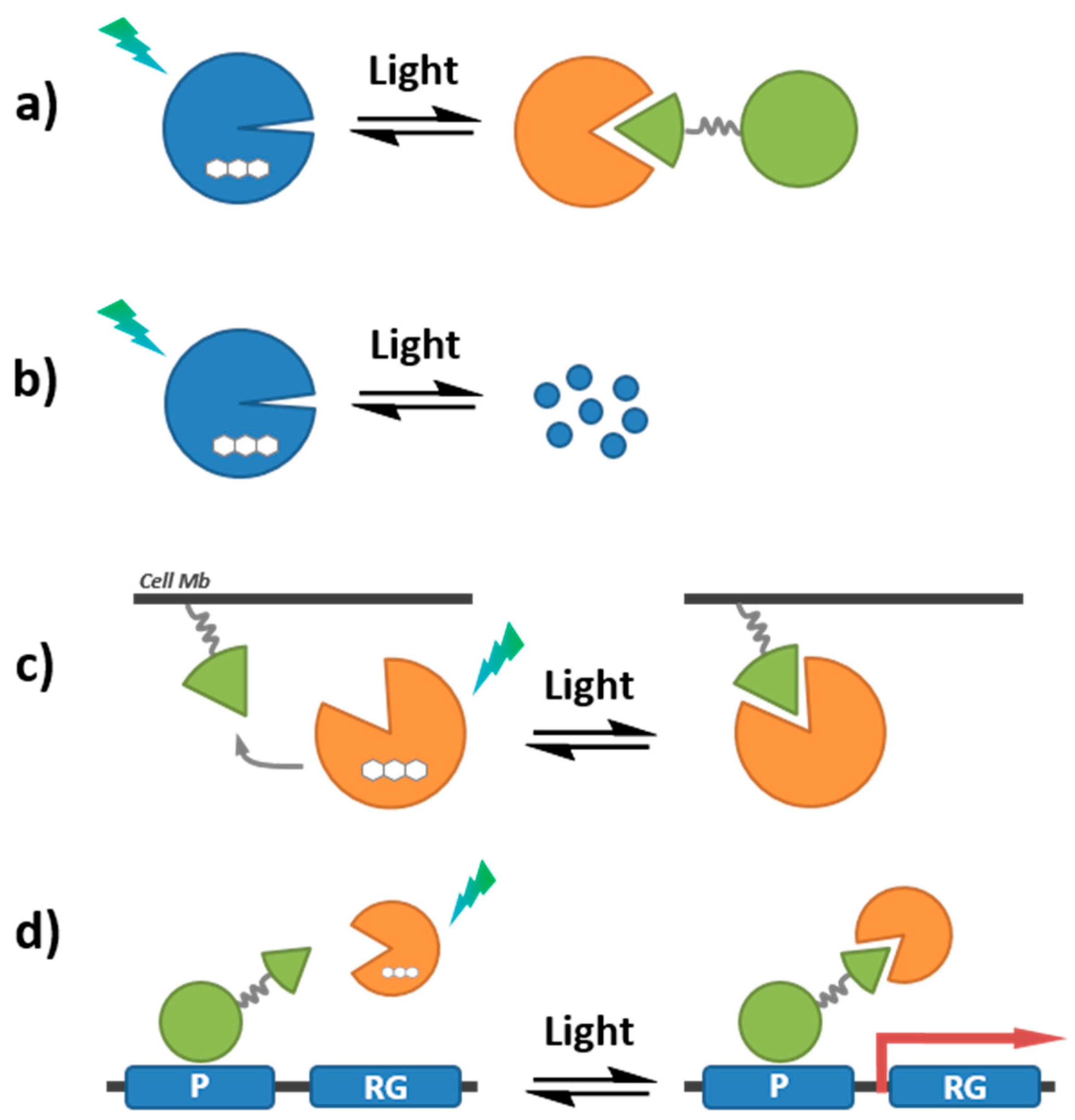
7.3. Quenching-Based Activation Strategies
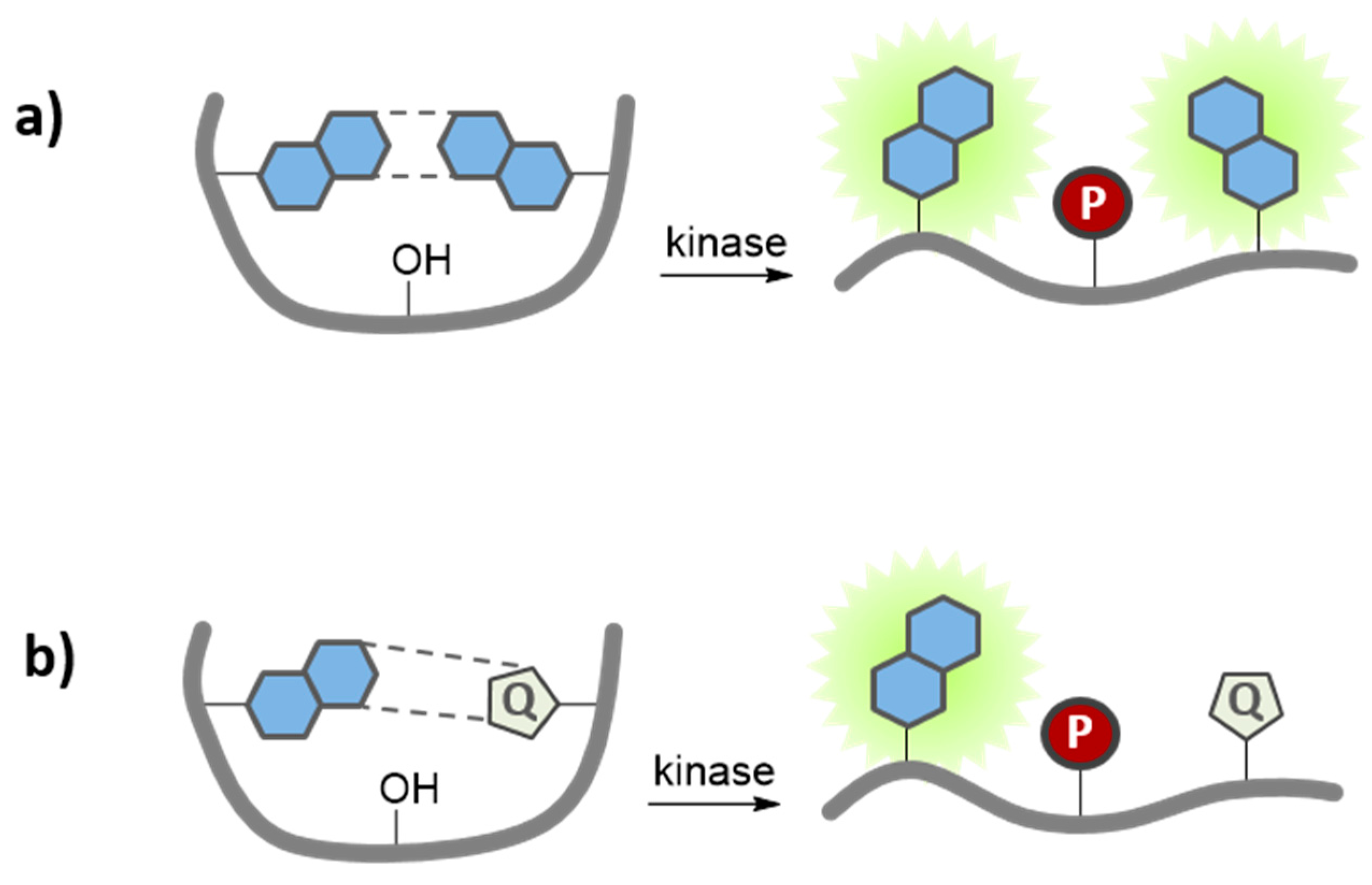
8. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hunter, T. Protein kinase classification. Methods Enzymol. 1991, 200, 3–37. [Google Scholar] [PubMed]
- Taylor, S.S.; Radzio-Andzelm, E. Three protein kinase structures define a common motif. Structure 1994, 2, 345–355. [Google Scholar] [CrossRef]
- Véron, M.; Radzio-Andzelm, E.; Tsigelny, I.; Taylor, S.S. Protein kinases share a common structural motif outside the conserved catalytic domain. Cell. Mol. Biol. 1994, 40, 587–596. [Google Scholar] [PubMed]
- Johnson, L.N.; Noble, M.E.; Owen, D.J. Active and inactive protein kinases: Structural basis for regulation. Cell 1996, 85, 149–158. [Google Scholar] [CrossRef]
- Huse, M.; Kuriyan, J. The conformational plasticity of protein kinases. Cell 2002, 109, 275–282. [Google Scholar] [CrossRef]
- Nolen, B.; Taylor, S.; Ghosh, G. Regulation of protein kinases: Controlling activity through activation segment conformation. Mol. Cell 2004, 15, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Endicott, J.A.; Noble, M.E.; Johnson, L.N. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 2012, 81, 587–613. [Google Scholar] [CrossRef] [PubMed]
- Kornev, A.P.; Haste, N.M.; Taylor, S.S.; Eyck, L.F. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA 2006, 103, 17783–17788. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Seeliger, M. Targeting Conformational Plasticity of Protein Kinases. ACS Chem. Biol. 2015, 10, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Casaletto, J.B.; McClatchey, A.I. Spatial regulation of receptor tyrosine kinases in development and cancer. Regulation of RTKs by heterodimerization. Nat. Rev. Cancer 2012, 12, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Porter, L.A.; Donoghue, D.J. Cyclin B1 and CDK1: Nuclear localization and upstream regulators. Prog. Cell Cycle Res. 2003, 5, 335–347. [Google Scholar] [PubMed]
- Jackman, M.; Kubota, Y.; den Elzen, N.; Hagting, A.; Pines, J. Cyclin A- and cyclin E-Cdk complexes shuttle between the nucleus and the cytoplasm. Mol. Biol. Cell 2002, 13, 1030–1045. [Google Scholar] [CrossRef] [PubMed]
- Kusakawa, G.; Saito, T.; Onuki, R.; Ishiguro, K.; Kishimoto, T.; Hisanaga, S. Calpain-dependent proteolytic cleavage of the p35 cyclin-dependent kinase 5 activator to p25. J. Biol. Chem. 2000, 275, 17166–17172. [Google Scholar] [CrossRef] [PubMed]
- Lemke, E.A.; Schultz, C. Principles for designing fluorescent sensors and reporters. Nat. Chem. Biol. 2011, 7, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C. Fluorescent Biosensors of Intracellular Targets from Genetically Encoded Reporters to Modular Polypeptide Probes. Cell Biochem. Biophys. 2010, 56, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Ibraheem, A.; Campbell, R.E. Designs and applications of fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2010, 14, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nakata, E.; Hamachi, I. Recent progress in strategies for the creation of protein-based fluorescent biosensors. Chembiochem 2009, 10, 2560–2577. [Google Scholar] [CrossRef] [PubMed]
- Tarrant, M.K.; Cole, P.A. The Chemical Biology of Protein Phosphorylation. Ann. Rev. Biochem. 2009, 78, 797–825. [Google Scholar] [CrossRef] [PubMed]
- González-Vera, J.A. Probing the kinome in real time with fluorescent peptides. Chem. Soc. Rev. 2011, 41, 1652–1664. [Google Scholar] [CrossRef] [PubMed]
- Pazos, E.; Vázquez, O.; Mascareñas, J.L.; Vázquez, M.E. Peptide-based fluorescent biosensors. Chem. Soc. Rev. 2009, 38, 3348–3359. [Google Scholar] [CrossRef] [PubMed]
- Van, T.N.N.; Morris, M.C. Fluorescent sensors of protein kinases: From basics to biomedical applications. Prog. Mol. Biol. Transl. Sci. 2013, 113, 217–274. [Google Scholar]
- Prével, C.; Pellerano, M.; Van, T.N.; Morris, M.C. Fluorescent biosensors for high throughput screening of protein kinase inhibitors. Biotechnol. J. 2014, 9, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C. Fluorescent Biosensors for Cancer Cell Imaging and Diagnostics. In Biosensors and Cancer; Preedy, V., Hunter, J., Eds.; CRC Press: London, UK, 2012; Chapter 6; pp. 101–124. [Google Scholar]
- Mizutani, T.; Kondo, T.; Darmanin, S.; Tsuda, M.; Tanaka, S.; Tobiume, M.; Asaka, M.; Ohba, Y. A novel FRET-based biosensor for the measurement of BCR-ABL activity and its response to drugs in living cells. Clin. Cancer Res. 2010, 16, 3964–3975. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, Y. Fluorescence Resonance Energy Transfer Biosensors for Cancer Detection and Evaluation of Drug Efficacy. Clin. Cancer Res. 2010, 16, 3822–3824. [Google Scholar] [CrossRef] [PubMed]
- Tunceroglu, A.; Matsuda, M.; Birge, R.B. Real-time fluorescent resonance energy transfer analysis to monitor drug resistance in chronic myelogenous leukemia. Mol. Cancer Ther. 2010, 9, 3065–3073. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Campbell, R.E.; Ting, A.Y.; Tsien, R.Y. Creating New Fluorescent Probes for Cell Biology. Nat. Rev. Mol. Cell Biol. 2002, 3, 906–918. [Google Scholar] [CrossRef] [PubMed]
