TCEPVDB: Artificial Intelligence-Based Proteome-Wide Screening of Antigens and Linear T-Cell Epitopes in the Poxviruses and the Development of a Repository
Abstract
1. Introduction
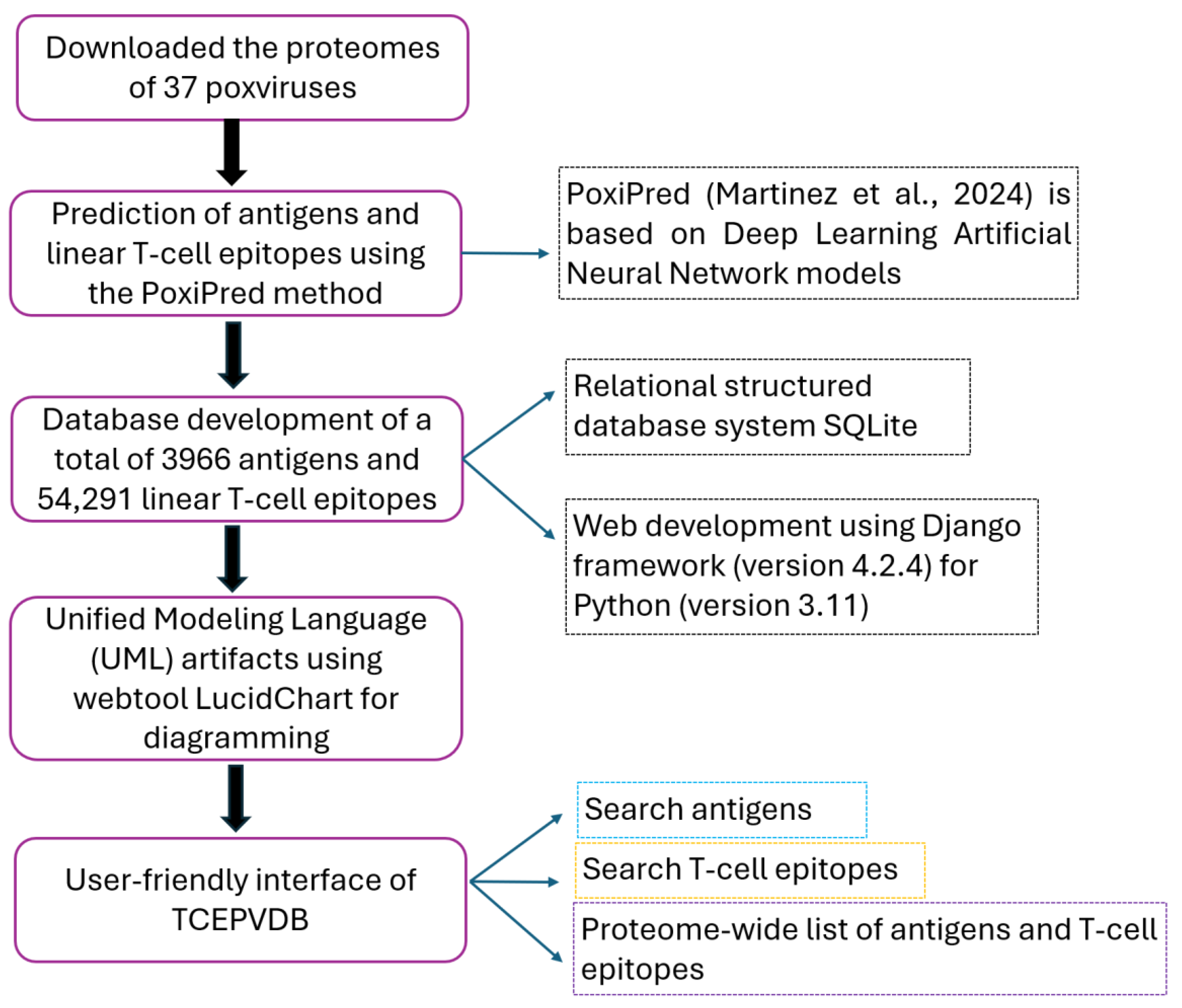
2. Materials and Methods
2.1. Obtention of Protein Répertoire
2.2. Predicting Antigens and LTCEs
2.3. Web Development
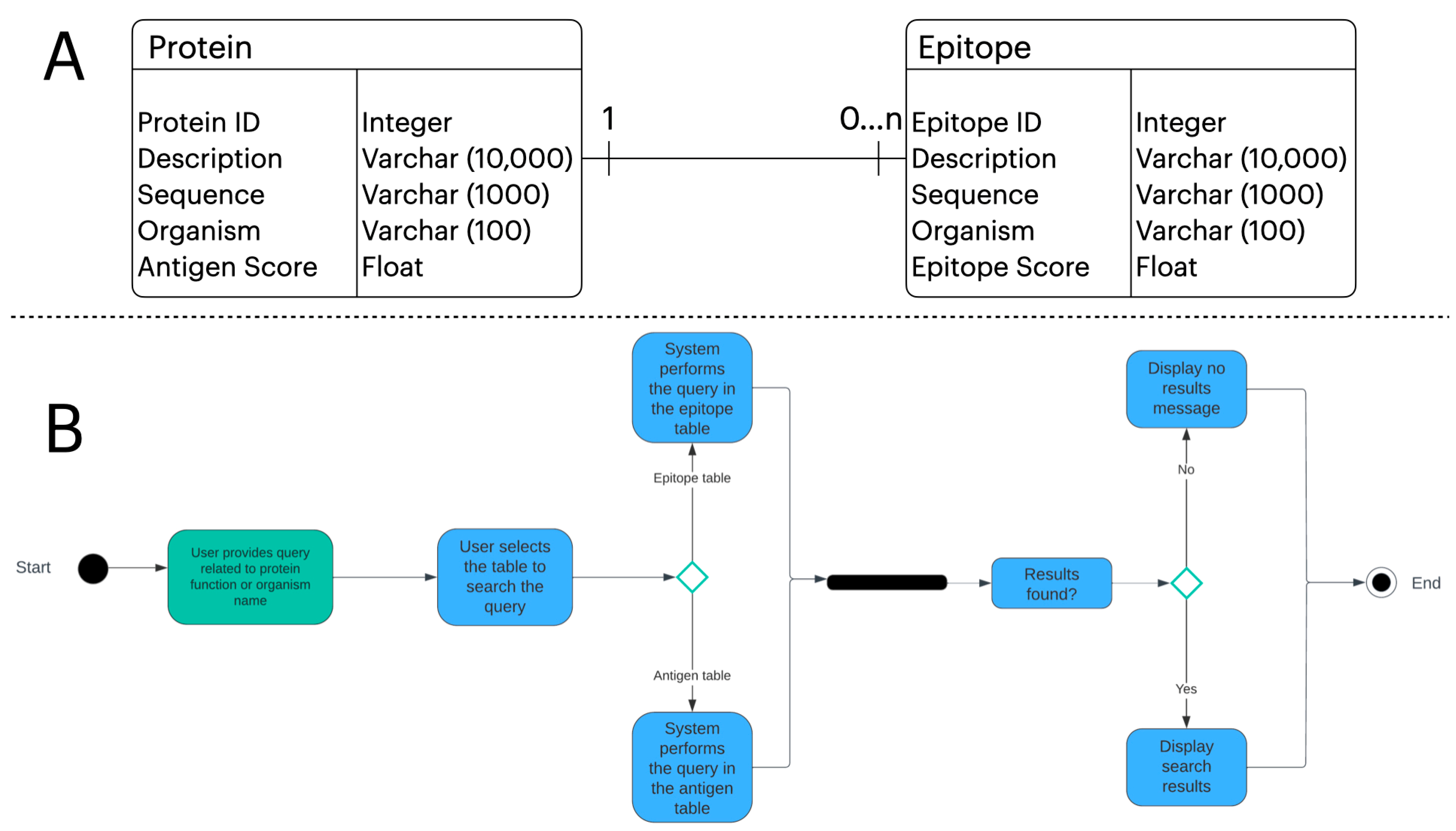
