Post-Translational Modifications in Histones and Their Role in Abiotic Stress Tolerance in Plants
Abstract
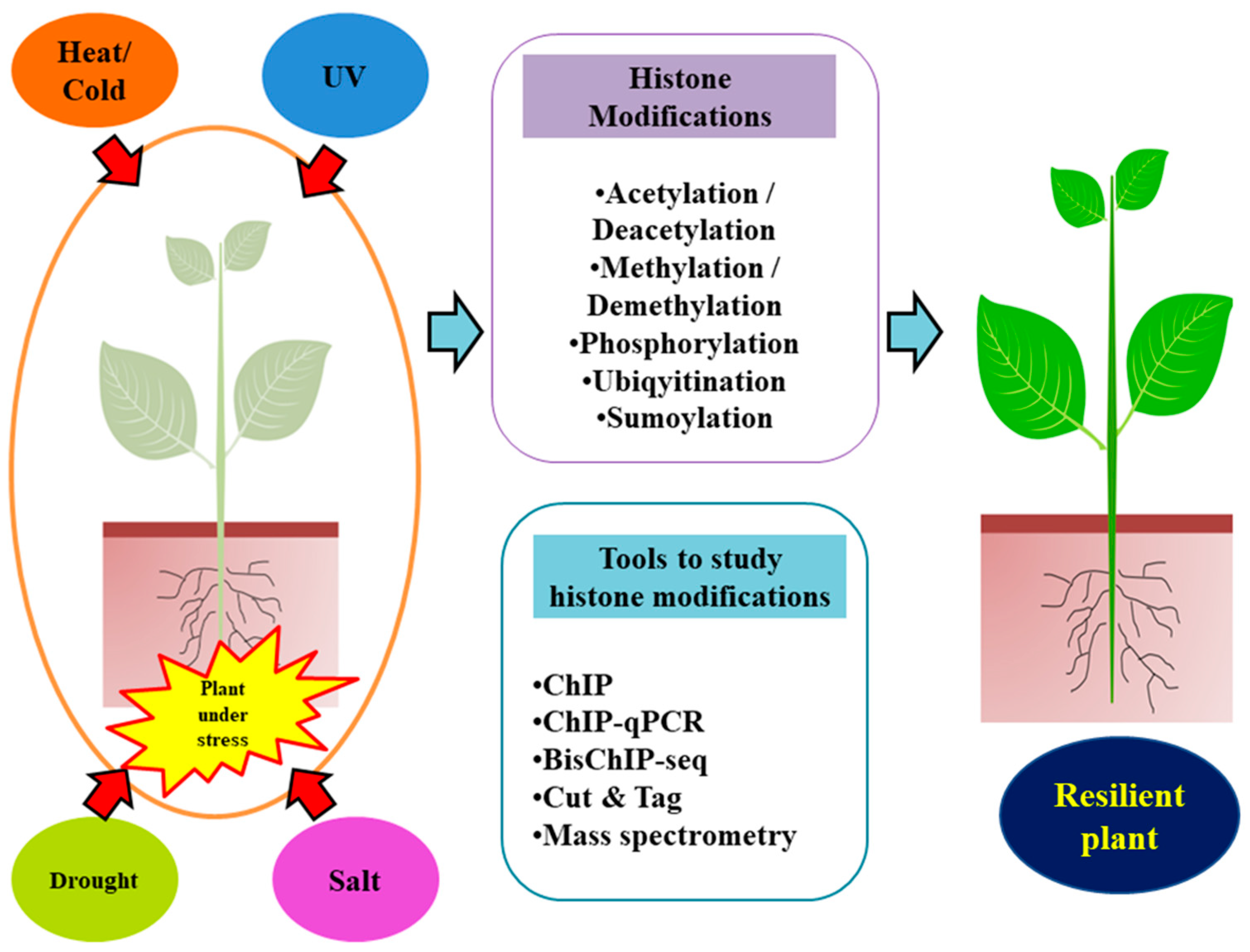
:1. Introduction
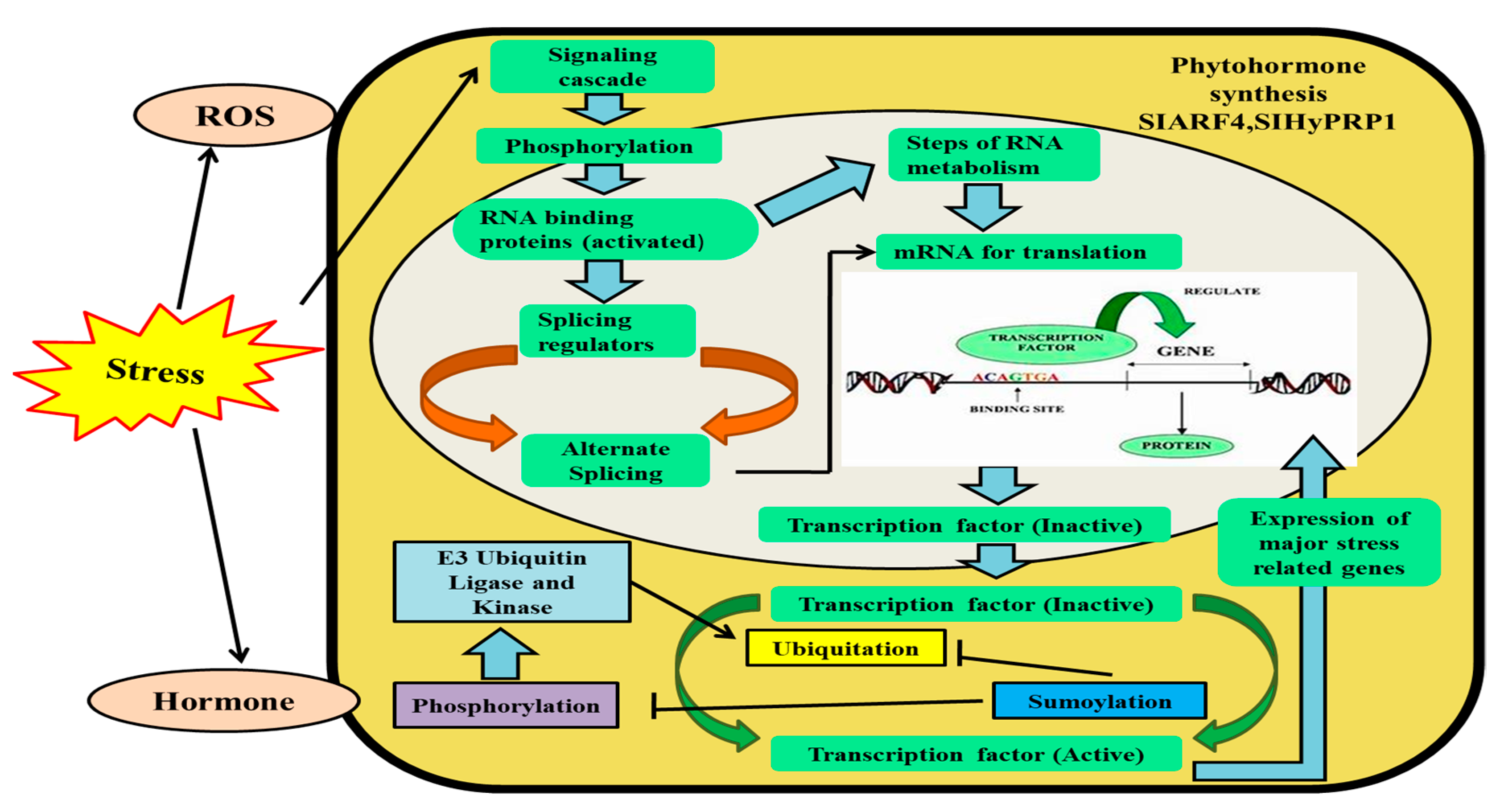
2. Epigenetic Memory and Chromatin Dynamics in Plant Stress Responses
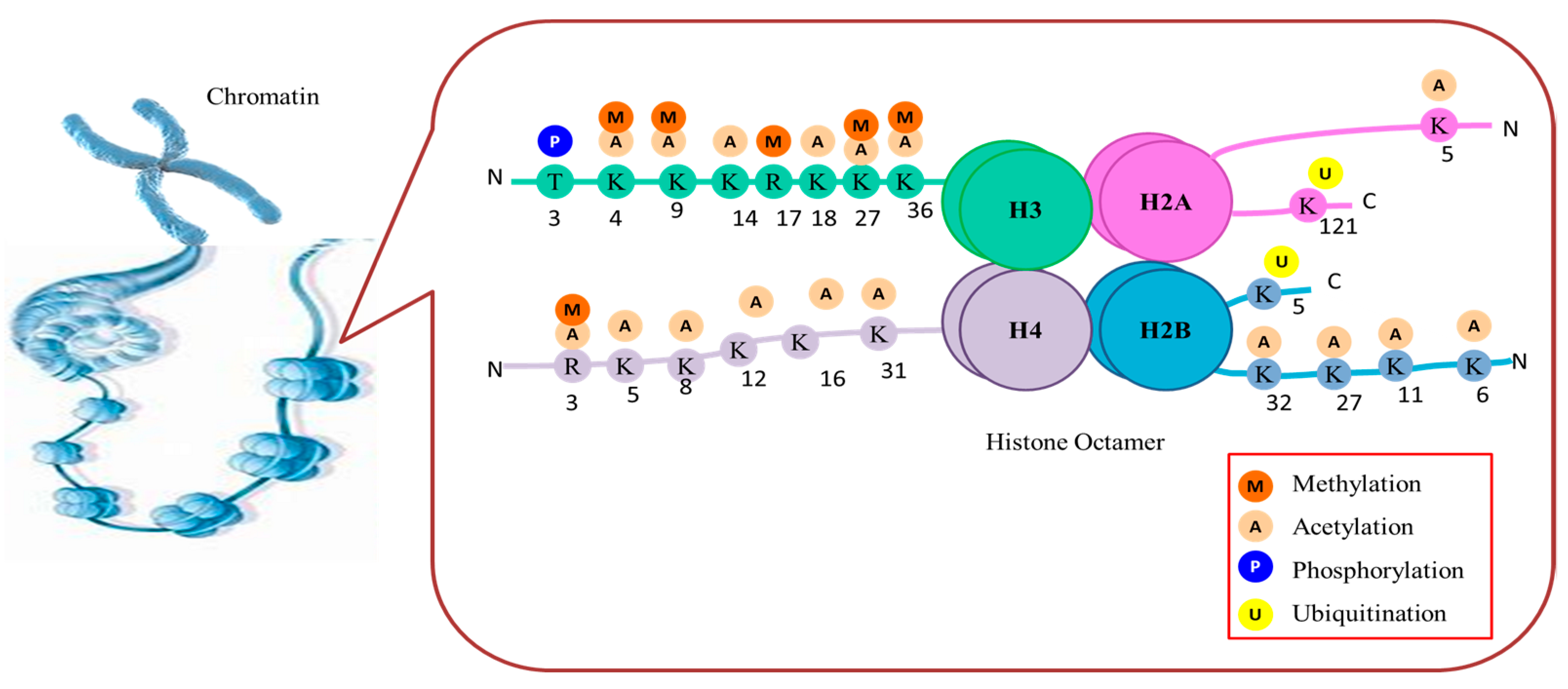
3. Molecular Sculptors: Types of Histone Post-Translational Modifications
3.1. Acetylation and Deacetylation
3.2. Methylation and Demethylation
3.3. Phosphorylation
3.4. Ubiquitination
3.5. Sumoylation
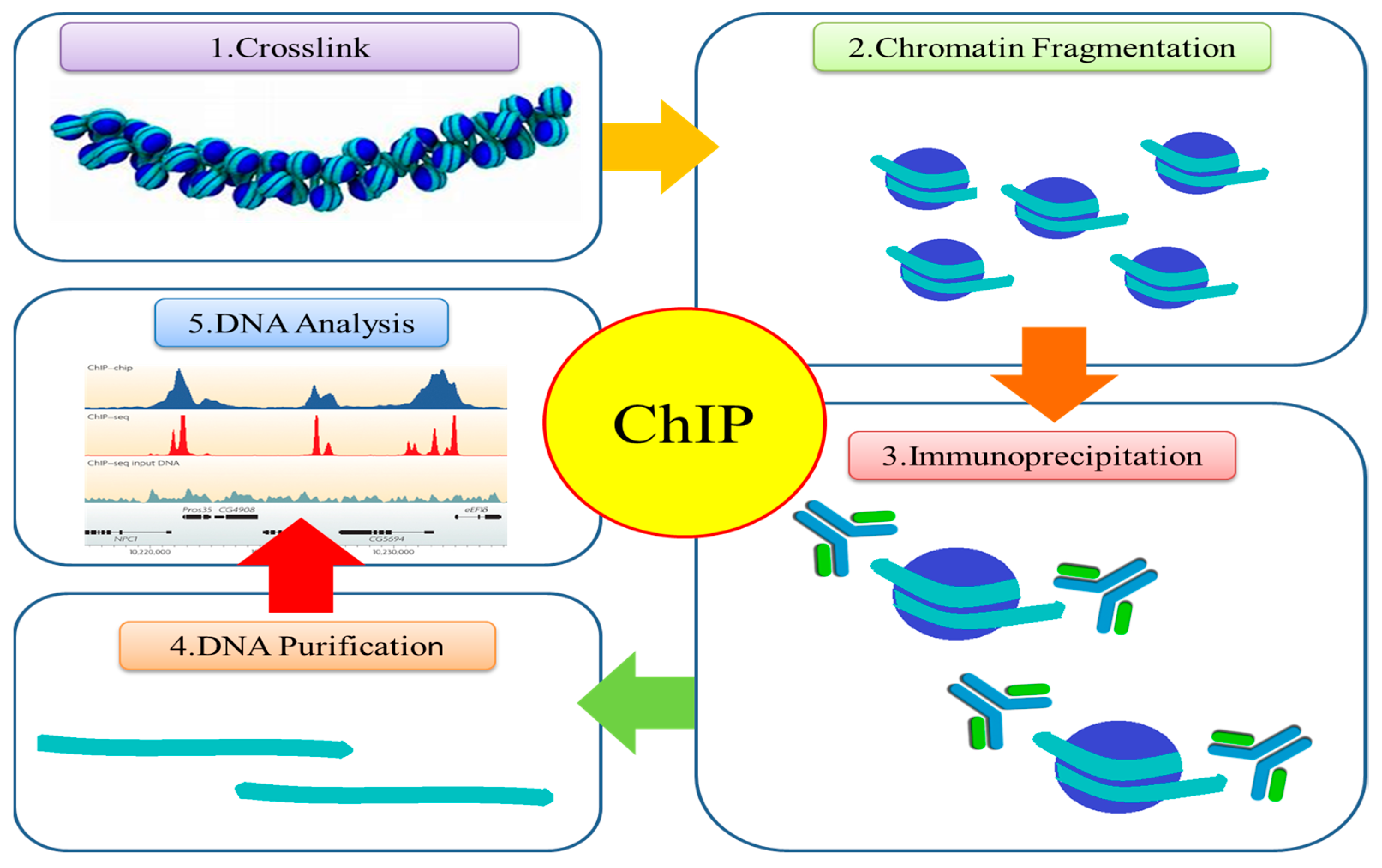
4. Tools to Study Histone Modifications
4.1. Chromatin Immunoprecipitation (ChIP) and ChIP-qPCR
4.2. Bisulphite-Treated Chromatin-Immunoprecipitated DNA (BisChIP-seq)
4.3. Cleavage under Targets and Tagmentation (CUT & Tag)
4.4. Mass Spectrometry
5. Role of PTM-Based Histone Modifications in Abiotic Stress Tolerance
5.1. Role in Heat Stress
5.2. Role in Cold Stress
5.3. Role in Salt Stress
5.4. Role in Drought Stress
5.5. Role in UV and Radiation Stress
6. Limitations and Path Ahead
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations of Genes
| (c-repeat binding factors) | CBF2 and CBF3 |
| 84KHDA903 | Tobacco Histone Deacetylase |
| AP2/EREBP TF | APETALA2/Ethylene-responsive Element-Binding Protein Transcription Factor |
| AP2/ERF | APETALA2/Ethylene-responsive Factor |
| AREB | ABA-Responsive Element-Binding Protein |
| ASF1 | Antisilencing Function 1 |
| At1g31280 (AtAgo2) | Argonaute 2 |
| At2g20880 | ERF53 Drought-Induced Transcription Factor |
| At3g03940 and At5g18190 | Ser/Thr protein kinases |
| At4g19130 (AtRPA1E) | Replication Factor-A protein 1-like protein |
| At5g20850 (AtRad51) | Homolog of Yeast RAD (Radiation repair gene) 51 |
| At5g24280 (AtGMI1) | Gamma Irradiation and Mitomycin C-Induced 1 |
| AtCOST1 | Arabidopsis gene Constitutively Stressed 1 |
| AtHAG1 | Arabidopsis Thaliana Histone Acetyltransferase |
| ATX1, ATX4/5 | Arabidopsis homolog of Trithorax1,4 and 5 (histone methyltransferase) |
| BdHD1 | Brachypodium distachyon Histone Deacetylase |
| BraHAC5 | Histone Acetyltransferase of the CBP Family 5 |
| BraHAG2 | Histone Acetyltransferase of the GNAT Family 2 |
| BZR1 | Brassinazole-resistant 1 |
| CBFs | Cold-Responsive Element-Binding Factors |
| CMT3 | Chromomethylase 3 |
| Cold-regulated genes (COR), such as COR15A, COR47, and COR78 | |
| COP1 | Constitutive Photomorphogenic 1 |
| CPKs | Calcium-Dependent Protein Kinases |
| CRT/DRE | Cold-Responsive Element/Dehydration Responsive Element |
| CTL | Chitinase-like |
| CYP707A1, CYP707A2 | Cytochrome P450 family 707-ABA 8′-Hydroxylase |
| DET1 | De-etiolated 1 |
| DOGL4 | Delay of Germination 1-LIKE 4 |
| DRE/CRT | Dehydration-responsive element/C-repeat |
| DREB | Dehydration-Responsive Element-Binding Protein |
| DRM | Domain rearrangement methylase |
| DSB | Double-Strand Break |
| ELP3 | Elongator Acetyltransferase Complex Subunit 3 |
| ERF113 | Ethylene-Responsive Factor113 |
| GCN5 | General Control nondepressible 5 |
| GNAT-MYST | Gcn5-related N-acetyltransferase |
| HAG1 | Histone Acetyltransferase of the Gnat Family 1 |
| HATs | Histone Acetyltransferases |
| HDACs | Histone Deacetylases |
| HDMs | Histone Demethylases |
| HhBREB2 | Halimodendron halodendron |
| HMTs | Histone Methyltransferases |
| HOS15 | High Expression of Osmotically Responsive Gene15 |
| HSFA3/A4A | Heat Shock Transcription Factor A3/A4A |
| HSFC1 | Heat Shock Transcription Factor C1 |
| HSP10/HSP17.6A | Heat Shock Protein 10/17.6A |
| HUB1/2, AtHUB2,OsHUB2, SlHUB1, SlHUB2 | Histone Monoubiquitination1/2 |
| HY5 | Elongated Hypocotyl 5, |
| IDS | Iduronate 2-Sulfatase |
| IDS1 | Indeterminate Spikelet1 |
| JMJ17 | Jumonji Domain-Containing Protein 17 |
| LBD-16 | Lateral Organ Boundaries Domain |
| MAPKs | Mitogen-Activated Protein Kinases |
| MET1 | Methyltransferase 1 |
| MLK1/2 | MUT9p-Like Kinase1/2 |
| MPK3/4/6 | Mitogen-Activated Protein Kinase 3/4/6 |
| MYB96 | myb Domain Protein 96 |
| MYST | MOZ, Ybf2/Sas3, Sas2, and Tip60 |
| MYST | MYST-Type Domain-Containing Lysine Acetyltransferase |
| OsAIR1 | Oryza sativa arsenic-induced RING E3 ligase 1 |
| OsbZIP46CA1 | Basic leucine zipper Transcription Factor |
| OsbZIP74 | Leucine Zipper Transcription Factor |
| OsCTR | Oryza sativa chloroplast targeting RING E3 ligase 1 |
| OsDIRP1 | Oryza sativa Drought-Induced RING Protein 1 |
| OsDIS1 | Oryza Sativa E3 ubiquitin–protein ligase 1 |
| OsHAC703, OsHAF701, OsHAG703, and OsHAM701 | |
| OsHAG702 | Oryza sativa Histone Acetyltransferase |
| OsHCl1 | Ring Finger E3 Ligase |
| OsHIRP1 | Heat-Induced Ring Finger protein |
| OsHTAS | Heat Tolerance at Seedling Stage |
| OsNAPL6 | Oryza Sativa histone chaperone of NAP superfamily |
| OsNTL3 | Membrane-Associated NAC Transcription Factor |
| OsPUB67 | Oryza Sativa U-box E3 ubiquitin ligase |
| OsRZP34 | Oryza Sativa Ring Zinc Finger Protein 34 |
| PIAL1/2 | Protein inhibitor of activated STAT-like 1 |
| PP1 | Protein Phosphatases1 |
| PRC2 | Polycomb-Repressive Complex 2 |
