Proteomic Characterization of Primary Human Pancreatic Cancer Cell Lines Following Long-Term Exposure to Gemcitabine
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Long-Term Exposure to Gemcitabine
2.3. Gemcitabine Sensitivity
2.4. Cell Morphology and Growth
2.5. Extracellular Lactate Content
2.6. Proteomic Analysis
2.7. Immunoblotting
2.8. Statistical Analysis
3. Results
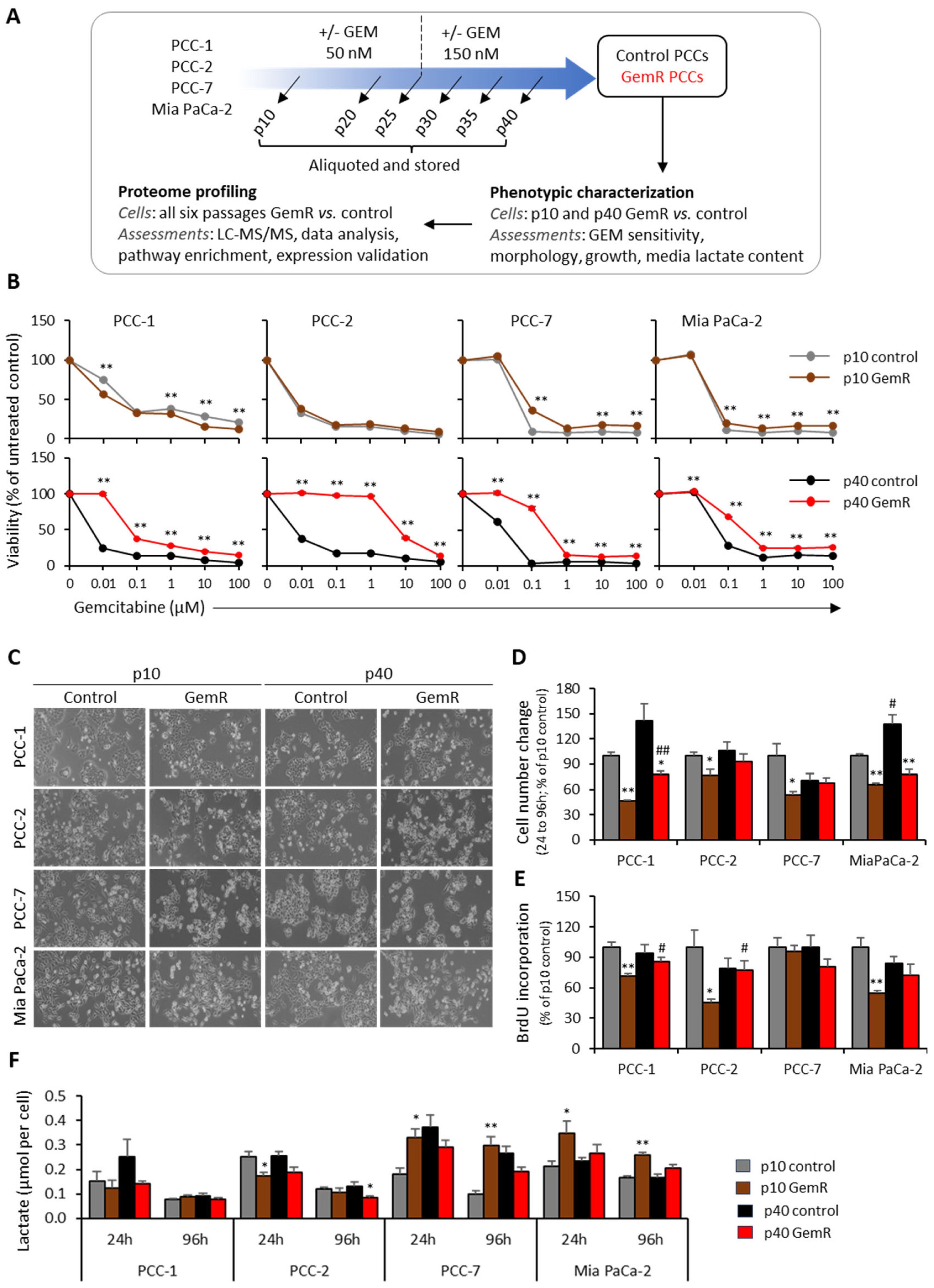
3.1. GEM Sensitivity
3.2. Phenotypic Differences
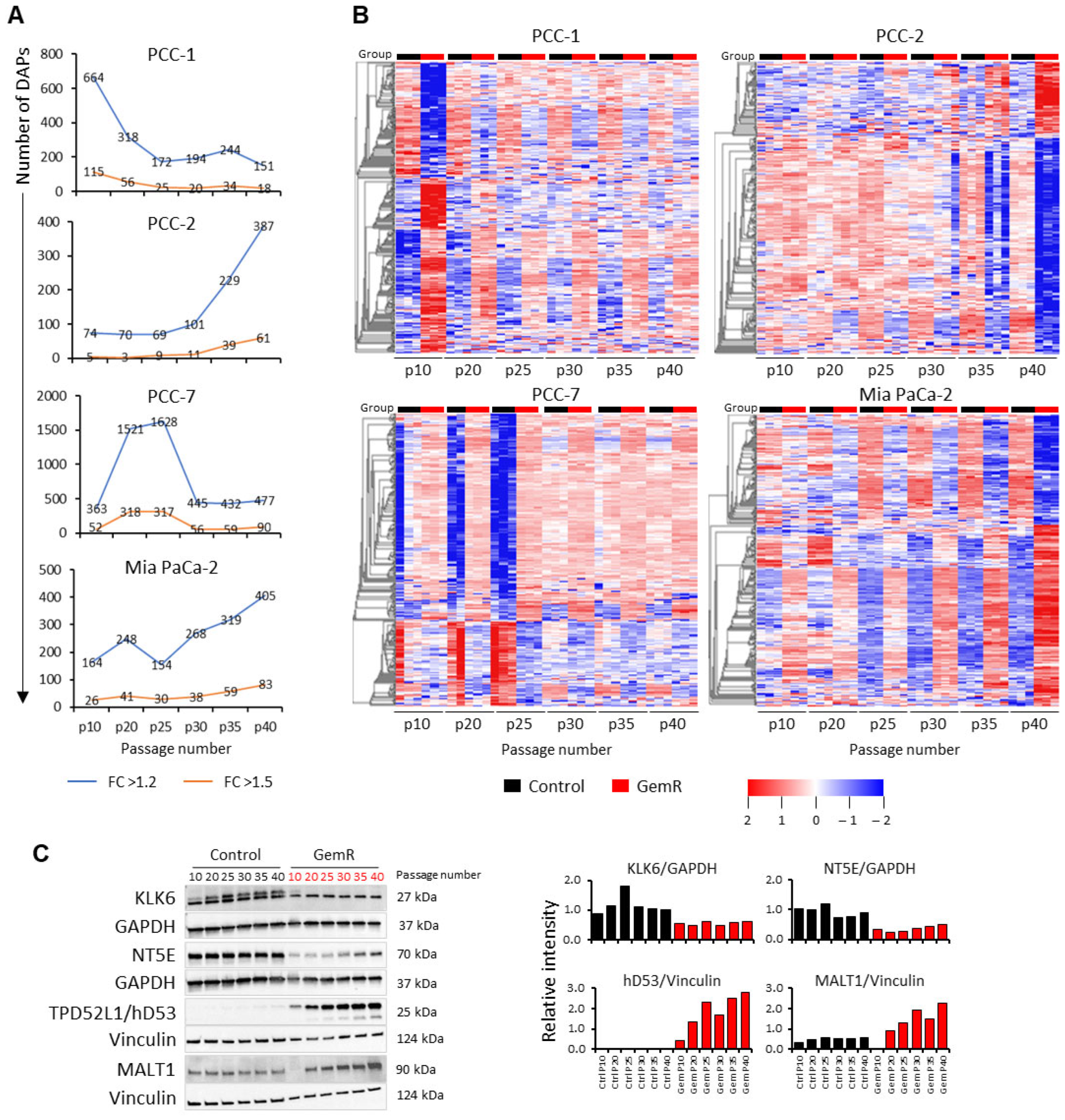
3.3. Overview of Proteome Profiles
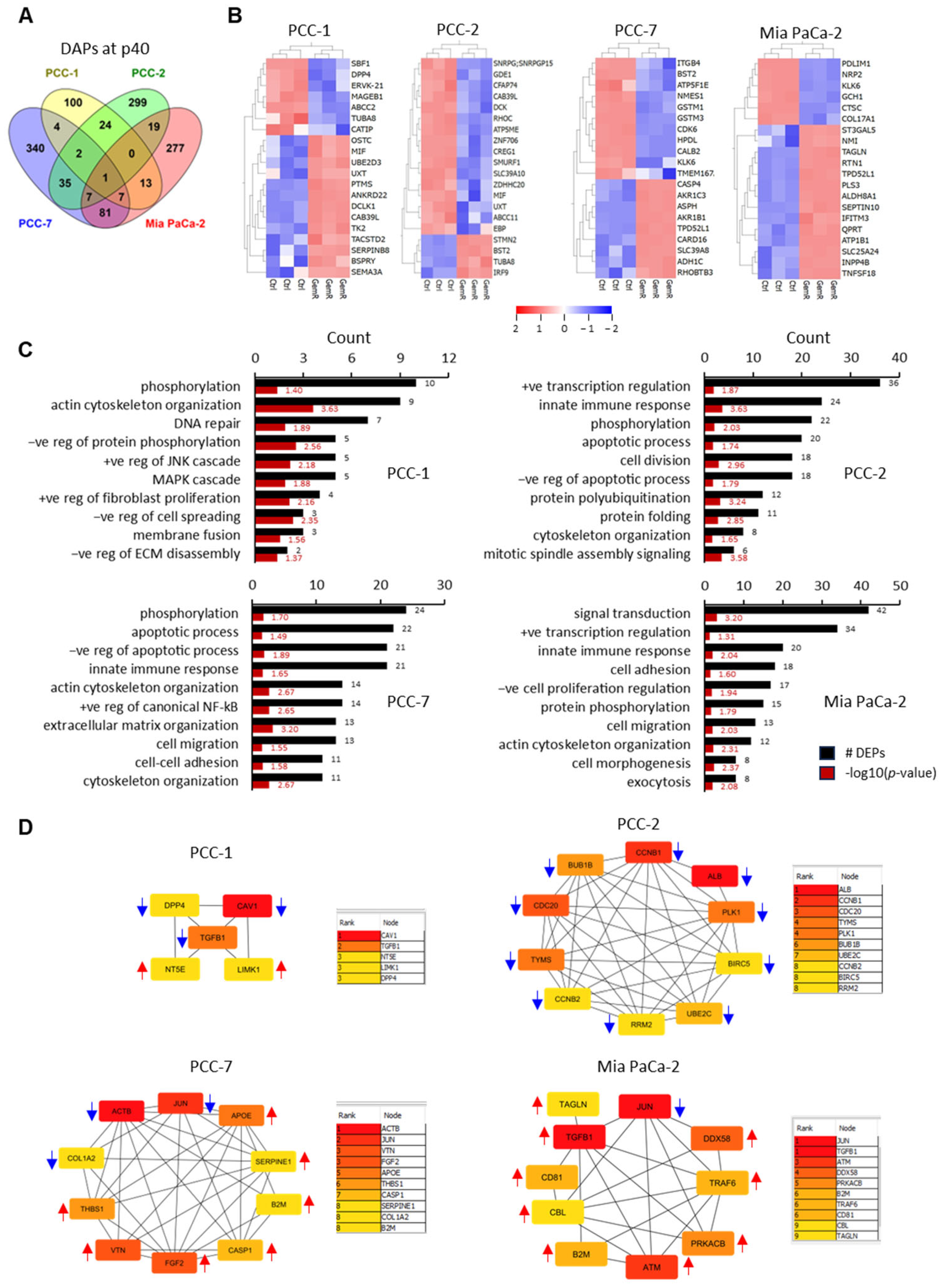
3.4. Differentially Abundant Proteins (DAPs) in GemR Versus Control Cells
3.5. Proteins Affected by GEM Exposure
3.6. Enrichment Analysis
3.7. Protein–Protein Interaction (PPI) Networks and Identification of Hub Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DAPs | Differentially abundant proteins |
| FC | Fold change |
| GemR | Gemcitabine-treated cells |
| GO | Gene ontology |
| KLK6 | Kallikrein-6 |
| MALT1 | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 |
| MS | Mass spectrometry |
| NGM | Normal growth medium |
| PCA | Principal component analysis |
| PCC | Pancreatic cancer cell |
| PDAC | Pancreatic ductal adenocarcinoma |
| PPI | Protein–protein interaction |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Park, W.; Chawla, A.; O’Reilly, E.M. Pancreatic Cancer: A Review. JAMA 2021, 326, 851–862. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardière, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Burris, H.A.; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Cripps, M.C.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P.; et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. 1997, 15, 2403–2413. [Google Scholar] [CrossRef]
- Muranaka, T.; Kuwatani, M.; Komatsu, Y.; Sawada, K.; Nakatsumi, H.; Kawamoto, Y.; Yuki, S.; Kubota, Y.; Kubo, K.; Kawahata, S.; et al. Comparison of efficacy and toxicity of FOLFIRINOX and gemcitabine with nab-paclitaxel in unresectable pancreatic cancer. J. Gastrointest. Oncol. 2017, 8, 566–571. [Google Scholar] [CrossRef]
- Klein-Brill, A.; Amar-Farkash, S.; Lawrence, G.; Collisson, E.A.; Aran, D. Comparison of FOLFIRINOX vs Gemcitabine Plus Nab-Paclitaxel as First-Line Chemotherapy for Metastatic Pancreatic Ductal Adenocarcinoma. JAMA Netw. Open 2022, 5, e2216199. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Tanno, S.; Koizumi, K.; Nishikawa, T.; Nakamura, K.; Minoguchi, M.; Izawa, T.; Mizukami, Y.; Okumura, T.; Kohgo, Y. Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br. J. Cancer 2007, 96, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Beutel, A.K.; Halbrook, C.J. Barriers and opportunities for gemcitabine in pancreatic cancer therapy. Am. J. Physiol. Cell Physiol. 2023, 324, C540–C552. [Google Scholar] [CrossRef] [PubMed]