- VanEngelenburg, S.B.; Palmer, A.E. Fluorescent biosensors of protein function. Curr. Opin. Chem. Biol. 2008, 12, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Aye-Han, N.N.; Ni, Q.; Zhang, J. Fluorescent biosensors for real-time tracking of post-translational modification dynamics. Curr. Opin. Chem. Biol. 2009, 13, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Lavis, L.; Raines, R. Bright Ideas for Chemical Biology. ACS Chem. Biol. 2008, 3, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Loving, G.; Sainlos, M.; Imperiali, B. Monitoring protein interactions and dynamics with solvatochromic fluorophores. Trends Biotechnol. 2010, 28, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Wang, Q.; Lawrence, D.S. Peptide-based fluorescent sensors of protein kinase activity: Design and applications. Biochim. Biophys. Acta 2007, 1784, 94–99. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lawrence, D.S.; Wang, Q. Seeing Is Believing: Peptide-Based Fluorescent Sensors of Protein Tyrosine Kinase Activity. Chembiochem 2007, 8, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-A.; Yeh, R.-H.; Yan, X.; Lawrence, D.S. Biosensors of protein kinase action: From in vitro assays to living cells. Biochim. Biophys. Acta 1697, 3951, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Yeh, R.-H.; Yan, X.; Cammer, M.; Bresnick, A.; Lawrence, D.S. Real time visualization of protein kinase activity in living cells. J. Biol. Chem. 2002, 277, 11527–11532. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lawrence, D.S. Phosphorylation-driven protein-protein interactions: A protein kinase sensing system. J. Am. Chem. Soc. 2005, 127, 7684–7685. [Google Scholar] [CrossRef] [PubMed]
- Van, T.N.; Pellerano, M.; Lykaso, S.; Morris, M.C. Fluorescent protein biosensor for probing CDK/cyclin activity in vitro and in living cells. Chembiochem 2014, 15, 2298–2305. [Google Scholar] [CrossRef] [PubMed]
- Gulyani, A.; Vitriol, E.; Allen, R.; Wu, J.; Gremyachinskiy, D.; Lewis, S.; Dewar, B.; Graves, L.M.; Kay, B.K.; Kuhlman, B.; et al. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nat. Chem. Biol. 2011, 7, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cahill, S.M.; Blumenstein, M.; Lawrence, D.S. Self-reporting fluorescent substrates of protein tyrosine kinases. J. Am. Chem. Soc. 2006, 128, 1808–1809. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Dai, Z.; Cahill, S.; Blumenstein, M.; Lawrence, D.S. Light-regulated sampling of protein tyrosine kinase activity. J. Am. Chem. Soc. 2006, 128, 14016–14017. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zimmerman, E.; Toutchkine, A.; Martin, T.; Graves, L.; Lawrence, D.S. Multicolor monitoring of dysregulated protein kinases in chronic myelogenous leukemia. ACS Chem. Biol. 2010, 5, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Agnes, R.; Lawrence, D.S. Deep quench: An expanded dynamic range for protein kinase sensors. J. Am. Chem. Soc. 2007, 129, 2742–2743. [Google Scholar] [CrossRef] [PubMed]
- Agnes, R.; Jernigan, F.; Shell, J.; Sharma, V.; Lawrence, D.S. Suborganelle Sensing of Mitochondrial cAMP-Dependent Protein Kinase Activity. J. Am. Chem. Soc. 2010, 132, 6075–6080. [Google Scholar] [CrossRef] [PubMed]
- Oien, N.P.; Nguyen, L.T.; Jernigan, F.E.; Priestman, M.A.; Lawrence, D.S. Long-wavelength fluorescent reporters for monitoring protein kinase activity. Angew. Chem. Int. Ed. 2014, 53, 3975–3978. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-A.; Yeh, R.-H.; Lawrence, D.S. Design and synthesis of a fluorescent reporter of protein kinase activity. J. Am. Chem. Soc. 2002, 124, 3840–3841. [Google Scholar] [CrossRef] [PubMed]
- Shults, M.; Imperiali, B. Versatile fluorescence probes of protein kinase activity. J. Am. Chem. Soc. 2003, 125, 14248–14249. [Google Scholar] [CrossRef] [PubMed]
- Shults, M.; Janes, K.; Lauffenburger, D.; Imperiali, B. A multiplexed homogeneous fluorescence-based assay for protein kinase activity in cell lysates. Nat. Methods 2005, 2, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Shults, M.; Carrico-Moniz, D.; Imperiali, B. Optimal Sox-based fluorescent chemosensor design for serine/threonine protein kinases. Anal. Biochem. 2006, 352, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Luković, E.; González-Vera, J.A.; Imperiali, B. Recognition-Domain Focused Chemosensors: Versatile and Efficient Reporters of Protein Kinase Activity. J. Am. Chem. Soc. 2008, 130, 12821–12827. [Google Scholar] [CrossRef] [PubMed]
- Luković, E.; Taylor, E.V.; Imperiali, B. Monitoring protein kinases in cellular media with highly selective chimeric reporters. Angew. Chem. Int. Ed. 2009, 48, 6828–6831. [Google Scholar] [CrossRef] [PubMed]
- Stains, C.; Luković, E.; Imperiali, B. A p38α-Selective Chemosensor for use in Unfractionated Cell Lysates. ACS Chem. Biol. 2011, 6, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Stains, C.; Tedford, N.; Walkup, T.; Luković, E.; Goguen, B.; Griffith, L.; Lauffenburger, D.; Imperiali, B. Interrogating Signaling Nodes Involved in Cellular Transformations Using Kinase Activity Probes. Chem. Biol. 2012, 19, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, M.; Lee, M.; Sames, D. A luminescent sensor for tyrosine phosphorylation. Org. Lett. 2008, 10, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Veldhuyzen, W.F.; Nguyen, Q.; McMaster, G.; Lawrence, D.S. A light-activated probe of intracellular protein kinase activity. J. Am. Chem. Soc. 2003, 125, 13358–13359. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Dulyaninova, N.G.; Kumar, S.; Bresnick, A.R.; Lawrence, D.S. Visual snapshots of intracellular kinase activity at the onset of mitosis. Chem. Biol. 2007, 14, 1254–1260. [Google Scholar] [CrossRef] [PubMed]