| PtrNAC006 | Populus truchocarpa NAC Domain-Containing Protein 6 |
| RAP2.6 | Related to AP2 6 |
| RD20/29A/29B | Responsive to Desiccation 20/29A/29B |
| RDM4 | (RNA-directed DNA Methylation-4). |
| RLKs | Receptor-Like Kinases |
| RTS1 | Repressor of Transcriptional Silencing 1 |
| SDIR1 | Salt- and Drought-Induced Ring Finger1 |
| SIARF4 | Solanum lycopersicum Auxin-Response Factor 4 |
| SIHyPRP1 | Solanum lycopersicum Hybrid Proline Rich Protein 1 |
| SIZ1/OsSIZ1/GmSIZ1/AtSIZ1/SlSIZ1 | SAP and MIZ 1 Domain containing Ligase 1 |
| SlHsfA1a | Heat Shock Transcription Factor A1a |
| SNAC1 | Stress-responsive NAC1 gene |
| SNC1 | Suppressor of NPR-1, Constitutive1 |
| SnRKs | Sucrose Nonfermenting-Related Kinases |
| WIND1 | Wound-Induced Dedifferentiation 1 |
| WRKY24/30 | Arabidopsis thaliana WRKY DNA-Binding Protein 24/30 |
| XBAT31 | XB3 Ortholog 1in Arabidopsis thaliana |
| ZAT12 | Zinc Finger of Arabidopsis thaliana 12 |
| ZmEXPB2 | Zea maize expansin-B2 |
| ZmXET1 | Zm Xyloglucan Endotransglucosylase Hydrolase 1 |
References
- Guerra, D.; Crosatti, C.; Khoshro, H.H.; Mastrangelo, A.M.; Mica, E.; Mazzucotelli, E. Post-transcriptional and post-translational regulations of drought and heat response in plants: A spider’s web of mechanisms. Front. Plant Sci. 2015, 6, 57. [Google Scholar] [CrossRef]
- Gupta, N.K.; Shavrukov, Y.; Singhal, R.K.; Borisjuk, N. (Eds.) Multiple Abiotic Stress Tolerances in Higher Plants: Addressing the Growing Challenges; CRC Press: Boca Raton, FL, USA, 2023. [Google Scholar]
- Ahammed, G.J.; Li, X.; Liu, A.; Chen, S. Brassinosteroids in plant tolerance to abiotic stress. J. Plant Growth Regul. 2020, 39, 1451–1464. [Google Scholar] [CrossRef]
- Jerome Jeyakumar, J.M.; Ali, A.; Wang, W.M.; Thiruvengadam, M. Characterizing the role of the miR156-SPL Network in plant development and stress response. Plants 2020, 9, 1206. [Google Scholar] [CrossRef]
- Lohani, N.; Jain, D.; Singh, M.B.; Bhalla, P.L. Engineering multiple abiotic stress tolerance in canola, Brassica napus. Front. Plant Sci. 2020, 11, 3. [Google Scholar] [CrossRef]
- Ghosh, U.K.; Islam, M.N.; Siddiqui, M.N.; Khan, M.A.R. Understanding the roles of osmolytes for acclimatizing plants to changing environment: A review of potential mechanism. Plant Signal. Behav. 2021, 16, 1913306. [Google Scholar] [CrossRef]
- Ramazi, S.; Zahiri, J. Posttranslational modifications in proteins: Resources, tools and prediction methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef]
- Ghimire, S.; Tang, X.; Zhang, N.; Liu, W.; Si, H. SUMO and SUMOylation in plant abiotic stress. Plant Growth Regul. 2020, 91, 317–325. [Google Scholar] [CrossRef]
- Ran, H.; Li, C.; Zhang, M.; Zhong, J.; Wang, H. Neglected PTM in Animal Adipogenesis: E3-mediated Ubiquitination. Gene 2023, 878, 147574. [Google Scholar] [CrossRef]
- Rehman, M.; Tanti, B. Understanding epigenetic modifications in response to abiotic stresses in plants. Biocatal. Agric. Biotechnol. 2020, 27, 101673. [Google Scholar] [CrossRef]
- Akhter, Z.; Bi, Z.; Ali, K.; Sun, C.; Fiaz, S.; Haider, F.U.; Bai, J. In Response to Abiotic Stress, DNA Methylation Confers Epigenetic Changes in Plants. Plants 2021, 10, 1096. [Google Scholar] [CrossRef]
- Vyse, K.; Faivre, L.; Romich, M.; Pagter, M.; Schubert, D.; Hincha, D.K.; Zuther, E. Transcriptional and post-transcriptional regulation and transcriptional memory of chromatin regulators in response to low temperature. Front. Plant Sci. 2020, 11, 39. [Google Scholar] [CrossRef]
- Grisan, V. Regulation of the Transcription Cycle by Co-Ordinate Interaction of Atp-Dependent Chromatin Remodeling and Histone Post-Translational Modifications. Doctoral Dissertation, University of Birmingham, Birmingham, UK, 2019. [Google Scholar]
- Luo, M.; Ríos, G.; Sarnowski, T.J.; Zhang, S.; Mantri, N.; Charron, J.B.; Libault, M. New insights into mechanisms of epigenetic modifiers in plant growth and development. Front. Plant Sci. 2020, 10, 1661. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Ali, F.; Dar, J.S.; Magray, A.R.; Ganai, B.A.; Chishti, M.Z. Posttranslational Modifications of Proteins and Their Role in Biological Processes and Associated Diseases. In Protein Modificomics; Academic Press: Cambridge, MA, USA, 2019; pp. 1–35. [Google Scholar]
- Duc, C.; Benoit, M.; Le Goff, S.; Simon, L.; Poulet, A.; Cotterell, S.; Tatout, C.; Probst, A.V. The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. Plant J. 2015, 81, 707–722. [Google Scholar] [CrossRef]
- Pardal, A.J.; Fernandes-Duarte, F.; Bowman, A.J. The histone chaperoning pathway: From ribosome to nucleosome. Essays Biochem. 2019, 63, 29–43. [Google Scholar]
- Chaudhry, U.K.; Gökçe, Z.N.; Gökçe, A.F. Salt stress and plant molecular responses. In Plant Defense Mechanisms; Intech Open: London, UK, 2022; p. 105. [Google Scholar]
- Lamke, J.; Baurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef]
- Friedrich, T.; Faivre, L.; Baurle, I.; Schubert, D. Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ. 2019, 42, 762–770. [Google Scholar] [CrossRef]