2.4. Unified Modeling Language (UML) Artifacts
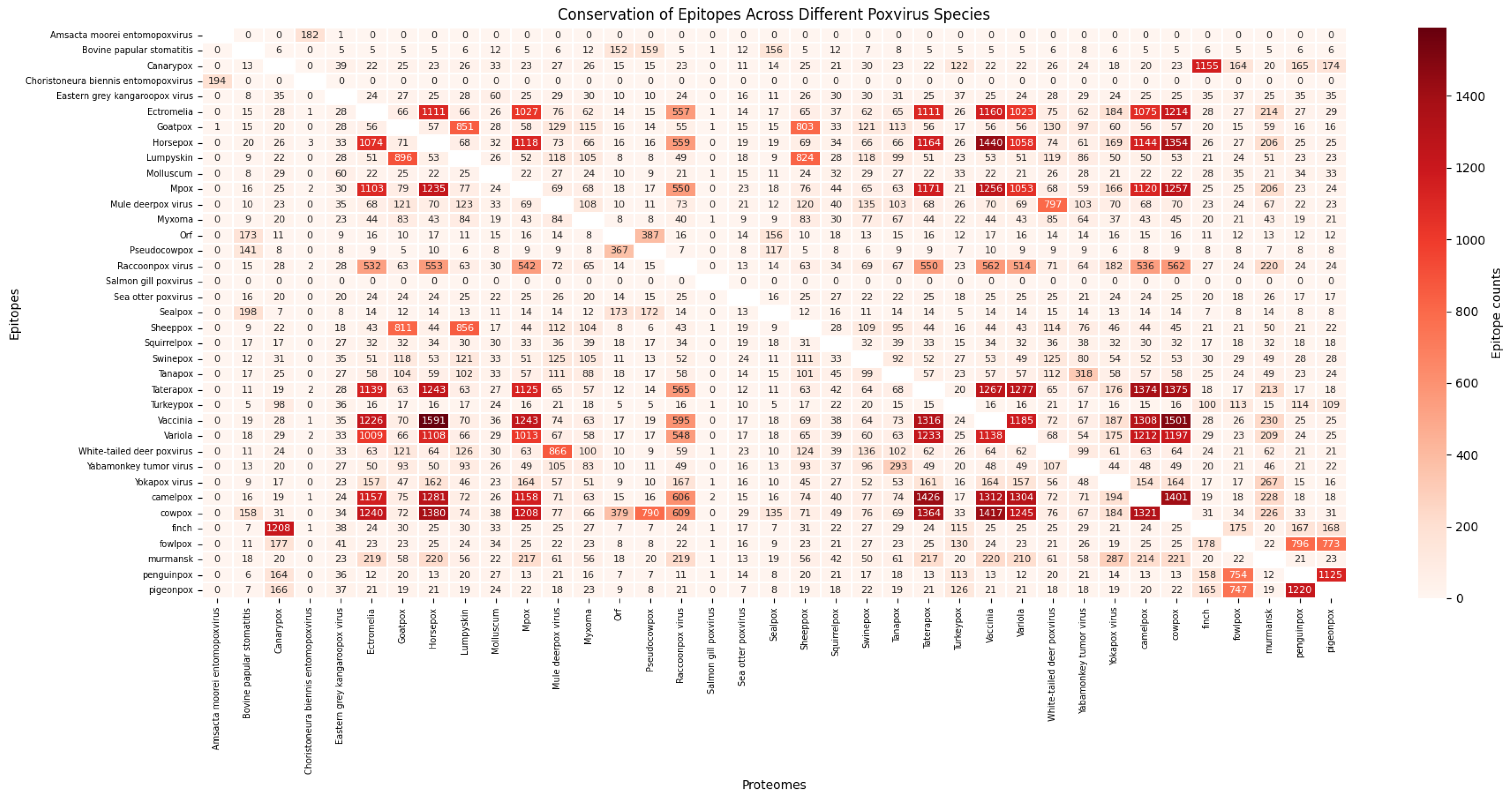
2.5. Conservation of Epitopes Across Poxvirus Species
3. Results
3.1. Organisms and Predictions
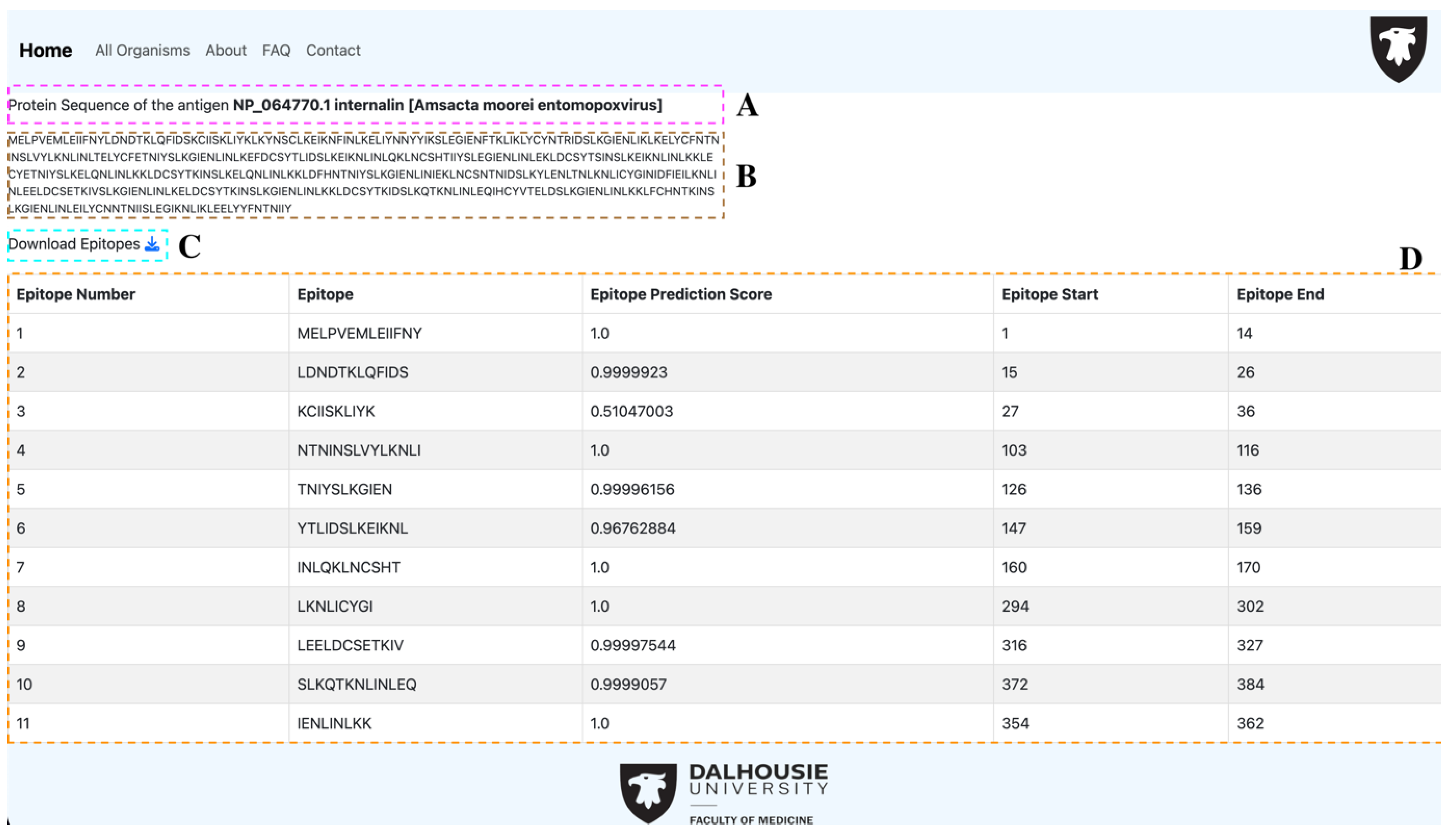
3.2. Webtool Functionalities
3.3. Conservation of Predicted Epitopes Across Poxvirus Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Günther, T.; Haas, L.; Alawi, M.; Wohlsein, P.; Marks, J.; Grundhoff, A.; Becher, P.; Fischer, N. Recovery of the first full-length genome sequence of a parapoxvirus directly from a clinical sample. Sci. Rep. 2017, 7, 3734. [Google Scholar] [CrossRef]
- Brüssow, H. Pandemic potential of poxviruses: From an ancient killer causing smallpox to the surge of monkeypox. Microb. Biotechnol. 2023, 16, 1723–1735. [Google Scholar] [CrossRef]
- Yang, Z.; Gray, M.; Winter, L. Why Do Poxviruses Still Matter? Cell Biosci. 2021, 11, 96. [Google Scholar] [CrossRef]
- 2022–2023 Mpox Outbreak Global Map. Available online: https://archive.cdc.gov/#/details?url=https://www.cdc.gov/poxvirus/mpox/response/2022/world-map.html (accessed on 21 September 2025).