- Klymchenko, A.; Mely, Y. Fluorescent environment-sensitive dyes as reporters of biomolecular interactions. Prog. Mol. Biol. Transl. Sci. 2013, 113, 35–58. [Google Scholar] [PubMed]
- Yaffe, M.B.; Elia, A.E. Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 2001, 13, 131–138. [Google Scholar] [CrossRef]
- Yaffe, M.B. Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell Biol. 2002, 3, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Seet, B.T.; Dikic, I.; Zhou, M.M.; Pawson, T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 2006, 7, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Pawson, T.; Kofler, M. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Curr. Opin. Cell Biol. 2009, 21, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.J.; Zhou, X.Z.; Shen, M.; Lu, K.P. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 1999, 283, 1325–1328. [Google Scholar] [CrossRef] [PubMed]
- Lowery, D.M.; Mohammad, D.H.; Elia, A.E.; Yaffe, M.B. The Polo-box domain: A molecular integrator of mitotic kinase cascades and Polo-like kinase function. Cell Cycle 2004, 3, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Marme, N.; Knemeyer, J.P.; Sauer, M.; Wolfrum, J. Inter- and intramolecular fluorescence quenching of organic dyes by tryptophan. Bioconj. Chem. 2003, 14, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ahsan, S.S.; Santiago-Berrios, M.B.; Abruña, H.D.; Webb, W.W. Mechanisms of quenching of Alexa fluorophores by natural amino acids. J. Am. Chem. Soc. 2010, 132, 7244–7245. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, M.; Zhu, Q.; Martí, A.; Dyer, J.; Halim, M.; Jockusch, S.; Turro, N.; Sames, D. Phosphorylation State-Responsive Lanthanide Peptide Conjugates: A Luminescence Switch Based on Reversible Complex Reorganization. Org. Lett. 2006, 8, 2723–2726. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, S.; Zondlo, N.J. Design of a protein kinase-inducible domain. J. Am. Chem. Soc. 2006, 128, 5590–5591. [Google Scholar] [CrossRef] [PubMed]
- Shults, M.D.; Pearce, D.A.; Imperiali, B. Modular and tunable chemosensor scaffold for divalent zinc. J. Am. Chem. Soc. 2003, 125, 10591–10597. [Google Scholar] [CrossRef] [PubMed]
- González-Vera, J.A.; Luković, E.; Imperiali, B. Synthesis of red-shifted 8-hydroxyquinoline derivatives using click chemistry and their incorporation into phosphorylation chemosensors. J. Org. Chem. 2009, 74, 7309–7314. [Google Scholar] [CrossRef] [PubMed]
- González-Vera, J.A.; Luković, E.; Imperiali, B. A rapid method for generation of selective Sox-based chemosensors of Ser/Thr kinases using combinatorial peptide libraries. Bioorg. Med. Chem. Lett. 2009, 19, 1258–1260. [Google Scholar] [CrossRef] [PubMed]
- McNeil, P.L. Direct introduction of molecules into cells. Curr. Prot. Cell Biol. 2001. [Google Scholar] [CrossRef]
- Lönn, P.; Dowdy, S.F. Cationic PTD/CPP-mediated macromolecular delivery: Charging into the cell. Expert. Opin. Drug. Deliv. 2015, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Heitz, F.; Morris, M.C.; Divita, G. Twenty years of cell-penetrating peptides: From molecular mechanisms to therapeutics. Br. J. Pharmacol. 2009, 157, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Kurzawa, L.; Pellerano, M.; Morris, M.C. PEP and CADY-mediated delivery of fluorescent peptides and proteins into living cells. Biochim. Biophys. Acta 2010, 1798, 2274–2285. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.B.; Pereira, M.P.; Kelley, S.O. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv. Drug. Deliv. Rev. 2009, 61, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Kim, S.J.; Kwag, D.S.; Kim, S.; Park, J.; Youn, Y.S.; Bae, Y.H.; Lee, E.S. Multifunctional delivery systems for advanced oral uptake of peptide/protein drugs. Curr. Pharm. Des. 2015, 21, 3097–3110. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, M.; Zhao, J.X. Recent Development of Silica Nanoparticles as Delivery Vectors for Cancer Imaging and Therapy. Nanomedicine 2014, 10, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Tabakman, S.; Welsher, K.; Dai, H. Carbon Nanotubes in Biology and Medicine: In vitro and in vivo Detection, Imaging and Drug Delivery. Nano Res. 2009, 2, 85–120. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R.Y. Breeding and building molecules to spy on cells and tumors. FEBS Lett. 2005, 579, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Daugherty, P. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 2005, 23, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Giepmans, B.; Adams, S.; Ellisman, M.; Tsien, R.Y. The Fluorescent Toolbox for Assessing Protein Location and Function. Science 2006, 312, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Chudakov, D.; Matz, M.; Lukyanov, S.; Lukyanov, K. Fluorescent Proteins and Their Applications in Imaging Living Cells and Tissues. Physiol. Rev. 2010, 90, 1103–1163. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, A. Proteins on the move: insights gained from fluorescent protein technologies. Nat. Rev. Mol. Cell. Biol. 2011, 12, 656–668. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Piatkevich, K.; Lionnet, T.; Singer, R.; Verkhusha, V. Modern fluorescent proteins and imaging technologies to study gene expression, nuclear localization, and dynamics. Curr. Opin. Cell Biol. 2011, 23, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Dean, K.M.; Palmer, A.E. Advances in fluorescence labelling strategies for dynamic cellular imaging. Nat. Chem. Biol. 2014, 10, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Betolngar, D.B.; Erard, M.; Pasquier, H.; Bousmah, Y.; Diop-Sy, A.; Guiot, E.; Vincent, P.; Mérola, F. pH sensitivity of FRET reporters based on cyan and yellow fluorescent proteins. Anal. Bioanal. Chem. 2015, 407, 4183–4193. [Google Scholar] [CrossRef] [PubMed]