- Avramova, Z. Transcriptional ‘memory’ of a stress: Transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015, 83, 149–159. [Google Scholar] [CrossRef]
- Hilker, M.; Schwachtje, J.; Baier, M.; Balazadeh, S.; Baurle, I.; Geiselhardt, S.; Hincha, D.K.; Kunze, R.; Mueller-Roeber, B.; Rillig, M.C.; et al. Priming and memory of stress responses in organisms lacking a nervous system. Biol. Rev. 2016, 91, 1118–1133. [Google Scholar] [CrossRef]
- Yang, H.; Berry, S.; Olsson, T.S.G.; Hartley, M.; Howard, M.; Dean, C. Distinct Phases of Polycomb Silencing to Hold Epigenetic Memory of Cold in Arabidopsis. Science 2017, 357, 1142–1145. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic Regulation in Plant Abiotic Stress Responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Zhao, T.; Zhan, Z.; Jiang, D. Histone Modifications and Their Regulatory Roles in Plant Development and Environmental Memory. J. Genet. Genom. 2019; in press. [Google Scholar] [CrossRef]
- Csizmok, V.; Forman-Kay, J.D. Complex Regulatory Mechanisms Mediated by the Interplay of Multiple Post-Translational Modifications. Curr. Opin. Struct. Biol. 2018, 48, 58–67. [Google Scholar] [CrossRef]
- Park, J.; Lim, C.J.; Shen, M.; Park, H.J.; Cha, J.Y.; Iniesto, E.; Rubio, V.; Mengiste, T.; Zhu, J.K.; Bressan, R.A.; et al. Epigenetic Switch from Repressive to Permissive Chromatin in Response to Cold Stress. Proc. Natl. Acad. Sci. USA 2018, 115, E5400–E5409. [Google Scholar] [CrossRef]
- Tahir, M.S.; Karagiannis, J.; Tian, L. HD2A and HD2C Co-Regulate Drought Stress Response by Modulating Stomatal Closure and Root Growth in Arabidopsis. Front. Plant Sci. 2022, 13, 1062722. [Google Scholar] [CrossRef]
- Luo, M.; Wang, Y.Y.; Liu, X.; Yang, S.; Lu, Q.; Cui, Y.; Wu, K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012, 63, 3297–3306. [Google Scholar] [CrossRef]
- Lin, J.; Song, N.; Liu, D.; Liu, X.; Chu, W.; Li, J.; Chang, S.; Liu, Z.; Chen, Y.; Yang, Q.; et al. Histone Acetyltransferase TaHAG1 Interacts with TaNACL to Promote Heat Stress Tolerance in Wheat. Plant Biotechnol. J. 2022, 20, 1645–1647. [Google Scholar] [CrossRef]
- Rymen, B.; Kawamura, A.; Lambolez, A.; Inagaki, S.; Takebayashi, A.; Iwase, A.; Sakamoto, Y.; Sako, K.; Favero, D.S.; Ikeuchi, M.; et al. Histone Acetylation Orchestrates Wound-Induced Transcriptional Activation and Cellular Reprogramming in Arabidopsis. Commun. Biol. 2019, 2, 404. [Google Scholar] [CrossRef]
- Kang, H.; Fan, T.; Wu, J.; Zhu, Y.; Shen, W.H. Histone Modification and Chromatin Remodeling in Plant Response to Pathogens. Front. Plant Sci. 2022, 13, 986940. [Google Scholar] [CrossRef]
- Forestan, C.; Farinati, S.; Zambelli, F.; Pavesi, G.; Rossi, V.; Varotto, S. Epigenetic Signatures of Stress Adaptation and Flowering Regulation in Response to Extended Drought and Recovery in Zea mays. Plant Cell Environ. 2020, 43, 55–75. [Google Scholar] [CrossRef]
- Zhong, R.; Cui, D.; Ye, Z.H. A group of Populus trichocarpa DUF231 proteins exhibit differential O-acetyltransferase activities toward xylan. PLoS ONE 2018, 13, e0194532. [Google Scholar] [CrossRef]
- Li, S.; Lin, Y.J.; Wang, P.; Zhang, B.; Li, M.; Chen, S.; Shi, R.; Tunlaya-Anukit, S.; Liu, X.; Wang, Z.; et al. The AREB1 Transcription Factor Influences Histone Acetylation to Regulate Drought Responses and Tolerance in Populus trichocarpa. Plant Cell. 2019, 31, 663–686. [Google Scholar] [CrossRef]
- Roca Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Melo, B.P.; de Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved Drought Stress Tolerance in Arabidopsis by CRISPR/dCas9 Fusion with a Histone Acetyl Transferase. Sci. Rep. 2019, 9, 8080. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, S.; Tao, W.; Zhang, X.; Liu, J.; Sun, J.; Zhang, H.; Pu, L.; Huang, R.; Chen, T. INDETERMINATE SPIKELET1 Recruits Histone Deacetylase and a Transcriptional Repression Complex to Regulate Rice Salt Tolerance. Plant Physiol. 2018, 178, 824–837. [Google Scholar] [CrossRef]
- Lee, H.G.; Seo, P.J. MYB96 Recruits the HDA15 Protein to Suppress Negative Regulators of ABA Signaling in Arabidopsis. Nat. Commun. 2019, 10, 1713. [Google Scholar] [CrossRef]
- Zheng, Y.; Fornelli, L.; Compton, P.D.; Sharma, S.; Canterbury, J.; Mullen, C.; Kelleher, N.L. Unabridged Analysis of Human Histone H3 by Differential Top-Down Mass Spectrometry Reveals Hypermethylated Proteoforms from MMSET/NSD2 Overexpression. Mol. Cell Proteom. 2016, 15, 776–790. [Google Scholar] [CrossRef]
- Song, Y.; Wang, R.; Li, L.W.; Liu, X.; Wang, Y.F.; Wang, Q.X.; Zhang, Q. Long Non-coding RNA HOTAIR Mediates the Switching of Histone H3 Lysine 27 Acetylation to Methylation to Promote Epithelial-to-Mesenchymal Transition in Gastric Cancer. Int. J. Oncol. 2019, 54, 77–86. [Google Scholar] [CrossRef]
- Buszewicz, D.; Archacki, R.; Palusiński, A.; Kotliński, M.; Fogtman, A.; Iwanicka-Nowicka, R.; Sosnowska, K.; Kuciński, J.; Pupel, P.; Olędzki, J.; et al. HD2C Histone Deacetylase and a SWI/SNF Chromatin Remodeling Complex Interact and Both Are Involved in Mediating the Heat Stress Response in Arabidopsis. Plant Cell Environ. 2016, 39, 2108–2122. [Google Scholar] [CrossRef]