- Amrutkar, M.; Gladhaug, I.P. Pancreatic Cancer Chemoresistance to Gemcitabine. Cancers 2017, 9, 157. [Google Scholar] [CrossRef] [PubMed]
- Amrutkar, M.; Vethe, N.T.; Verbeke, C.S.; Aasrum, M.; Finstadsveen, A.V.; Santha, P.; Gladhaug, I.P. Differential Gemcitabine Sensitivity in Primary Human Pancreatic Cancer Cells and Paired Stellate Cells Is Driven by Heterogenous Drug Uptake and Processing. Cancers 2020, 12, 3628. [Google Scholar] [CrossRef]
- Dash, S.; Ueda, T.; Komuro, A.; Amano, H.; Honda, M.; Kawazu, M.; Okada, H. MYC/Glutamine Dependency Is a Therapeutic Vulnerability in Pancreatic Cancer with Deoxycytidine Kinase Inactivation-Induced Gemcitabine Resistance. Mol. Cancer Res. 2023, 21, 444–457. [Google Scholar] [CrossRef]
- Amrutkar, M.; Berg, K.; Balto, A.; Skilbrei, M.G.; Finstadsveen, A.V.; Aasrum, M.; Gladhaug, I.P.; Verbeke, C.S. Pancreatic stellate cell-induced gemcitabine resistance in pancreatic cancer is associated with LDHA- and MCT4-mediated enhanced glycolysis. Cancer Cell Int. 2023, 23, 9. [Google Scholar] [CrossRef]
- Amrutkar, M.; Gladhaug, I.P. Stellate Cells Aid Growth-Permissive Metabolic Reprogramming and Promote Gemcitabine Chemoresistance in Pancreatic Cancer. Cancers 2021, 13, 601. [Google Scholar] [CrossRef]
- Amrutkar, M.; Aasrum, M.; Verbeke, C.S.; Gladhaug, I.P. Secretion of fibronectin by human pancreatic stellate cells promotes chemoresistance to gemcitabine in pancreatic cancer cells. BMC Cancer 2019, 19, 596. [Google Scholar] [CrossRef]
- Lin, Q.; Shen, S.; Qian, Z.; Rasam, S.S.; Serratore, A.; Jusko, W.J.; Kandel, E.S.; Qu, J.; Straubinger, R.M. Comparative Proteomic Analysis Identifies Key Metabolic Regulators of Gemcitabine Resistance in Pancreatic Cancer. Mol. Cell. Proteom. 2022, 21, 100409. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, Z.; Chen, Q.; Chen, K.; Liu, W.; Liu, G.; Chu, X.; Li, D.; Ma, Y.; Tian, X.; et al. Hypoxia-induced epigenetic regulation of miR-485-3p promotes stemness and chemoresistance in pancreatic ductal adenocarcinoma via SLC7A11-mediated ferroptosis. Cell Death Discov. 2024, 10, 262. [Google Scholar] [CrossRef]
- Emran, T.B.; Shahriar, A.; Mahmud, A.R.; Rahman, T.; Abir, M.H.; Siddiquee, M.F.; Ahmed, H.; Rahman, N.; Nainu, F.; Wahyudin, E.; et al. Multidrug Resistance in Cancer: Understanding Molecular Mechanisms, Immunoprevention and Therapeutic Approaches. Front. Oncol. 2022, 12, 891652. [Google Scholar] [CrossRef] [PubMed]
- Szakacs, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef]
- Zeng, S.; Pottler, M.; Lan, B.; Grutzmann, R.; Pilarsky, C.; Yang, H. Chemoresistance in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 4504. [Google Scholar] [CrossRef]
- Koltai, T.; Reshkin, S.J.; Carvalho, T.M.A.; Di Molfetta, D.; Greco, M.R.; Alfarouk, K.O.; Cardone, R.A. Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma: A Physiopathologic and Pharmacologic Review. Cancers 2022, 14, 2486. [Google Scholar] [CrossRef]