- Ting, A.Y.; Kain, K.H.; Klemke, R.L.; Tsien, R.Y. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc. Natl. Acad. Sci. USA 2001, 98, 15003–15008. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, K.; Mochizuki, N.; Ohba, Y.; Mizuno, H.; Miyawaki, A.; Matsuda, M. A pair of fluorescent resonance energy transfer-based probes for tyrosine phosphorylation of the CrkII adaptor protein in vivo. J. Biol. Chem. 2001, 276, 31305–31310. [Google Scholar] [CrossRef] [PubMed]
- Fuller, B.G.; Lampson, M.A.; Foley, E.A.; Rosasco-Nitcher, S.; Le, K.V.; Tobelmann, P.; Brautigan, D.L.; Stukenberg, P.T.; Kapoor, T.M. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature 2008, 453, 1132–1136. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol. Biol. Cell 2008, 19, 4366–4373. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Sato, M.; Umezawa, Y. Fluorescent indicators for Akt/protein kinase B and dynamics of Akt activity visualized in living cells. J. Biol. Chem. 2003, 278, 30945–30951. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, M.T.; Ni, Q.; Tsien, R.Y.; Zhang, J.; Newton, A.C. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J. Biol. Chem. 2005, 280, 5581–5587. [Google Scholar] [CrossRef] [PubMed]
- Tsou, P.; Zheng, B.; Hsu, C.H.; Sasaki, A.T.; Cantley, L.C. A fluorescent reporter of AMPK activity and cellular energy stress. Cell Metab. 2011, 13, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.A.; Zhongsheng, Y.; Hunter, T. Monitoring ATM kinase activity in living cells. DNA Repair 2007, 6, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Erickson, J.R.; Patel, R.; Ferguson, A.; Bossuyt, J.; Bers, D.M. Fluorescence resonance energy transfer-based sensor Camui provides new insight into mechanisms of calcium/calmodulin-dependent protein kinase II activation in intact cardiomyocytes. Circ. Res. 2011, 109, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Gavet, O.; Pines, J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell 2010, 18, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.D.; Ehrhardt, A.G.; Cellurale, C.; Zhong, H.; Yasuda, R.; Davis, R.J.; Svoboda, K. A genetically encoded fluorescent sensor of ERK activity. Proc. Natl. Acad. Sci. USA. 2008, 105, 19264–19269. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Kawai, Y.; Umezawa, Y. Genetically encoded fluorescent indicators to visualize protein phosphorylation by extracellular signal-regulated kinase in single living cells. Anal. Chem. 2007, 79, 2570–2575. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Ozawa, T.; Inukai, K.; Asano, T.; Umezawa, Y. Fluorescent indicators for imaging protein phosphorylation in single living cells. Nat. Biotechnol. 2002, 20, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Fosbrink, M.; Aye-Han, N.N.; Cheong, R.; Levchenko, A.; Zhang, J. Visualization of JNK activity dynamics with a genetically-encoded fluorescent biosensor. Proc. Natl. Acad. Sci. USA 2010, 107, 5459–5464. [Google Scholar] [CrossRef] [PubMed]
- Seong, J.; Ouyang, M.; Kim, T.; Sun, J.; Wen, P.C.; Lu, S.; Zhuo, Y.; Llewellyn, N.M.; Schlaepfer, D.D.; Guan, J.L.; et al. Detection of focal adhesion kinase activation at membrane microdomains by fluorescence resonance energy transfer. Nat. Commun. 2011, 2, 406. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Ting, A.Y. A genetically encoded fluorescent reporter of histone phosphorylation in living cells. Angew. Chem. Int. Ed. 2004, 43, 2940–2943. [Google Scholar] [CrossRef] [PubMed]
- Chew, T.L.; Wolf, W.A.; Gallagher, P.J.; Matsumura, F.; Chisholm, R.L. A fluorescent resonant energy transfer-based biosensor reveals transient and regional myosin light chain kinase activation in lamella and cleavage furrows. J. Cell Biol. 2002, 156, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Miyazaki, M.; Aoki, R.; Zama, T.; Inouye, S.; Hirose, K.; Iino, M.; Hagiwara, M. A fluorescent indicator for visualizing cAMP-induced phosphorylation in vivo. Nat. Biotechnol. 2000, 18, 313–316. [Google Scholar] [PubMed]
- Zhang, J.; Ma, Y.; Taylor, S.S.; Tsien, R.Y. Genetically encoded reporters of protein kinase activity reveal impact of substrate tethering. Proc. Natl. Acad. Sci. USA 2001, 98, 14997–15002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hupfeld, C.J.; Taylor, S.S.; Olefsky, J.M.; Tsien, R.Y. Insulin disrupts β-adrenergenic signalling to protein kinase activity in adipocytes. Nature 2005, 437, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.D.; Zhang, J. Subcellular dynamics of protein kinase activity visualized by FRET-based reporters. Biochem. Biophys. Res. Commun. 2006, 348, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, J.; Xiang, Y.K. FRET-based direct detection of dynamic protein kinase activity on the sarcoplasmic reticulum in cardiomyocytes. Biochem. Biophys. Res. Commun. 2011, 404, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Violin, J.D.; Zhang, J.; Tsien, R.Y.; Newton, A.C. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol. 2003, 161, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Kajimoto, T.; Sawamura, S.; Tohyama, Y.; Mori, Y.; Newton, A.C. Protein kinase C (Δ)-specific activity reporter reveals agonist-evoked nuclear activity controlled by Src family of kinases. J. Biol. Chem. 2010, 285, 41896–41910. [Google Scholar] [CrossRef] [PubMed]