- Li, H.; Yan, S.; Zhao, L.; Tan, J.; Zhang, Q.; Gao, F.; Wang, P.; Hou, H.; Li, L. Histone Acetylation Associated Up-Regulation of the Cell Wall Related Genes Is Involved in Salt Stress Induced Maize Root Swelling. BMC Plant Biol. 2014, 14, 105. [Google Scholar] [CrossRef]
- Eom, S.H.; Hyun, T.K. Histone Acetyltransferases (HATs) in Chinese Cabbage: Insights from Histone H3 Acetylation and Expression Profiling of HATs in Response to Abiotic Stresses. J. Am. Soc. Hortic. Sci. 2018, 143, 296–303. [Google Scholar] [CrossRef]
- Fang, H.; Liu, X.; Thorn, G.; Duan, J.; Tian, L. Expression Analysis of Histone Acetyltransferases in Rice under Drought Stress. Biochem. Biophys. Res. Commun. 2014, 443, 400–405. [Google Scholar] [CrossRef]
- Baek, D.; Shin, G.; Kim, M.C.; Shen, M.; Lee, S.Y.; Yun, D.J. Histone Deacetylase HDA9 with ABI4 Contributes to Abscisic Acid Homeostasis in Drought Stress Response. Front. Plant Sci. 2020, 11, 143. [Google Scholar] [CrossRef]
- Song, J.; Henry, H.A.L.; Tian, L. Brachypodium Histone Deacetylase BdHD1 Positively Regulates ABA and Drought Stress Responses. Plant Sci. 2019, 283, 355–365. [Google Scholar] [CrossRef]
- Ivanova, T.; Dincheva, I.; Badjakov, I.; Iantcheva, A. Transcriptional and Metabolic Profiling of Arabidopsis thaliana Transgenic Plants Expressing Histone Acetyltransferase HAC1 upon the Application of Abiotic Stress—Salt and Low Temperature. Metabolites 2023, 13, 994. [Google Scholar] [CrossRef]
- Papaefthimiou, D.; Likotrafiti, E.; Kapazoglou, A.; Bladenopoulos, K.; Tsaftaris, A. Epigenetic Chromatin Modifiers in Barley: III. Isolation and Characterization of the Barley GNAT-MYST Family of Histone Acetyltransferases and Responses to Exogenous ABA. Plant Physiol. Biochem. 2010, 48, 98–107. [Google Scholar] [CrossRef]
- Billah, M.; Aktar, S.; Brestic, M.; Zivcak, M.; Khaldun, A.B.M.; Uddin, M.S.; Hossain, A. Progressive Genomic Approaches to Explore Drought- and Salt-Induced Oxidative Stress Responses in Plants under Changing Climate. Plants 2021, 10, 1910. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, B.; Liu, C.; Tong, B.; Guan, T.; Xia, D. Expression of a Populus Histone Deacetylase Gene 84KHDA903 in Tobacco Enhances Drought Tolerance. Plant Sci. 2017, 265, 1–11. [Google Scholar] [CrossRef]
- Li, S.; He, X.; Gao, Y.; Zhou, C.; Chiang, V.L.; Li, W. Histone acetylation changes in plant response to drought stress. Genes 2021, 12, 1409. [Google Scholar] [CrossRef]
- Ding, J.; Shen, J.; Mao, H.; Xie, W.; Li, X.; Zhang, Q. RNA-Directed DNA Methylation Is Involved in Regulating Photoperiod-Sensitive Male Sterility in Rice. Mol. Plant. 2012, 5, 1210–1216. [Google Scholar] [CrossRef]
- Liu, J.; Shi, Y.; Yang, S. Insights into the Regulation of C-Repeat Binding Factors in Plant Cold Signaling. J. Integr. Plant Biol. 2018, 60, 780–795. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, A.; Jin, J.B.; Zhao, B.; Wang, T.J.; Wu, Y.; Xu, Z.Y. Arabidopsis Histone H3K4 Demethylase JMJ 17 Functions in Dehydration Stress Response. N. Phytol. 2019, 223, 1372–1387. [Google Scholar] [CrossRef]
- Mozgova, I.; Mikulski, P.; Pecinka, A.; Farrona, S. Epigenetic Mechanisms of Abiotic Stress Response and Memory in Plants. In Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications: Transcriptional Regulation and Chromatin Remodelling in Plants; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–64. [Google Scholar]
- Zhou, S.; Chen, Q.; Sun, Y.; Li, Y. Histone H2B Monoubiquitination Regulates Salt Stress-Induced Microtubule Depolymerization in Arabidopsis. Plant Cell Environ. 2017, 40, 1512–1530. [Google Scholar] [CrossRef]
- Zhou, Y.; Romero-Campero, F.J.; Gómez-Zambrano, Á.; Turck, F.; Calonje, M. H2A Mono ubiquitination in Arabidopsis thaliana Is Generally Independent of LHP1 and PRC2 Activity. Genome Biol. 2017, 18, 6. [Google Scholar] [CrossRef]
- Zarreen, F.; Karim, M.J.; Chakraborty, S. The Diverse Roles of Histone H2B Mono ubiquitination in the Life of Plants. J. Exp. Bot. 2022, 73, 3854–3865. [Google Scholar] [CrossRef]
- Chen, K.; Tang, W.S.; Zhou, Y.B.; Xu, Z.S.; Chen, J.; Ma, Y.Z.; Li, H.Y. Overexpression of GmUBC9 Gene Enhances Plant Drought Resistance and Affects Flowering Time via Histone H2B Mono ubiquitination. Front. Plant Sci. 2020, 11, 555794. [Google Scholar] [CrossRef]
- Ma, S.; Tang, N.; Li, X.; Xie, Y.; Xiang, D.; Fu, J.; Shen, J.; Yang, J.; Tu, H.; Li, X.; et al. Reversible Histone H2B Monoubiquitination Fine-Tunes Abscisic Acid Signaling and Drought Response in Rice. Mol. Plant. 2019, 12, 263–277. [Google Scholar] [CrossRef]
- Wang, Z.; Casas-Mollano, J.A.; Xu, J.; Riethoven, J.-J.M.; Zhang, C.; Cerutti, H. Osmotic Stress Induces Phosphorylation of Histone H3 at Threonine 3 in Pericentromeric Regions of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 8487–8492. [Google Scholar] [CrossRef]