- Kong, R.; Qian, X.; Ying, W. Pancreatic cancer cells spectral library by DIA-MS and the phenotype analysis of gemcitabine sensitivity. Sci. Data 2022, 9, 283. [Google Scholar] [CrossRef]
- Chen, Y.W.; Liu, J.Y.; Lin, S.T.; Li, J.M.; Huang, S.H.; Chen, J.Y.; Wu, J.Y.; Kuo, C.C.; Wu, C.L.; Lu, Y.C.; et al. Proteomic analysis of gemcitabine-induced drug resistance in pancreatic cancer cells. Mol. Biosyst. 2011, 7, 3065–3074. [Google Scholar] [CrossRef] [PubMed]
- Le Large, T.Y.S.; El Hassouni, B.; Funel, N.; Kok, B.; Piersma, S.R.; Pham, T.V.; Olive, K.P.; Kazemier, G.; van Laarhoven, H.W.M.; Jimenez, C.R.; et al. Proteomic analysis of gemcitabine-resistant pancreatic cancer cells reveals that microtubule-associated protein 2 upregulation associates with taxane treatment. Ther. Adv. Med. Oncol. 2019, 11, 1758835919841233. [Google Scholar] [CrossRef] [PubMed]
- Krasny, L.; Huang, P.H. Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology. Mol. Omics 2021, 17, 29–42. [Google Scholar] [CrossRef]
- Lou, R.; Shui, W. Acquisition and Analysis of DIA-Based Proteomic Data: A Comprehensive Survey in 2023. Mol. Cell. Proteom. 2024, 23, 100712. [Google Scholar] [CrossRef] [PubMed]
- Amrutkar, M.; Larsen, E.K.; Aasrum, M.; Finstadsveen, A.V.; Andresen, P.A.; Verbeke, C.S.; Gladhaug, I.P. Establishment and Characterization of Paired Primary Cultures of Human Pancreatic Cancer Cells and Stellate Cells Derived from the Same Tumor. Cells 2020, 9, 227. [Google Scholar] [CrossRef]
- Batth, T.S.; Tollenaere, M.X.; Ruther, P.; Gonzalez-Franquesa, A.; Prabhakar, B.S.; Bekker-Jensen, S.; Deshmukh, A.S.; Olsen, J.V. Protein Aggregation Capture on Microparticles Enables Multipurpose Proteomics Sample Preparation. Mol. Cell. Proteom. 2019, 18, 1027–1035. [Google Scholar] [CrossRef]
- Demichev, V.; Szyrwiel, L.; Yu, F.; Teo, G.C.; Rosenberger, G.; Niewienda, A.; Ludwig, D.; Decker, J.; Kaspar-Schoenefeld, S.; Lilley, K.S.; et al. dia-PASEF data analysis using FragPipe and DIA-NN for deep proteomics of low sample amounts. Nat. Commun. 2022, 13, 3944. [Google Scholar] [CrossRef]
- Gorska, A.M.; Santos-Garcia, I.; Eiriz, I.; Bruning, T.; Nyman, T.; Pahnke, J. Evaluation of cerebrospinal fluid (CSF) and interstitial fluid (ISF) mouse proteomes for the validation and description of Alzheimer’s disease biomarkers. J. Neurosci. Methods 2024, 411, 110239. [Google Scholar] [CrossRef]
- Amrutkar, M.; Verbeke, C.S.; Finstadsveen, A.V.; Dorg, L.; Labori, K.J.; Gladhaug, I.P. Neoadjuvant chemotherapy is associated with an altered metabolic profile and increased cancer stemness in patients with pancreatic ductal adenocarcinoma. Mol. Oncol. 2023, 17, 59–81. [Google Scholar] [CrossRef]