- Dhar, A.D.; Werchniak, A.E.; Li, Y.; Brennick, J.B.; Goldsmith, C.S.; Kline, R.; Damon, I.; Klaus, S.N. Tanapox infection in a college student. New Engl. J. Med. 2004, 350, 361–366. [Google Scholar] [CrossRef]
- Ginzburg, V.E.; Liauchonak, I. Human orf: A typical rash in an urban medical practice. Can. Fam. Physician 2017, 63, 769–771. [Google Scholar]
- Hebert, A.A.; Bhatia, N.; Del Rosso, J.Q. Molluscum Contagiosum: Epidemiology, considerations, treatment options, and therapeutic gaps. J. Clin. Aesthetic Dermatol. 2023, 16 (Suppl. 1), S4–S11. [Google Scholar]
- Balamurugan, V.; Venkatesan, G.; Bhanuprakash, V.; Singh, R.K. Camelpox, an emerging orthopox viral disease. Indian J. Virol. 2013, 24, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Bolte, A.L.; Meurer, J.; Kaleta, E.F. Avian host spectrum of avipoxviruses. Avian Pathol. 1999, 28, 415–432. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A., Jr.; Travers, P.; Walport, M. Immunobiology: The Immune System in Health and Disease, 5th ed.; T-cell Receptor Gene Rearrangement; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Delany, I.; Rappuoli, R.; Seib, K.L. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a012476. [Google Scholar] [CrossRef]
- Sette, A.; Rappuoli, R. Reverse vaccinology: Developing vaccines in the era of genomics. Immunity 2010, 33, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.S.; Perez-Rueda, E.; Kumar, A.; Dutt, M.; Maya, C.R.; Ledesma-Dominguez, L.; Casa, P.L.; Kumar, A.; de Avila e Silva, S.; Kelvin, D.J. CDBProm: The Comprehensive Directory of Bacterial Promoters. NAR Genom. Bioinform. 2024, 6, lqae018. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.S.; Pérez-Rueda, E.; Sarkar, S.; Kumar, A.; de Ávila e Silva, S. Machine learning and statistics shape a novel path in archaeal promoter annotation. BMC Bioinform. 2022, 23, 171. [Google Scholar] [CrossRef]
- Yang, Z.; Bogdan, P.; Nazarian, S. An in silico deep learning approach to multi-epitope vaccine design: A SARS-CoV-2 case study. Sci. Rep. 2021, 11, 3238. [Google Scholar] [CrossRef]
- Keshavarzi Arshadi, A.; Webb, J.; Salem, M.; Cruz, E.; Calad-Thomson, S.; Ghadirian, N.; Collins, J.; Diez-Cecilia, E.; Kelly, B.; Goodarzi, H.; et al. Artificial Intelligence for COVID-19 Drug Discovery and Vaccine Development. Front. Artif. Intell. 2020, 3, 65. [Google Scholar] [CrossRef]
- Webby, R.J.; Perez, D.R.; Coleman, J.S.; Guan, Y.; Knight, J.H.; Govorkova, E.A.; McClain-Moss, L.R.; Peiris, J.S.; Rehg, J.E.; Tuomanen, E.I.; et al. Responsiveness to a pandemic alert: Use of reverse genetics for rapid development of influenza vaccines. Lancet 2004, 363, 1099–1103. [Google Scholar] [CrossRef]
- Masignani, V.; Pizza, M.; Moxon, E.R. The development of a vaccine against Meningococcus B using reverse vaccinology. Front. Immunol. 2019, 10, 751. [Google Scholar] [CrossRef]
- Moxon, R.; Reche, P.A.; Rappuoli, R. Editorial: Reverse Vaccinology. Front. Immunol. 2019, 10, 2776. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.S.; Dutt, M.; Kelvin, D.J.; Kumar, A. PoxiPred: An Artificial-Intelligence-Based Method for the Prediction of Potential Antigens and Epitopes to Accelerate Vaccine Development Efforts against Poxviruses. Biology 2004, 13, 125. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- He, Y.; Xiang, Z.; Mobley, H.L. Vaxign: The first web-based vaccine design program for reverse vaccinology and applications for vaccine development. BioMed Res. Int. 2010, 2010, 297505. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Cuypers, B.; Rappuoli, R.; Brozzi, A. A Lean Reverse Vaccinology Pipeline with Publicly Available Bioinformatic Tools. Methods Mol. Biol. 2023, 2673, 341–356. [Google Scholar] [CrossRef]