- Wang, Y.; Cai, S.; Yin, L.; Shi, K.; Xia, X.; Zhou, Y.; Yu, J.; Zhou, J. Tomato HsfA1a Plays a Critical Role in Plant Drought Tolerance by Activating ATG Genes and Inducing Autophagy. Autophagy 2015, 11, 2033–2047. [Google Scholar] [CrossRef]
- Ueda, M.; Seki, M. Histone Modifications Form Epigenetic Regulatory Networks to Regulate Abiotic Stress Response. Plant Physiol. 2020, 182, 15–26. [Google Scholar] [CrossRef]
- Cai, H.; Wang, H.; Zhou, L.; Li, B.; Zhang, S.; He, Y.; Xu, Y. Time-Series Transcriptomic Analysis of Contrasting Rice Materials under Heat Stress Reveals a Faster Response in the Tolerant Cultivar. Int. J. Mol. Sci. 2023, 24, 9408. [Google Scholar] [CrossRef]
- Mishra, N.; Srivastava, A.P.; Esmaeili, N.; Hu, W.; Shen, G. Overexpression of the Rice Gene OsSIZ1 in Arabidopsis Improves Drought-, Heat-, and Salt-Tolerance Simultaneously. PLoS ONE 2018, 13, e0201716. [Google Scholar] [CrossRef]
- Zhang, S.S.; Yang, H.; Ding, L.; Song, Z.T.; Ma, H.; Chang, F.; Liu, J.X. Tissue-Specific Transcriptomics Reveals an Important Role of the Unfolded Protein Response in Maintaining Fertility upon Heat Stress in Arabidopsis. Plant Cell. 2017, 29, 1007–1023. [Google Scholar] [CrossRef]
- Saad, A.S.I.; Li, X.; Li, H.P.; Huang, T.; Gao, C.S.; Guo, M.W.; Liao, Y.C. A Rice Stress-Responsive NAC Gene Enhances Tolerance of Transgenic Wheat to Drought and Salt Stresses. Plant Sci. 2013, 203, 33–40. [Google Scholar] [CrossRef]
- Liang, Q.; Geng, Q.; Jiang, L.; Liang, M.; Li, L.; Zhang, C.; Wang, W. Protein Methylome Analysis in Arabidopsis Reveals Regulation in RNA-Related Processes. J. Proteom. 2020, 213, 103601. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, K.; Yin, L.; Yu, Y.; Qi, J.; Shen, W.H.; Zhu, J.; Zhang, Y.; Dong, A. H3K4me2 Functions as a Repressive Epigenetic Mark in Plants. Epigenetics Chromatin. 2019, 12, 1–14. [Google Scholar] [CrossRef]
- Fiorucci, A.S.; Bourbousse, C.; Concia, L.; Rougée, M.; Deton-Cabanillas, A.F.; Zabulon, G.; Layat, E.; Latrasse, D.; Kim, S.K.; Chaumont, N.; et al. Arabidopsis S2Lb Links AtCOMPASS-like and SDG2 Activity in H3K4me3 Independently from Histone H2B Monoubiquitination. Genome Biol. 2019, 20, 1–21. [Google Scholar] [CrossRef]
- Zheng, B.; Chen, X. Dynamics of Histone H3 Lysine 27 Trimethylation in Plant Development. Curr. Opin. Plant Biol. 2011, 14, 123–129. [Google Scholar] [CrossRef]
- Xiao, J.; Lee, U.S.; Wagner, D. Tug of War: Adding and Removing Histone Lysine Methylation in Arabidopsis. Curr. Opin. Plant Biol. 2016, 34, 41–53. [Google Scholar] [CrossRef]
- Cheng, K.; Xu, Y.; Yang, C.; Ouellette, L.; Niu, L.; Zhou, X.; Chu, L.; Zhuang, F.; Liu, J.; Wu, H.; et al. Histone Tales: Lysine Methylation, a Protagonist in Arabidopsis Development. J. Exp. Bot. 2020, 71, 793–807. [Google Scholar] [CrossRef]
- Wang, T.S.; Cheng, J.K.; Lei, Q.Y.; Wang, Y.P. A Switch for Transcriptional Activation and Repression: Histone Arginine Methylation. In The DNA, RNA, and Histone Methylomes; Springer: Berlin/Heidelberg, Germany, 2019; pp. 521–541. [Google Scholar]
- Liu, W.; Tanasa, B.; Tyurina, O.V.; Zhou, T.Y.; Gassmann, R.; Liu, W.T.; Ohgi, K.A.; Benner, C.; Garcia-Bassets, I.; Aggarwal, A.K.; et al. PHF8 Mediates Histone H4 Lysine 20 Demethylation Events Involved in Cell Cycle Progression. Nature 2010, 466, 508–512. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent Advances in Utilizing Transcription Factors to Improve Plant Abiotic Stress Tolerance by Transgenic Technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef]
- Berger, S.L. The Complex Language of Chromatin Regulation during Transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb Gene Silencing: Knowns and Unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb Complex PRC2 and Its Mark in Life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Vijayanathan, M.; Trejo-Arellano, M.G.; Mozgová, I. Polycomb Repressive Complex 2 in Eukaryotes—An Evolutionary Perspective. Epigenomes 2022, 6, 3. [Google Scholar] [CrossRef]
- Baile, F.; Gómez-Zambrano, A.; Calonje, M. Roles of Polycomb Complexes in Regulating Gene Expression and Chromatin Structure in Plants. Plant Commun. 2021, 3, 100267. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Liu, B.; Xu, Z.Y. Dynamic regulation of DNA methylation and histone modifications in response to abiotic stresses in plants. J. Integr. Plant Biol. 2022, 64, 2252–2274. [Google Scholar] [CrossRef]
- Pandey, G.; Sharma, N.; Pankaj Sahu, P.; Prasad, M. Chromatin-based epigenetic regulation of plant abiotic stress response. Curr. Genom. 2016, 17, 490–498. [Google Scholar] [CrossRef]
- Cui, X.; Zheng, Y.; Lu, Y.; Issakidis-Bourguet, E.; Zhou, D.X. Metabolic Control of Histone Demethylase Activity Involved in Plant Response to High Temperature. Plant Physiol. 2021, 185, 1813–1828. [Google Scholar] [CrossRef]
- Lama, P. Identification and Functional Characterization of GmMYB176-Specific Protein Kinases in Soybean; The University of Western Ontario: London, ON, Canada, 2016. [Google Scholar]