- Li, Y.; Amrutkar, M.; Finstadsveen, A.V.; Dalen, K.T.; Verbeke, C.S.; Gladhaug, I.P. Fatty acids abrogate the growth-suppressive effects induced by inhibition of cholesterol flux in pancreatic cancer cells. Cancer Cell Int. 2023, 23, 276. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Gugenheim, J.; Crovetto, A.; Petrucciani, N. Neoadjuvant therapy for pancreatic cancer. Updates Surg. 2022, 74, 35–42. [Google Scholar] [CrossRef]
- Imafuji, H.; Matsuo, Y.; Ueda, G.; Omi, K.; Hayashi, Y.; Saito, K.; Tsuboi, K.; Morimoto, M.; Koide, S.; Ogawa, R.; et al. Acquisition of gemcitabine resistance enhances angiogenesis via upregulation of IL-8 production in pancreatic cancer. Oncol. Rep. 2019, 41, 3508–3516. [Google Scholar] [CrossRef]
- Fujiwara-Tani, R.; Sasaki, T.; Takagi, T.; Mori, S.; Kishi, S.; Nishiguchi, Y.; Ohmori, H.; Fujii, K.; Kuniyasu, H. Gemcitabine Resistance in Pancreatic Ductal Carcinoma Cell Lines Stems from Reprogramming of Energy Metabolism. Int. J. Mol. Sci. 2022, 23, 7824. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Peng, Y.; Zhu, Y.; Xu, D.; Zhu, F.; Xu, W.; Chen, Q.; Zhu, X.; Liu, T.; Hou, C.; et al. Glycolysis promotes the progression of pancreatic cancer and reduces cancer cell sensitivity to gemcitabine. Biomed. Pharmacother. 2020, 121, 109521. [Google Scholar] [CrossRef]
- Zhou, Y.; Lih, T.M.; Pan, J.; Hoti, N.; Dong, M.; Cao, L.; Hu, Y.; Cho, K.C.; Chen, S.Y.; Eguez, R.V.; et al. Proteomic signatures of 16 major types of human cancer reveal universal and cancer-type-specific proteins for the identification of potential therapeutic targets. J. Hematol. Oncol. 2020, 13, 170. [Google Scholar] [CrossRef]
- Wei, X.; Zhou, X.; Zhao, Y.; He, Y.; Weng, Z.; Xu, C. A 14-gene gemcitabine resistance gene signature is significantly associated with the prognosis of pancreatic cancer patients. Sci. Rep. 2021, 11, 6087. [Google Scholar] [CrossRef]
- Jiang, Y.; Ren, X.; Zhao, J.; Liu, G.; Liu, F.; Guo, X.; Hao, M.; Liu, H.; Liu, K.; Huang, H. Exploring the Molecular Therapeutic Mechanisms of Gemcitabine through Quantitative Proteomics. J. Proteome Res. 2024, 23, 2343–2354. [Google Scholar] [CrossRef]
- Mu, L.; Han, Z.; Yu, S.; Wang, A.; Chen, D.; Kong, S.; Gu, Y.; Xu, L.; Liu, A.; Sun, R.; et al. Pan-cancer analysis of ASB3 and the potential clinical implications for immune microenvironment of glioblastoma multiforme. Front. Immunol. 2022, 13, 842524. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Yoo, H.M. Emerging role of protein modification by UFM1 in cancer. Biochem. Biophys. Res. Commun. 2022, 633, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Zhang, C.; Liu, B.; Gao, G.; Tang, Y.; Lu, Y.; Pan, Z.; Wang, G.; Feng, W. Ribosomal protein L22-like1 promotes prostate cancer progression by activating PI3K/Akt/mTOR signalling pathway. J. Cell. Mol. Med. 2023, 27, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Chang-Liu, C.M.; Woloschak, G.E. Effect of passage number on cellular response to DNA-damaging agents: Cell survival and gene expression. Cancer Lett. 1997, 113, 77–86. [Google Scholar] [CrossRef]