- Schleifenbaum, A.; Stier, G.; Gasch, A.; Sattler, M.; Schultz, C. Genetically encoded FRET probe for PKC activity based on pleckstrin. J. Am. Chem. Soc. 2004, 126, 11786–11787. [Google Scholar] [CrossRef] [PubMed]
- Brumbaugh, J.; Schleifenbaum, A.; Gasch, A.; Sattler, M.; Schultz, C. A dual parameter FRET probe for measuring PKC and PKA activity in living cells. J. Am. Chem. Soc. 2006, 128, 24–25. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, M.T.; Toker, A.; Tsien, R.Y.; Newton, A.C. Calcium-dependent regulation of protein kinase D revealed by a genetically encoded kinase activity reporter. J. Biol. Chem. 2007, 282, 6733–6742. [Google Scholar] [CrossRef] [PubMed]
- Macůrek, L.; Lindqvist, A.; Lim, D.; Lampson, M.A.; Klompmaker, R.; Freire, R.; Clouin, C.; Taylor, S.S.; Yaffe, M.B.; Medema, R.H. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 2008, 455, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Tomida, T.; Takekawa, M.; O’Grady, P.; Saito, H. Stimulus-specific distinctions in spatial and temporal dynamics of stress-activated protein kinase revealed by a fluorescence resonance energy transfer biosensor. Mol. Cell Biol. 2009, 29, 6117–6127. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Botvinick, E.L.; Zhao, Y.; Berns, M.W.; Usami, S.; Tsien, R.Y.; Chien, S. Visualizing the mechanical activation of Src. Nature 2005, 434, 1040–1045. [Google Scholar] [CrossRef] [PubMed]
- Hitosugi, T.; Sasaki, K.; Sato, M.; Suzuki, Y.; Umezawa, Y. Epidermal growth factor directs sex-specific steroid signaling through Src activation. J. Biol. Chem. 2007, 282, 10697–10706. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Sun, J.; Wu, J.; He, H.T.; Wang, Y.; Zhu, C. A FRET-based biosensor for imaging Syk activities in living cells. Cell Mol. Bioeng. 2011, 4, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Randriamampita, C.; Mouchacca, P.; Malissen, B.; Marguet, D.; Trautmann, A.; Lellouch, A.C. Dependent FRET Based Biosensor Reveals Kinase Activity at both the Immunological Synapse and the Antisynapse. PLoS ONE 2008, 3, e1521. [Google Scholar] [CrossRef] [PubMed]
- Timm, T.; von Kries, J.P.; Li, X.; Zempel, H.; Mandelkow, E.; Mandelkow, E.M. Microtubule affinity regulating kinase activity in living neurons was examined by a genetically encoded fluorescence resonance energy transfer/fluorescence lifetime imaging-based biosensor: inhibitors with therapeutic potential. J. Biol. Chem. 2011, 286, 41711–41722. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, N.; Aoki, K.; Yamada, M.; Yukinaga, H.; Fujita, Y.; Kamioka, Y.; Matsuda, M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 2011, 22, 4647–4656. [Google Scholar] [CrossRef] [PubMed]
- Depry, C.; Zhang, J. Using FRET-based reporters to visualize subcellular dynamics of protein kinase activity. Methods. Mol. Biol. 2011, 756, 285–294. [Google Scholar] [PubMed]
- Smith, J.A.; Poteet-Smith, C.E.; Malarkey, K.; Sturgill, T.W. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J. Biol. Chem. 1999, 274, 2893–2898. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Pana, S.; Luoa, Q.; Zhang, Z. Monitoring of dual bio-molecular events using FRET biosensors based on mTagBFP/sfGFP and mVenus/mKOk fluorescent protein pairs. Biosens. Bioelectron. 2013, 46, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Piljic, A.; Schultz, C. Simultaneous recording of multiple cellular events by FRET. ACS Chem. Biol. 2008, 3, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.W.; Hazelwood, K.L.; Davidson, M.W.; Campbell, R.E. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat. Methods. 2008, 5, 401–403. [Google Scholar]
- Carlson, H.J.; Campbell, R.E. Genetically encoded FRET-based biosensors for multiparameter fluorescence imaging. Curr. Opin. Biotechnol. 2009, 20, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Welch, C.M.; Elliott, H.; Danuser, G.; Hahn, K.M. Imaging the coordination of multiple signalling activities in living cells. Nat. Rev. Mol. Cell Biol. 2011, 12, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, K.A.; Taylor, D.L. Fluorescent-protein biosensors: New tools for drug discovery. Trends. Biotechnol. 1998, 16, 135–140. [Google Scholar] [CrossRef]
- Costantini, L.M.; Baloban, M.; Markwardt, M.L.; Rizzo, M.; Guo, F.; Verkhusha, V.V.; Snapp, E.L. A palette of fluorescent proteins optimized for diverse cellular environments. Nat. Commun. 2015, 6, 7670. [Google Scholar] [CrossRef] [PubMed]
- Ellenberg, J.; Lippincott-Schwartz, J.; Presley, J.F. Dual-colour imaging with GFP variants. Trends Cell Biol. 1999, 9, 52–56. [Google Scholar] [CrossRef]
- Bastiaens, P.I.; Pepperkok, R. Observing proteins in their natural habitat: The living cell. Trends Biochem. Sci. 2000, 25, 631–637. [Google Scholar] [CrossRef]