- Yuan, L.; Liu, X.; Luo, M.; Yang, S.; Wu, K. Involvement of Histone Modifications in Plant Abiotic Stress Responses. J. Integr. Plant Biol. 2013, 55, 892–901. [Google Scholar] [CrossRef]
- Houben, A.; Demidov, D.; Caperta, A.D.; Karimi, R.; Agueci, F.; Vlasenko, L. Phosphorylation of Histone H3 in Plants-A Dynamic Affair. Biochim. Biophys. Acta (BBA)—Gene Struct. Expr. 2007, 1769, 308–315. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Satish, L.; Kalendar, R.; Narayanan, M.; Kandasamy, S.; Sharma, A.; Emamverdian, A.; Wei, Q.; Zhou, M. The dynamism of transposon methylation for plant development and stress adaptation. Int. J. Mol. Sci. 2021, 22, 11387. [Google Scholar] [CrossRef]
- Sawicka, A.; Seiser, C. Sensing Core Histone Phosphorylation—A Matter of Perfect Timing. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2014, 1839, 711–718. [Google Scholar] [CrossRef]
- Wang, F.; Higgins, J.M.G. Histone Modifications and Mitosis: Countermarks, Landmarks, and Bookmarks. Trends Cell Biol. 2013, 23, 175–184. [Google Scholar] [CrossRef]
- Su, Y.; Wang, S.; Zhang, F.; Zheng, H.; Liu, Y.; Huang, T.; Ding, Y. Phosphorylation of Histone H2A at Serine 95: A Plant-Specific Mark Involved in Flowering Time Regulation and H2A.Z Deposition. Plant Cell 2017, 29, 2197–2213. [Google Scholar] [CrossRef]
- Stadler, J.; Richly, H. Regulation of DNA Repair Mechanisms: How the Chromatin Environment Regulates the DNA Damage Response. Int. J. Mol. Sci. 2017, 18, 1715. [Google Scholar] [CrossRef]
- Dubrez, L.; Causse, S.; Bonan, N.B.; Dumétier, B.; Garrido, C. Heat-Shock Proteins: Chaperoning DNA Repair. Oncogene 2020, 39, 516–529. [Google Scholar] [CrossRef]
- Aleksandrov, R.; Hristova, R.; Stoynov, S.; Gospodinov, A. The Chromatin Response to Double-Strand DNA Breaks and Their Repair. Cells 2020, 9, 1853. [Google Scholar] [CrossRef]
- Kerk, D.; Templeton, G.; Moorhead, G.B. Evolutionary Radiation Pattern of Novel Protein Phosphatases Revealed by Analysis of Protein Data from the Completely Sequenced Genomes of Humans, Green Algae, and Higher Plants. Plant Physiol. 2008, 146, 351–367. [Google Scholar] [CrossRef]
- Andrási, N.; Rigó, G.; Zsigmond, L.; Pérez-Salamó, I.; Papdi, C.; Klement, E.; Pettkó-Szandtner, A.; Baba, A.I.; Ayaydin, F.; Dasari, R. The Mitogen-Activated Protein Kinase 4-Phosphorylated Heat Shock Factor A4A Regulates Responses to Combined Salt and Heat Stresses. J. Exp. Bot. 2019, 70, 4903–4918. [Google Scholar] [CrossRef]
- Ibáñez, C.; Delker, C.; Martinez, C.; Bürstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M. Brassinosteroids Dominate Hormonal Regulation of Plant Thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310.e3. [Google Scholar] [CrossRef]
- Huang, T.; Zhang, H.; Zhou, Y.; Su, Y.; Zheng, H.; Ding, Y. Phosphorylation of Histone H2A at Serine 95 Is Essential for Flowering Time and Development in Arabidopsis. Front. Plant Sci. 2021, 12, 761008. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin Modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Oss-Ronen, L.; Sarusi, T.; Cohen, I. Histone Mono-Ubiquitination in Transcriptional Regulation and Its Mark on Life: Emerging Roles in Tissue Development and Disease. Cells 2022, 11, 2404. [Google Scholar] [CrossRef]
- Yu, F.; Wu, Y.; Xie, Q. Ubiquitin-Proteasome System in ABA Signaling: From Perception to Action. Mol. Plant. 2016, 9, 21–33. [Google Scholar] [CrossRef]
- Qin, Q.; Wang, Y.; Huang, L.; Du, F.; Zhao, X.; Li, Z.; Wang, W.; Fu, B. A U-Box E3 Ubiquitin Ligase OsPUB67 Is Positively Involved in Drought Tolerance in Rice. Plant Mol. Biol. 2020, 102, 89–107. [Google Scholar] [CrossRef]
- Ning, Y.; Jantasuriyarat, C.; Zhao, Q.; Zhang, H.; Chen, S.; Liu, J.; Liu, L.; Tang, S.; Park, C.H.; Wang, X.; et al. The SINA E3 Ligase OsDIS1 Negatively Regulates Drought Response in Rice. Plant Physiol. 2011, 157, 242–255. [Google Scholar] [CrossRef]
- Hsu, K.H.; Liu, C.C.; Wu, S.J.; Kuo, Y.Y.; Lu, C.A.; Wu, C.R.; Lian, P.J.; Hong, C.Y.; Ke, Y.T.; Huang, J.H.; et al. Expression of a Gene Encoding a Rice RING Zinc-Finger Protein, OsRZFP34, Enhances Stomata Opening. Plant Mol. Biol. 2014, 86, 125–137. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Pareek, A.; Singla-Pareek, S.L. A NAP-family histone chaperone functions in abiotic stress response and adaptation. Plant Physiol. 2016, 171, 2854–2868. [Google Scholar] [CrossRef]
- Lyzenga, W.J.; Booth, J.K.; Stone, S.L. The Arabidopsis RING-Type E3 Ligase XBAT32 Mediates the Proteasomal Degradation of the Ethylene Biosynthetic Enzyme, 1-Aminocyclopropane-1-Carboxylate Synthase 7. Plant J. 2012, 71, 23–34. [Google Scholar] [CrossRef]
- Bratzel, F.; López-Torrejón, G.; Koch, M.; Del Pozo, J.C.; Calonje, M. Keeping Cell Identity in Arabidopsis Requires PRC1 RING-Finger Homologs That Catalyze H2A Monoubiquitination. Curr. Biol. 2010, 20, 1853–1862. [Google Scholar] [CrossRef]
- Zhou, M.; Paša-Tolić, L.; Stenoien, D.L. Profiling of Histone Post-Translational Modifications in Mouse Brain with High-Resolution Top-Down Mass Spectrometry. J. Proteome Res. 2017, 16, 599–608. [Google Scholar] [CrossRef]