- Kato, S.; Espinoza, N.; Lange, S.; Villalon, M.; Cuello, M.; Owen, G.I. Characterization and phenotypic variation with passage number of cultured human endometrial adenocarcinoma cells. Tissue Cell 2008, 40, 95–102. [Google Scholar] [CrossRef]
- Zhao, H.; Duan, Q.; Zhang, Z.; Li, H.; Wu, H.; Shen, Q.; Wang, C.; Yin, T. Up-regulation of glycolysis promotes the stemness and EMT phenotypes in gemcitabine-resistant pancreatic cancer cells. J. Cell Mol. Med. 2017, 21, 2055–2067. [Google Scholar] [CrossRef]
- Zhou, W.; Capello, M.; Fredolini, C.; Piemonti, L.; Liotta, L.A.; Novelli, F.; Petricoin, E.F. Proteomic analysis of pancreatic ductal adenocarcinoma cells reveals metabolic alterations. J. Proteome Res. 2011, 10, 1944–1952. [Google Scholar] [CrossRef]
- Grasso, C.; Jansen, G.; Giovannetti, E. Drug resistance in pancreatic cancer: Impact of altered energy metabolism. Crit. Rev. Oncol. Hematol. 2017, 114, 139–152. [Google Scholar] [CrossRef]
- Chatterjee, M.; Ben-Josef, E.; Thomas, D.G.; Morgan, M.A.; Zalupski, M.M.; Khan, G.; Andrew Robinson, C.; Griffith, K.A.; Chen, C.S.; Ludwig, T.; et al. Caveolin-1 is Associated with Tumor Progression and Confers a Multi-Modality Resistance Phenotype in Pancreatic Cancer. Sci. Rep. 2015, 5, 10867. [Google Scholar] [CrossRef]
- Borsoi, C.; Leonard, F.; Lee, Y.; Zaid, M.; Elganainy, D.; Alexander, J.F.; Kai, M.; Liu, Y.T.; Kang, Y.; Liu, X.; et al. Gemcitabine enhances the transport of nanovector-albumin-bound paclitaxel in gemcitabine-resistant pancreatic ductal adenocarcinoma. Cancer Lett. 2017, 403, 296–304. [Google Scholar] [CrossRef]
- Luo, Q.; Hu, Z.; Zhao, H.; Fan, Y.; Tu, X.; Wang, Y.; Liu, X. The role of TGF-beta in the tumor microenvironment of pancreatic cancer. Genes Dis. 2023, 10, 1513–1524. [Google Scholar] [CrossRef]
- Wang, X.; Su, W.; Qin, C.; Gao, R.; Shao, S.; Xu, X.; Zhang, Z.; Gao, J. Knockdown of TGF-beta in Pancreatic Cancer Helps Ameliorate Gemcitabine Resistance. Front. Biosci. 2024, 29, 269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Y.Y.; Chen, Y.; Wang, J.; Wang, Q.; Lu, H. TGF-beta Signaling and Resistance to Cancer Therapy. Front. Cell Dev. Biol. 2021, 9, 786728. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Xiong, L.; Wei, Y.; Wei, X.; Zhou, X.; Zhao, J.; Tang, H. A pancancer analysis of the oncogenic role of cyclin B1 (CCNB1) in human tumors. Sci. Rep. 2023, 13, 16226. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Li, X.; Meng, W.B.; Bai, Z.T.; Rui, S.Z.; Wang, Z.F.; Zhou, W.C.; Jin, X.D. Effect of CCNB1 silencing on cell cycle, senescence, and apoptosis through the p53 signaling pathway in pancreatic cancer. J. Cell. Physiol. 2018, 234, 619–631. [Google Scholar] [CrossRef]