- D’Mello, A.; Ahearn, C.P.; Murphy, T.F.; Tettelin, H. ReVac: A reverse vaccinology computational pipeline for prioritization of prokaryotic protein vaccine candidates. BMC Genom. 2019, 20, 981. [Google Scholar] [CrossRef]
- Ras-Carmona, A.; Lehmann, A.A.; Lehmann, P.V.; Reche, P.A. Prediction of B cell epitopes in proteins using a novel sequence similarity-based method. Sci. Rep. 2022, 12, 13739. [Google Scholar] [CrossRef] [PubMed]
- Snyder, E.E.; Kampanya, N.; Lu, J.; Nordberg, E.K.; Karur, H.R.; Shukla, M.; Soneja, J.; Tian, Y.; Xue, T.; Yoo, H.; et al. PATRIC: The VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007, 35 (Suppl. 1), D401–D406. [Google Scholar] [CrossRef]
- Doneva, N.; Dimitrov, I. Viral immunogenicity prediction by machine learning methods. Int. J. Mol Sci. 2024, 25, 2949. [Google Scholar] [CrossRef]
- Kiyotani, K.; Toyoshima, Y.; Nemoto, K.; Nakamura, Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet. 2020, 65, 569–575. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. Toward the Quantitative Prediction of T-Cell Epitopes: CoMFA and CoMSIA Studies of Peptides with Affinity for the Class I MHC Molecule HLA-A*0201. J. Med. Chem. 2001, 44, 3572–3581. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Ye, X.; Sakurai, T.; Mu, Z.; Wei, L. ToxIBTL: Prediction of peptide toxicity based on information bottleneck and transfer learning. Bioinformatics 2022, 38, 1514–1524. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.S.; Amorim, V.M.F.; Soares, E.P.; de Souza, R.F.; Guzzo, C.R. Antagonistic trends between binding affinity and drug-likeness in SARS-CoV-2 Mpro inhibitors revealed by machine learning. Viruses 2025, 17, 935. [Google Scholar] [CrossRef] [PubMed]
- Kacen, A.; Javitt, A.; Kramer, M.P.; Morgenstern, D.; Tsaban, T.; Shmueli, M.D.; Teo, G.C.; Leprevost, F.D.V.; Barnea, E.; Yu, F.; et al. Post-translational modifications reshape the antigenic landscape of the MHC I immunopeptidome in tumors. Nat. Biotechnol. 2023, 41, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Pritam, M. Exploring the whole proteome of monkeypox virus to design B cell epitope-based oral vaccines using immunoinformatics approaches. Int. J. Biol. Macromol. 2023, 252, 126498. [Google Scholar] [CrossRef] [PubMed]
| Organism | Genome Accession | Proteins (n) | Predicted Antigens (n) | Predicted Linear T-Cell Epitopes (n) |
|---|---|---|---|---|
| Amsacta moorei entomopox virus | GCF_000837185.1 | 294 | 157 | 1891 |
| Bovine papular stomatitis virus | GCF_000844045.1 | 130 | 61 | 932 |
| Canarypox virus | GCF_000841685.1 | 322 | 167 | 2387 |
| Choristoneura biennis entomopoxvirus | GCF_000909015.1 | 334 | 179 | 2341 |
| Eastern grey kangaroopox virus | GCF_006450915.1 | 162 | 82 | 1167 |
| Ectromelia virus | GCF_000841905.1 | 180 | 127 | 1714 |
| Goatpox virus | GCF_000840165.1 | 149 | 68 | 1092 |
| Horsepox virus | GCF_000860085.1 | 228 | 154 | 1844 |
| Lumpy skin disease virus | GCF_000839805.1 | 156 | 77 | 1193 |
| Molluscum contagiosum virus | GCF_000843325.1 | 163 | 59 | 895 |
| Mpox virus | GCF_000857045.1 | 183 | 134 | 1777 |
| Mule deerpox virus | GCF_000861985.1 | 169 | 81 | 1113 |
| Myxoma virus | GCF_000843685.1 | 158 | 73 | 924 |
| Orf virus | GCF_000844845.1 | 130 | 52 | 737 |
| Pseudocowpox virus | GCF_000886295.1 | 125 | 55 | 775 |
| Raccoonpox virus | GCF_001029045.1 | 207 | 128 | 1750 |
| Salmon gill poxvirus | GCF_001271235.1 | 210 | 105 | 1490 |
| Sea otter poxvirus | GCF_003260795.1 | 132 | 70 | 1074 |
| Sealpox virus | GCF_002219465.1 | 119 | 52 | 820 |