- Geng, F.; Wenzel, S.; Tansey, W.P. Ubiquitin and Proteasomes in Transcription. Annu. Rev. Biochem. 2012, 81, 177–201. [Google Scholar] [CrossRef]
- Zou, B.; Yang, D.L.; Shi, Z.; Dong, H.; Hua, J. Monoubiquitination of Histone 2B at the Disease Resistance Gene Locus Regulates Its Expression and Impacts Immune Responses in Arabidopsis. Plant Physiol. 2014, 165, 309–318. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Zhang, H.; Hong, Y.; Huang, L.; Liu, S.; Li, X.; Ouyang, Z.; Song, F. Tomato Histone H2B Monoubiquitination Enzymes SlHUB1 and SlHUB2 Contribute to Disease Resistance against Botrytis cinerea through Modulating the Balance between SA- and JA/ET-Mediated Signaling Pathways. BMC Plant Biol. 2015, 15, 252. [Google Scholar] [CrossRef]
- Kim, J.H.; Lim, S.D.; Jang, C.S. Oryza sativa Drought-, Heat-, and Salt-Induced RING Finger Protein 1 (OsDHSRP1) Negatively Regulates Abiotic Stress-Responsive Gene Expression. Plant Mol. Biol. 2020, 103, 235–252. [Google Scholar] [CrossRef]
- Kim, J.H.; Khan, I.U.; Kim, M.S.; Seo, Y.W. Functional Characterization of Wheat Histone H2B Monoubiquitination Enzyme TaHUB2 in Response to Vernalization in Keumkang (Triticum aestivum L.). J. Plant Interact. 2021, 16, 93–103. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, X.; Yang, S.; Ren, H.; Yuan, J.; Jin, H.; Shi, C.; Lai, Z.; Xia, G. Functional Analysis of Tomato CHIP Ubiquitin E3 Ligase in Heat Tolerance. Sci. Rep. 2021, 11, 1713. [Google Scholar] [CrossRef]
- Wang, P.; Guo, K.; Su, Q.; Deng, J.; Zhang, X.; Tu, L. Histone Ubiquitination Controls Organ Size in Cotton (Gossypium hirsutum). Plant J. 2022, 110, 1005–1020. [Google Scholar] [CrossRef]
- Shiio, Y.; Eisenman, R.N. Histone Sumoylation Is Associated with Transcriptional Repression. Proc. Natl. Acad. Sci. USA 2003, 100, 13225–13230. [Google Scholar] [CrossRef]
- Miller, M.J.; Barrett-Wilt, G.A.; Hua, Z.; Vierstra, R.D. Proteomic Analyses Identify a Diverse Array of Nuclear Processes Affected by Small Ubiquitin-like Modifier Conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 16512–16517. [Google Scholar] [CrossRef]
- To, T.K.; Kim, J.M.; Matsui, A.; Kurihara, Y.; Morosawa, T.; Ishida, J.; Tanaka, M.; Endo, T.; Kakutani, T.; Toyoda, T.; et al. Arabidopsis HDA6 Regulates Locus-Directed Heterochromatin Silencing in Cooperation with MET1. PLoS Genet. 2011, 7, e1002055. [Google Scholar] [CrossRef]
- Han, D.; Chen, C.; Xia, S.; Liu, J.; Shu, J.; Nguyen, V.; Lai, J.; Cui, Y.; Yang, C. Chromatin-Associated SUMOylation Controls the Transcriptional Switch between Plant Development and Heat-Stress Responses. Plant Commun. 2020, 2, 100091. [Google Scholar] [CrossRef]
- Han, G.; Qiao, Z.; Li, Y.; Yang, Z.; Wang, C.; Zhang, Y.; Liu, L.; Wang, B. RING Zinc Finger Proteins in Plant Abiotic Stress Tolerance. Front. Plant Sci. 2022, 13, 877011. [Google Scholar] [CrossRef]
- Budhiraja, R.; Hermkes, R.; Müller, S.; Schmidt, J.; Colby, T.; Panigrahi, K.; Coupland, G.; Bachmair, A. Substrates Related to Chromatin and to RNA-Dependent Processes Are Modified by Arabidopsis SUMO Isoforms That Differ in a Conserved Residue with Influence on Desumoylation. Plant Physiol. 2009, 149, 1529–1540. [Google Scholar] [CrossRef]
- Augustine, R.C.; Vierstra, R.D. SUMOylation: Re-wiring the Plant Nucleus during Stress and Development. Curr. Opin. Plant Biol. 2018, 45, 143–154. [Google Scholar] [CrossRef]
- Roy, D.; Sadanandom, A. SUMO Mediated Regulation of Transcription Factors as a Mechanism for Transducing Environmental Cues into Cellular Signaling in Plants. Cell. Mol. Life Sci. 2021, 78, 2641–2664. [Google Scholar] [CrossRef]
- Wawrzyn’ska, A.; Sirko, A. Proteasomal Degradation of Proteins Is Important for the Proper Transcriptional Response to Sulfur Deficiency Conditions in Plants. Plant Cell Physiol. 2020, 61, 1548–1564. [Google Scholar] [CrossRef]
- Marand, A.P.; Zhang, T.; Zhu, B.; Jiang, J. Towards Genome-Wide Prediction and Characterization of Enhancers in Plants. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 131–139. [Google Scholar] [CrossRef]
- Municio Diaz, C.M. Characterization of CAP-D2 and CAP-D3 Condensin Subunits in Arabidopsis thaliana. Ph.D. Thesis, Martin-Luther-Universität Halle-Wittenberg, Halle, Germany, 2019. [Google Scholar]
- Saleh, A.; Alvarez-Venegas, R.; Avramova, Z. An Efficient Chromatin Immunoprecipitation (ChIP) Protocol for Studying Histone Modifications in Arabidopsis Plants. Nat. Protoc. 2008, 3, 1018–1025. [Google Scholar] [CrossRef]
- Zhu, Z.; Dong, A.; Shen, W.H. Histone Variants and Chromatin Assembly in Plant Abiotic Stress Responses. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 343–348. [Google Scholar] [CrossRef]
- Debbarma, J.; Sarki, Y.N.; Saikia, B.; Boruah, H.P.D.; Singha, D.L.; Chikkaputtaiah, C. Ethylene Response Factor (ERF) Family Proteins in Abiotic Stresses and CRISPR–Cas9 Genome Editing of ERFs for Multiple Abiotic Stress Tolerance in Crop Plants: A Review. Mol. Biotechnol. 2019, 61, 153–172. [Google Scholar] [CrossRef]