- Wouters, F.S.; Verveer, P.J.; Bastiaens, P.I. Imaging biochemistry inside cells. Trends Cell Biol. 2001, 11, 203–211. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J.; Snapp, E.; Kenworthy, A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2001, 2, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Lippincott-Schwartz, J.; Patterson, G.H. Development and Use of Fluorescent Protein Markers in Living Cells. Science 2003, 300, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Kurzawa, L.; Morris, M.C. Cell-Cycle Markers and Biosensors. Chembiochem 2010, 11, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Jackman, M.; Lindon, C.; Nigg, E.A.; Pines, J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003, 5, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.C.; Xu, N.; Luo, K.Q. Degradation of cyclin B is required for the onset of anaphase in Mammalian cells. J. Biol. Chem. 2003, 278, 37865–37873. [Google Scholar] [CrossRef] [PubMed]
- Rizzuto, R.; Brini, M.; De Giorgi, F.; Rossi, R.; Heim, R.; Tsien, R.Y.; Pozzan, T. Double labelling of subcellular structures with organelle-targeted GFP mutants in vivo. Curr. Biol. 1996, 6, 183–188. [Google Scholar] [CrossRef]
- De Giorgi, F.; Ahmed, Z.; Bastianutto, C.; Brini, M.; Jouaville, L.S.; Marsault, R.; Murgia, M.; Pinton, P.; Pozzan, T.; Rizzuto, R. Targeting GFP to organelles. Methods Cell Biol. 1999, 58, 75–85. [Google Scholar] [PubMed]
- Jones, J.; Myers, J.; Ferrell, J.; Meyer, T. Probing the precision of the mitotic clock with a live-cell fluorescent biosensor. Nat. Biotechnol. 2004, 22, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Thomas, N. Lighting the circle of life: Fluorescent sensors for covert surveillance of the cell cycle. Cell Cycle 2003, 2, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Sakaue-Sawano, A.; Kurokawa, H.; Morimura, T.; Hanyu, A.; Hama, H.; Osawa, H.; Kashiwagi, S.; Fukami, K.; Miyata, T.; Miyoshi, H.; et al. Visualizing Spatiotemporal Dynamics of Multicellular Cell-Cycle Progression. Cell 2007, 132, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Jing, C.; Cornish, V.W. Chemical tags for labeling proteins inside living cells. Accounts Chem. Res. 2011, 44, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.; Cornish, V. Selective chemical labeling of proteins in living cells. Curr. Opin. Chem. Biol. 2005, 9, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.; Ting, A.Y. Site-specific labeling of proteins with small molecules in live cells. Curr. Opin. Biotechnol. 2005, 16, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Keppler, A.; Gendreizig, S.; Gronemeyer, T.; Pick, H.; Vogel, H.; Johnsson, K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003, 21, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Keppler, A.; Pick, H.; Arrivoli, C.; Vogel, H.; Johnsson, K. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. USA 2004, 101, 9955–9959. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.; Howarth, M.; Lin, W.; Ting, A.Y. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods 2005, 2, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Jäger, M.; Nir, E.; Weiss, S. Site-specific labeling of proteins for single-molecule FRET by combining chemical and enzymatic modification. Protein Sci. 2006, 15, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Ting, A. Transglutaminase-Catalyzed Site-Specific Conjugation of Small-Molecule Probes to Proteins in Vitro and on the Surface of Living Cells. J. Am. Chem. Soc. 2006, 128, 4542–4543. [Google Scholar] [CrossRef] [PubMed]
- Uttamapinant, C.; White, K.A.; Baruah, H.; Thompson, S.; Fernández-Suárez, M.; Puthenveetil, S.; Ting, A.Y. A fluorophore ligase for site-specific protein labeling inside living cells. Proc. Natl. Acad. Sci. USA 2010, 107, 10914–10919. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Thompson, S.; Ting, A.Y. Structure-guided engineering of a Pacific Blue fluorophore ligase for specific protein imaging in living cells. Biochemistry 2011, 50, 8221–8225. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.S.; Nivón, L.G.; Richter, F.; Goldman, P.J.; Deerinck, T.J.; Yao, J.Z.; Richardson, D.; Phipps, W.S.; Ye, A.Z.; Ellisman, M.H.; et al. Computational design of a red fluorophore ligase for site-specific protein labeling in living cells. Proc. Natl. Acad. Sci. USA 2014, 111, E4551–E4559. [Google Scholar] [CrossRef] [PubMed]
- Simard, J.R.; Getlik, M.; Grütter, C.; Pawar, V.; Wulfert, S.; Rabiller, M.; Rauh, D. Development of a Fluorescent-tagged kinase assay system for detection and characterization of allosteric kinase inhibitors. J. Am. Chem. Soc. 2009, 131, 13286–13296. [Google Scholar] [PubMed]
- Simard, J.R.; Rauh, D. FLiK: A direct-binding assay for the identification and kinetic characterization of stabilizers of inactive kinase conformations. Methods Enzymol. 2014, 548, 147–171. [Google Scholar] [PubMed]
- Simard, J.R.; Grütter, C.; Pawar, V.; Aust, B.; Wolf, A.; Rabiller, M.; Wulfert, S.; Robubi, A.; Klüter, S.; Ottmann, C.; et al. High-throughput screening to identify inhibitors which stabilize inactive kinase conformations in p38α. J. Am. Chem. Soc. 2009, 131, 18478–18488. [Google Scholar] [CrossRef] [PubMed]
- Simard, J.R.; Getlik, M.; Grütter, C.; Schneider, R.; Wulfert, S.; Rauh, D. Fluorophore labelling of the glycine-rich loop as a method of identifying inhibitors that bind to active and inactive kinase conformations. J. Am. Chem. Soc. 2010, 132, 4152–4160. [Google Scholar] [CrossRef] [PubMed]