| Sheeppox virus | GCF_000840205.1 | 147 | 72 | 1113 |
| Squirrelpox virus | GCF_000913615.1 | 141 | 63 | 1083 |
| Swinepox virus | GCF_000839965.1 | 146 | 75 | 1183 |
| Tanapox virus | GCF_000847185.1 | 155 | 79 | 1140 |
| Taterapox virus | GCF_000869985.1 | 220 | 140 | 1761 |
| Turkeypox virus | GCF_001431935.1 | 170 | 92 | 1400 |
| Vaccinia virus | GCF_000860085.1 | 214 | 150 | 1851 |
| Variola virus | GCF_000859885.1 | 211 | 142 | 1663 |
| White-tailed deer poxvirus | MF966153 | 171 | 85 | 1167 |
| Yaba monkey tumor virus | GCF_000845705.1 | 140 | 59 | 902 |
| Yokapox virus | GCF_000892975.1 | 186 | 105 | 1410 |
| Camelpox virus | GCF_000839105.1 | 261 | 164 | 1887 |
| Cowpox virus | GCF_000839185.1 | 214 | 142 | 1959 |
| Finch poxvirus | OM869482 | 335 | 186 | 2414 |
| Fowlpox virus | GCF_000838605.1 | 251 | 146 | 1973 |
| Murmansk poxvirus | GCF_002270885.1 | 206 | 115 | 1647 |
| Penguinpox virus | GCF_000923135.1 | 242 | 137 | 1914 |
| Pigeonpox virus | GCF_000922075.1 | 224 | 133 | 1908 |
| Name | Focus Organism | Type | Antigen Prediction Method | Epitope Type | Specificity to Poxviruses | Structural Integration | Output Option | User Interface |
|---|---|---|---|---|---|---|---|---|
| TCEPVDB | Poxviruses (n = 37) | Database (epitopes + antigen repository) | PoxiPred (ML-based, proteome-wide) | Linear T-cell epitopes | Yes | No, but output can be utilized for structure modeling | Tabular + downloadable .fasta | Web-based, with custom search |
| IEDB Epitope Tools | Broad (4700+ species) | Prediction + curated experimental database | Multiple ML-based tools (e.g., NetMHCpan) | T-cell, B-cell, MHC ligands | No, but information on multiple poxviruses can be extracted | Partial (some 3D epitope mapping) | Epitope lists, binding scores, and plots | Web-based, modular tools |
| Vaxign | Broad (bacteria, viruses, parasites) | Pipeline + database | Reverse vaccinology (genomic + ML filters | T-cell (MHC I/II), B-cell | No | Partial (subcellular localization) | Ranked antigen list + epitope predictions based on the scores | Web-based, form-driven |
| VaxiJen | Broad (pathogen-agnostic) | Standalone prediction tool | Alignment-independent auto cross-covariance | No, only antigenicity scores | Can be used to predict antigenicity scores for individual proteins of poxviruses | No | Antigenicity score | Web-based, simple input |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dutt, M.; Kumar, A.; Ostadgavahi, A.T.; Kelvin, D.J.; Martinez, G.S. TCEPVDB: Artificial Intelligence-Based Proteome-Wide Screening of Antigens and Linear T-Cell Epitopes in the Poxviruses and the Development of a Repository. Proteomes 2025, 13, 58. https://doi.org/10.3390/proteomes13040058
Dutt M, Kumar A, Ostadgavahi AT, Kelvin DJ, Martinez GS. TCEPVDB: Artificial Intelligence-Based Proteome-Wide Screening of Antigens and Linear T-Cell Epitopes in the Poxviruses and the Development of a Repository. Proteomes. 2025; 13(4):58. https://doi.org/10.3390/proteomes13040058
Chicago/Turabian StyleDutt, Mansi, Anuj Kumar, Ali Toloue Ostadgavahi, David J. Kelvin, and Gustavo Sganzerla Martinez. 2025. "TCEPVDB: Artificial Intelligence-Based Proteome-Wide Screening of Antigens and Linear T-Cell Epitopes in the Poxviruses and the Development of a Repository" Proteomes 13, no. 4: 58. https://doi.org/10.3390/proteomes13040058
APA StyleDutt, M., Kumar, A., Ostadgavahi, A. T., Kelvin, D. J., & Martinez, G. S. (2025). TCEPVDB: Artificial Intelligence-Based Proteome-Wide Screening of Antigens and Linear T-Cell Epitopes in the Poxviruses and the Development of a Repository. Proteomes, 13(4), 58. https://doi.org/10.3390/proteomes13040058