- Lukey, M.J.; Greene, K.S.; Erickson, J.W.; Wilson, K.F.; Cerione, R.A. The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat. Commun. 2016, 7, 11321. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, S.; Zhao, X.; Cao, L.; Karnad, A.; Kumar, A.P.; Freeman, J.W. Gemcitabine resistance of pancreatic cancer cells is mediated by IGF1R dependent upregulation of CD44 expression and isoform switching. Cell Death Dis. 2022, 13, 682. [Google Scholar] [CrossRef] [PubMed]
- Mullen, N.J.; Singh, P.K. Nucleotide metabolism: A pan-cancer metabolic dependency. Nat. Rev. Cancer 2023, 23, 275–294. [Google Scholar] [CrossRef]
- Carbonara, K.; Andonovski, M.; Coorssen, J.R. Proteomes Are of Proteoforms: Embracing the Complexity. Proteomes 2021, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Zhang, L.; Chang, X.; Chen, K.; He, T.; Shi, J.; Lv, F.; Pan, L.; Wu, Y.; Cheng, Q.; et al. Integrative proteogenomic characterization of Wilms tumor. Nat. Commun. 2025, 16, 7715. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Zhang, Y.; Feng, X.; Li, N.; Chen, L.; Zhan, X. Proteoformics: Current status and future perspectives. J. Proteom. 2025, 321, 105524. [Google Scholar] [CrossRef] [PubMed]



| Gene (Protein ID) | Description | Molecular Function | Biological Process |
|---|---|---|---|
| DAPs common to all four PCC lines | |||
| KLK6 (Q92876) | Kallikrein-6 | Hydrolase, protease | CNS development, collagen catabolism, differentiation, invasion |
| DAPs common to three primary PCC lines | |||
| AFP (P02771) | Alpha-fetoprotein | Metal-ion binding | Ovulation, progesterone metabolic process |
| ALG5 (Q9Y673) | Dolichyl-phosphate beta-glucosyltransferase | Glycosyl-transferase | Glycosylation |
| ASB3 (Q9Y575) | Ankyrin repeat and SOCS box protein 3 | Protein modification | Protein ubiquitination |
| C1QTNF3 (Q9BXJ4) | Complement C1q tumor necrosis factor-related protein 3 | Identical protein binding | Cellular triglyceride homeostasis, NF-kB signaling, cytokine production, gluconeogenesis |
| MT-ATP6 (P00846) | ATP synthase subunit a | Transmembrane transporter | ATP synthesis, ion transport |
| NDUFA4 (O00483) | Cytochrome c oxidase subunit NDUFA4 | NADH activity | Electron transport, respiration |
| NUFIP1 (Q9UHK0) | Nuclear fragile X mental retardation-interacting protein 1 | RNA-binding | Transcription regulation, RNA processing |
| PRR14L (Q5THK1) | Protein PRR14L | Unknown | Cell division, skeletal myogenesis, tumorigenesis |
| TMA7 (Q9Y2S6) | Translation machinery-associated protein 7 | Cytoplasmic translation | Tumor progression, proliferation |
| TNIK (Q9UKE5) | TRAF2 and NCK-interacting protein kinase | Kinase activity, transferase | Neurogenesis, Wnt signaling pathway |
| UFM1 (P61960) | Ubiquitin-fold modifier 1 | Ubiquitylation | Apoptosis, ufmylation, Ubl conjugation pathway |
| UXT (Q9UBK9) | Protein UXT | Chaperone | Apoptosis, transcription regulation, chromatin/microtubule binding |
| DAPs common to PCC-2, PCC-7, and Mia PaCa-2 | |||
| CCDC71L (Q8N9Z2) | Coiled-coil domain-containing protein 71L | Lipid metabolic process | Fat cell differentiation |