- Simard, J.R.; Klüter, S.; Grütter, C.; Getlik, M.; Rabiller, M.; Rode, H.B.; Rauh, D. A new screening assay for allosteric inhibitors of cSrc. Nat. Chem. Biol. 2009, 5, 394–396. [Google Scholar] [CrossRef] [PubMed]
- Getlik, M.; Simard, J.R.; Termathe, M.; Grütter, C.; Rabiller, M.; van Otterlo, W.A.; Rauh, D. Fluorophore labelled kinase detects ligands that bind within the MAPK insert of p38a kinase. PLoS ONE 2012, 7, e39713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Adrian, F.J.; Jahnke, W.; Cowan-Jacob, S.W.; Li, A.G.; Iacob, R.E.; Sim, T.; Powers, J.; Dierks, C.; Sun, F.; et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature 2010, 463, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Becker, R.; Simard, J.R.; Getlik, M.; Bohlke, N.; Janning, P.; Rauh, D. Direct Binding Assay for the Detection of Type IV allosteric Inhibitors of Abl. J. Am. Chem. Soc. 2012, 134, 9138–9141. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Suárez, M.; Ting, A.Y. Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol. 2008, 9, 929–943. [Google Scholar] [CrossRef] [PubMed]
- Waggoner, A. Fluorescent labels for proteomics and genomics. Curr. Opin. Chem. Biol. 2006, 10, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Wysocki, L.M.; Lavis, L.D. Advances in the chemistry of small molecule fluorescent probes. Curr. Opin. Chem. Biol. 2011, 15, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Xiao, Y.; Xu, Y.; Guo, X.; Qian, J.; Zhu, W. “Alive” dyes as fluorescent sensors: Fluorophore, mechanism, receptor and images in living cells. Chem. Commun. 2010, 46, 6418–6436. [Google Scholar] [CrossRef] [PubMed]
- Frangioni, J.V. In vivo near-infrared fluorescence imaging. Curr. Opin. Chem. Biol. 2003, 7, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Frangioni, J.V. New technologies for human cancer imaging. J. Clin. Oncol. 2008, 26, 4012–4021. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, V. Fluorescence molecular imaging. Annu. Rev. Biomed. Eng. 2006, 8, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Weissleder, R.; Pittet, M.J. Imaging in the era of molecular oncology. Nature 2008, 452, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Rao, J.; Dragulescu-Andrasi, A.; Yao, H. Fluorescence imaging in vivo: Recent advances. Curr. Opin. Biotechnol. 2007, 18, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Razgulin, A.; Ma, N.; Rao, J. Strategies for in vivo imaging of enzyme activity: An overview and recent advances. Chem. Soc. Rev. 2011, 40, 4186–4216. [Google Scholar] [CrossRef] [PubMed]
- Shcherbo, D.; Shemiakina, A.; Ryabova, A.V.; Luker, K.E.; Schmidt, B.T.; Souslova, E.A.; Gorodnicheva, T.V.; Strukova, L.; Shidlovskiy, K.M.; Britanova, O.V.; et al. Near-infrared fluorescent proteins. Nat. Methods 2010, 7, 827–829. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Verkhusha, V.V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 2013, 10, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Baloban, M.; Verkhusha, V.V. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr. Opin. Chem. Biol. 2015, 27, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Ho, N.; Weissleder, R.; Tung, C.H. Development of water-soluble far-red fluorogenic dyes for enzyme sensing. Tetrahedron 2006, 62, 578–585. [Google Scholar] [CrossRef]
- Marx, V. Probes: Paths to photostability. Nat. Methods 2015, 12, 187–190. [Google Scholar] [CrossRef]
- Peng, X.; Yang, Z.; Wang, J.; Fan, J.; He, Y.; Song, F.; Wang, B.; Sun, S.; Qu, J.; Qi, J.; et al. Fluorescence ratiometry and fluorescence lifetime imaging: Using a single molecular sensor for dual mode imaging of cellular viscosity. J. Am. Chem. Soc. 2011, 133, 6626–6635. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Behera, R.K.; Behera, P.K.; Mishra, B.K.; Behera, G.B. Cyanines during the 1990s: A Review. Chem. Rev. 2000, 100, 1973–2012. [Google Scholar] [CrossRef] [PubMed]
- Lukinavičius, G.; Umezawa, K.; Olivier, N.; Honigmann, A.; Yang, G.; Plass, T.; Mueller, V.; Reymond, L.; Corrêa, I.R.; Luo, Z.G.; et al. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem. 2013, 5, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Koide, Y.; Urano, Y.; Hanaoka, K.; Piao, W.; Kusakabe, M.; Saito, N.; Terai, T.; Okabe, T.; Nagano, T. Development of NIR Fluorescent Dyes Based on Si-rhodamine for in vivo Imaging. J. Am. Chem. Soc. 2012, 134, 5029–5031. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Q.; Liu, J.; Lv, X.; Liu, Y.; Zhao, Y.; Guo, W. Rhodamine-inspired far-red to near-infrared dyes and their application as fluorescence probes. Angew. Chem. Int. Ed. 2012, 51, 7634–7636. [Google Scholar] [CrossRef] [PubMed]
- Koide, Y.; Urano, Y.; Hanaoka, K.; Terai, T.; Nagano, T. Evolution of Group 14 Rhodamines as Platforms for Near-Infrared Fluorescence Probes Utilizing Photoinduced Electron Transfer. ACS Chem. Biol. 2011, 6, 600–608. [Google Scholar] [CrossRef] [PubMed]
- McCann, T.; Kosaka, N.; Koide, Y.; Mitsunaga, M.; Choyke, P.; Nagano, T.; Urano, Y.; Kobayashi, H. Activatable Optical Imaging with a Silica-Rhodamine Based Near Infrared (SiR700) Fluorophore: A comparison with cyanine based dyes. Bioconj. Chem. 2011, 22, 2531–2538. [Google Scholar] [CrossRef] [PubMed]
- Resch-Genger, U.; Grabolle, M.; Cavalière-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, M.; Le Goffand, A.; Cosnier, S. Nanomaterials for biosensing applications: A review. Front. Chem. 2014, 2, 63. [Google Scholar] [CrossRef] [PubMed]
- Leia, J.; Ju, H. Signal amplification using functional nanomaterials for biosensing. Chem. Soc. Rev. 2012, 41, 2122–2134. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Larson, D.R.; Lawrence, D.S. Illuminating the chemistry of life: Design, synthesis, and applications of “caged” and related photoresponsive compounds. ACS Chem. Biol. 2009, 4, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G.; Heckel, A. Biologically active molecules with a “light switch”. Angew. Chem. Int. Ed. 2006, 45, 4900–4921. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Xing, B. Photoactive molecules for applications in molecular imaging and cell biology. Chem. Soc. Rev. 2010, 39, 2835–2846. [Google Scholar] [CrossRef] [PubMed]
- Brieke, C.; Rohrbach, F.; Gottschalk, A.; Mayer, G.; Heckel, A. Light-Controlled Tools. Angew. Chem. Int. Ed. 2012, 51, 8446–8476. [Google Scholar] [CrossRef] [PubMed]
- Blanc, A.; Bochet, C.G. Wavelength-controlled orthogonal photolysis of protecting groups. J. Org. Chem. 2002, 67, 5567–5577. [Google Scholar] [CrossRef] [PubMed]
- Klán, P.; Šolomek, T.; Bochet, C.G.; Blanc, A.; Givens, R.; Rubina, M.; Popik, V.; Kostikov, A.; Wirz, J. Photoremovable protecting groups in chemistry and biology: Reaction mechanisms and efficacy. Chem. Rev. 2013, 113, 119–191. [Google Scholar] [CrossRef] [PubMed]
- Shell, T.; Shell, J.; Rodgers, Z.; Lawrence, D.S. Tunable Visible and Near-IR Photoactivation of Light-Responsive Compounds by Using Fluorophores as Light-Capturing Antennas. Angew. Chem. Int. Ed. 2014, 53, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Gautier, A.; Gauron, C.; Volovitch, M.; Bensimon, D.; Jullien, L.; Vriz, S. How to control proteins with light in living systems. Nat. Chem. Biol. 2014, 10, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Sengupta, P.; Lippincott-Schwartz, J.; Verkhusha, V.V. Photocontrollable fluorescent proteins for superresolution imaging. Annu. Rev. Biophys. 2014, 43, 303–329. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Shemetov, A.A.; Kaberniuk, A.A.; Verkhusha, V.V. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu. Rev. Biochem. 2015, 84, 519–550. [Google Scholar] [CrossRef] [PubMed]
- Szobota, S.; Isacoff, E.Y. Optical control of neuronal activity. Annu. Rev. Biophys. 2010, 39, 329–348. [Google Scholar] [CrossRef] [PubMed]
- Fenno, L.; Yizhar, O.; Deisseroth, K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011, 34, 389–412. [Google Scholar] [CrossRef] [PubMed]
- Airan, R.D.; Thompson, K.R.; Fenno, L.E.; Bernstein, H.; Deisseroth, K. Temporally precise in vivo control of intracellular signalling. Nature 2009, 458, 1025–1029. [Google Scholar] [CrossRef] [PubMed]
- Möglich, A.; Moffat, K. Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 2010, 9, 1286–1300. [Google Scholar] [CrossRef] [PubMed]
- Levskaya, A.; Weiner, O.D.; Lim, W.A.; Voigt, C.A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009, 461, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Pathak, G.P.; Vrana, J.D.; Tucker, C.L. Optogenetic control of cell function using engineered photoreceptors. Biol. Cell 2013, 105, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Levitz, J.; Pantoja, C.; Gaub, B.; Janovjak, H.; Reiner, A.; Hoagland, A.; Schoppik, D.; Kane, B.; Stawski, P.; Schier, A.F.; et al. Optical control of metabotropic glutamate receptors. Nat. Neurosci. 2013, 16, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.; Weber, W. Optogenetic tools for mammalian systems. Mol. Biosyst. 2013, 9, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Grusch, M.; Schelch, K.; Riedler, R.; Reichhart, E.; Differ, C.; Berger, W.; Inglés-Prieto, Á.; Janovjak, H. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 2014, 33, 1713–1726. [Google Scholar] [CrossRef] [PubMed]
- Wend, S.; Wagner, H.J.; Müller, K.; Zurbriggen, M.D.; Weber, W.; Radziwill, G. Optogenetic control of protein kinase activity in mammalian cells. ACS Synth. Biol. 2014, 3, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Choyke, P.L. Target-cancer-cell-specific activatable fluorescence imaging probes: Rational design and in vivo applications. Acc. Chem. Res. 2011, 44, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Xie, J.; Chen, X. Activatable Molecular Probes for Cancer Imaging. Curr. Top. Med. Chem. 2010, 10, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Kosaka, N.; Choyke, P.L.; Kobayashi, H. H-type dimer formation of fluorophores: A mechanism for activatable, in vivo optical molecular imaging. ACS Chem. Biol. 2009, 4, 535–546. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Vera, J.A.; Morris, M.C. Fluorescent Reporters and Biosensors for Probing the Dynamic Behavior of Protein Kinases. Proteomes 2015, 3, 369-410. https://doi.org/10.3390/proteomes3040369
González-Vera JA, Morris MC. Fluorescent Reporters and Biosensors for Probing the Dynamic Behavior of Protein Kinases. Proteomes. 2015; 3(4):369-410. https://doi.org/10.3390/proteomes3040369
Chicago/Turabian StyleGonzález-Vera, Juan A., and May C. Morris. 2015. "Fluorescent Reporters and Biosensors for Probing the Dynamic Behavior of Protein Kinases" Proteomes 3, no. 4: 369-410. https://doi.org/10.3390/proteomes3040369
APA StyleGonzález-Vera, J. A., & Morris, M. C. (2015). Fluorescent Reporters and Biosensors for Probing the Dynamic Behavior of Protein Kinases. Proteomes, 3(4), 369-410. https://doi.org/10.3390/proteomes3040369