| IGFBP6 (P24592) | Insulin-like growth factor-binding protein 6 | Growth factor binding | Cell growth, migration, signal transduction, MAPK cascade |
| INPP4B (O15327) | Inositol polyphosphate 4-phosphatase type II | Hydrolase | Lipid metabolism |
| MALT1 (Q9UDY8) | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | Hydrolase, protease | Inflammatory response, immunity, Ubl conjugation pathway |
| PSMB9 (P28065) | Proteasome subunit beta type 9 | Hydrolase, protease | Host-virus interaction, immunity, endopeptidase activity |
| TGM2 (P21980) | Protein-glutamine gamma-glutamyltransferase 2 | Acyltransferase, hydrolase | Apoptotic process, inflammation, cell adhesion |
| Cell Line | Change | List of Proteins |
|---|---|---|
| Altered expression in GemR versus ‘unchanged control’ | ||
| PCC-1 | ↑ | ACACA, ALDH1A3, CAPN1, CKAP4, CLIP1, DPYSL2, DUSP4, DYNC1H1, ENO2, GDA, GSN, IQGAP1, KRT19, LAMB3, MACF1, PGD, PKM, PPP1R12A, SERPINB5, VCAN, VPS13C |
| ↓ | ELAC2, FABP5, FLNC, HECTD1, MANF, MPI, PFKP, PTK2, RRBP1, UBE2S | |
| PCC-2 | ↑ | BST2, DECR1, MBOAT7, PPL, RUFY1, SAMD9, TKFC, TM7SF2, TRIM21 |
| ↓ | DNMT1, EZR, HMMR, MALT1, MCAM, NSUN2, PBK, SDHA, SNRPG, TACC3, TMA7, TTK, UFM1, UXT | |
| PCC-7 | ↑ | AKR1B1, AKR1C1, ALDH1A3, GBP2 |
| ↓ | ATAD3B, CKAP4, DENND5B, EPS8, FAM114A1, FDXR, GOLGA3, GUSB, HPDL, NT5E, ORC6, P3H3, RRP1B, SEPTIN10, SLC4A2, SVIL, UACA | |
| Mia PaCa-2 | ↑ | ARHGEF12, ATM, CARS1, COL3A1, CTNND1, DCPS, PTPRF, RDX, RRM1, TNKS1BP1, UTRN |
| ↓ | DOCK11, FDXR, IDH1 | |
| Passage-associated successive change in expression in GemR group | ||
| PCC-1 | ↑ | ALDH16A1, ARFIP2, CD109, CKAP4, MID1, MPI, MTUS1, PGD, PPP1R12A, SQOR |
| ↓ | PDP1, PFKP | |
| PCC-2 | ↑ | MACROD1, MAOB, MBOAT7, PPL, SAMD9, SQSTM1, TM7SF2, TUBA8 |
| ↓ | ANLN, ASB3, AURKA, CCT3, CCT5, CCT7, CFAP74, CLPTM1L, DNMT1, EZR, FAM92A, GOLPH3, HMMR, ITGA6, KIF2C, MALT1, MTRR, NSUN2, NUP155, PBL, PLK1, RBM23, SNRPG, TACC3, TARS1, TMA7, TTK, UFM1, UXT, ZNF622 | |
| PCC-7 | ↑ | AK3, ASPH, FBXO2, FMNL2, HKDC1, NAMPT, NBAS, NT5E, PDP1, PSMB8, SEC62, SLC22A18, SYTL2, TPD52L1 |
| ↓ | ACOT7, ANXA6, ATAD3B, DPYD, FAM114A1, FDXR, FKBP5, HPDL, PDCD4 | |
| Mia PaCa-2 | ↑ | AKR1B1, CARS1, DCPS, EPHA4, FHL1, MALT1, RNH1, RRM1, SEPTIN10, SWAP70, TALDO1, TRIM21 |
| ↓ | ANXA2, ATP13A3, DOCK11, DPYD, IDH1, ITPR3, KRT80, MYOF, NAMPT, NQO2, PDLIM1, PLEC, PODXL, PYGB, RAP2B, SERPINB6, SLC25A13, SNCG, STAT3, TP63, TRIP12, UAP1, YWHAQ | |
| Cell Line | Pathway ID | Description | Count | % | p-Value | Adj. p-Value |
|---|---|---|---|---|---|---|
| PCC-1 | hsa01100 | Metabolic pathways | 25 | 16.6 | <0.01 | 0.03 |
| hsa00240 | Pyrimidine metabolism | 6 | 4.0 | <0.01 | 0.02 | |
| hsa01232 | Nucleotide metabolism | 6 | 4.0 | <0.01 | 0.03 | |
| hsa01240 | Biosynthesis of cofactors | 5 | 3.3 | <0.05 | 1.00 | |
| PCC-2 | hsa01100 | Metabolic pathways | 42 | 10.9 | <0.05 | 1.00 |
| hsa04110 | Cell cycle | 15 | 3.9 | <0.01 | 0.00 | |
| hsa04120 | Ubiquitin mediated proteolysis | 11 | 2.8 | <0.01 | 0.07 | |
| hsa04218 | Cellular senescence | 9 | 2.3 | <0.01 | 0.60 | |
| hsa01232 | Nucleotide metabolism | 8 | 2.1 | <0.01 | 0.14 | |
| hsa00240 | Pyrimidine metabolism | 6 | 1.6 | <0.01 | 0.32 | |
| hsa03420 | Nucleotide excision repair | 5 | 1.3 | <0.04 | 1.00 | |
| PCC-7 | hsa01100 | Metabolic pathways | 61 | 12.8 | <0.01 | 0.06 |
| hsa05200 | Pathways in cancer | 22 | 4.6 | <0.05 | 0.47 | |
| hsa05205 | Proteoglycans in cancer | 17 | 3.6 | <0.01 | 0.01 | |
| hsa04151 | PI3K-Akt signaling pathway | 17 | 3.6 | <0.05 | 0.37 | |
| hsa04510 | Focal adhesion | 15 | 3.1 | <0.01 | 0.06 | |
| hsa04512 | ECM-receptor interaction | 10 | 2.1 | <0.01 | 0.05 | |
| hsa04218 | Cellular senescence | 10 | 2.1 | <0.05 | 0.33 | |
| hsa04142 | Lysosome | 9 | 1.9 | <0.05 | 0.33 | |
| hsa04625 | C-type lectin receptor signaling pathway | 8 | 1.7 | <0.05 | 0.33 | |
| hsa04066 | HIF-1 signaling pathway | 8 | 1.7 | <0.05 | 0.33 | |
| Mia PaCa-2 | hsa01100 | Metabolic pathways | 55 | 13.6 | <0.01 | 0.05 |
| hsa04010 | MAPK signaling pathway | 16 | 4.0 | <0.01 | 0.07 | |
| hsa05205 | Proteoglycans in cancer | 15 | 3.7 | <0.01 | 0.03 | |
| hsa04022 | cGMP-PKG signaling pathway | 14 | 3.5 | <0.01 | 0.02 | |
| hsa04024 | cAMP signaling pathway | 11 | 2.7 | <0.05 | 0.27 | |
| hsa04210 | Apoptosis | 10 | 2.5 | <0.01 | 0.07 | |
| hsa04218 | Cellular senescence | 10 | 2.5 | <0.01 | 0.14 | |
| hsa04625 | C-type lectin receptor signaling pathway | 9 | 2.2 | <0.01 | 0.06 | |
| hsa04142 | Lysosome | 9 | 2.2 | <0.01 | 0.14 | |
| hsa00230 | Purine metabolism | 8 | 2.0 | <0.05 | 0.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amrutkar, M.; Li, Y.; Finstadsveen, A.V.; Verbeke, C.S.; Gladhaug, I.P. Proteomic Characterization of Primary Human Pancreatic Cancer Cell Lines Following Long-Term Exposure to Gemcitabine. Proteomes 2025, 13, 48. https://doi.org/10.3390/proteomes13040048
Amrutkar M, Li Y, Finstadsveen AV, Verbeke CS, Gladhaug IP. Proteomic Characterization of Primary Human Pancreatic Cancer Cell Lines Following Long-Term Exposure to Gemcitabine. Proteomes. 2025; 13(4):48. https://doi.org/10.3390/proteomes13040048
Chicago/Turabian StyleAmrutkar, Manoj, Yuchuan Li, Anette Vefferstad Finstadsveen, Caroline S. Verbeke, and Ivar P. Gladhaug. 2025. "Proteomic Characterization of Primary Human Pancreatic Cancer Cell Lines Following Long-Term Exposure to Gemcitabine" Proteomes 13, no. 4: 48. https://doi.org/10.3390/proteomes13040048
APA StyleAmrutkar, M., Li, Y., Finstadsveen, A. V., Verbeke, C. S., & Gladhaug, I. P. (2025). Proteomic Characterization of Primary Human Pancreatic Cancer Cell Lines Following Long-Term Exposure to Gemcitabine. Proteomes, 13(4), 48. https://doi.org/10.3390/proteomes13040048