- Chen, X.; Bhadauria, V.; Ma, B. ChIP-seq: A Powerful Tool for Studying Protein-DNA Interactions in Plants. Curr. Issues Mol. Biol. 2018, 27, 171–180. [Google Scholar] [CrossRef]
- Park, P.J. ChIP–seq: Advantages and Challenges of a Maturing Technology. Nat. Rev. Genet. 2009, 10, 669–680. [Google Scholar] [CrossRef]
- Flensburg, C.; Kinkel, S.A.; Keniry, A.; Blewitt, M.E.; Oshlack, A. A Comparison of Control Samples for ChIP-seq of Histone Modifications. Front. Genet. 2014, 5, 329. [Google Scholar] [CrossRef]
- Nakato, R.; Sakata, T. Methods for ChIP-seq Analysis: A Practical Workflow and Advanced Applications. Methods 2021, 187, 44–53. [Google Scholar] [CrossRef]
- Solomon, E.R.; Caldwell, K.K.; Allan, A.M. A Novel Method for the Normalization of ChIP-qPCR Data. MethodsX 2021, 8, 101504. [Google Scholar] [CrossRef]
- Asp, P. How to Combine ChIP with qPCR. In Chromatin Immunoprecipitation: Methods and Protocols; Hamana Press: New York, NY, USA, 2018; pp. 29–42. [Google Scholar]
- Kuhn, A.; Østergaard, L. Chromatin Immunoprecipitation (ChIP) to Assess Histone Marks in Auxin-Treated Arabidopsis thaliana Inflorescence Tissue. Bio-Protoc. 2020, 10, e3832. [Google Scholar] [CrossRef]
- Laurent, L.; Wong, E.; Li, G.; Huynh, T.; Tsirigos, A.; Ong, C.T.; Low, H.M.; Kin Sung, K.W.; Rigoutsos, I.; Loring, J.; et al. Dynamic Changes in the Human Methylome during Differentiation. Genome Res. 2010, 20, 320–331. [Google Scholar] [CrossRef]
- Stirzaker, C.; Song, J.Z.; Statham, A.L.; Clark, S.J. Bisulphite Sequencing of Chromatin Immunoprecipitated DNA (BisChIP-seq). In DNA Methylation Protocols; Hamana Press: New York, NY, USA, 2018; pp. 285–302. [Google Scholar]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for Efficient Epigenomic Profiling of Small Samples and Single Cells. Nat. Commun. 2019, 10, 1930. [Google Scholar]
- Tao, X.Y.; Guan, X.Y.; Hong, G.J.; He, Y.Q.; Li, S.J.; Feng, S.L.; Wang, J.; Chen, G.; Xu, F.; Wang, J.W.; et al. Biotinylated Tn5 Transposase-Mediated CUT & Tag Efficiently Profiles Transcription Factor-DNA Interactions in Plants. Plant Biotechnol. J. 2023; in press. [Google Scholar]
- Bartosovic, M.; Kabbe, M.; Castelo-Branco, G. Single-Cell CUT&Tag Profiles Histone Modifications and Transcription Factors in Complex Tissues. Nat. Biotechnol. 2021, 39, 825–835. [Google Scholar]
- Kaya-Okur, H.S.; Janssens, D.H.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. Efficient Low-Cost Chromatin Profiling with CUT&Tag. Nat. Protoc. 2020, 15, 3264–3283. [Google Scholar]
- Tao, X.; Feng, S.; Zhao, T.; Guan, X. Efficient Chromatin Profiling of H3K4me3 Modification in Cotton Using CUT&Tag. Plant Methods. 2020, 16, 1–15. [Google Scholar]
- Wu, S.J.; Furlan, S.N.; Mihalas, A.B.; Kaya-Okur, H.S.; Feroze, A.H.; Emerson, S.N.; Zheng, Y.; Carson, K.; Cimino, P.J.; Keene, C.D.; et al. Single-Cell CUT&Tag Analysis of Chromatin Modifications in Differentiation and Tumor Progression. Nat. Biotechnol. 2021, 39, 819–824. [Google Scholar]
- Sidoli, S.; Garcia, B.A. Characterization of Individual Histone Posttranslational Modifications and Their Combinatorial Patterns by Mass Spectrometry-Based Proteomics Strategies. In Histones; Humana Press: New York, NY, USA, 2017; pp. 121–148. [Google Scholar]
- Huang, H.; Lin, S.; Garcia, B.A.; Zhao, Y. Quantitative Proteomic Analysis of Histone Modifications. Chem. Rev. 2015, 115, 2376–2418. [Google Scholar] [CrossRef]
- Khan, A.; Eikani, C.K.; Khan, H.; Iavarone, A.T.; Pesavento, J.J. Characterization of Chlamydomonasreinhardtii Core Histones by Top-Down Mass Spectrometry Reveals Unique Algae-Specific Variants and Post-Translational Modifications. J. Proteome Res. 2018, 17, 23–32. [Google Scholar] [CrossRef]
- Anderson, L.C.; Karch, K.R.; Ugrin, S.A.; Coradin, M.; English, A.M.; Sidoli, S.; Shabanowitz, J.; Garcia, B.A.; Hunt, D.F. Analyses of Histone Proteoforms Using Front-End Electron Transfer Dissociation-Enabled Orbitrap Instruments. Mol. Cell. Proteom. 2016, 15, 975–988. [Google Scholar] [CrossRef]
- Zhou, M.; Malhan, N.; Ahkami, A.H.; Engbrecht, K.; Myers, G.; Dahlberg, J.; Hollingsworth, J.; Sievert, J.A.; Hutmacher, R.; Madera, M. Top-Down Mass Spectrometry of Histone Modifications in Sorghum Reveals Potential Epigenetic Markers for Drought Acclimation. Methods 2020, 184, 29–39. [Google Scholar] [CrossRef]
- Miryeganeh, M. Plants’ Epigenetic Mechanisms and Abiotic Stress. Genes 2021, 12, 1106. [Google Scholar] [CrossRef]
- Ahmad, N.; Naeem, M.; Ali, H.; Alabbosh, K.F.; Hussain, H.; Khan, I.; Siddiqui, S.A.; Khan, A.A.; Iqbal, B. From Challenges to Solutions: The Impact of Melatonin on Abiotic Stress Synergies in Horticultural Plants via Redox Regulation and Epigenetic Signaling. Sci. Hortic. 2023, 321, 112369. [Google Scholar] [CrossRef]
- Song, Z.T.; Sun, L.; Lu, S.J.; Tian, Y.; Ding, Y.; Liu, J.X. Transcription Factor Interaction with COMPASS-Like Complex Regulates Histone H3K4 Trimethylation for Specific Gene Expression in Plants. Proc. Natl. Acad. Sci. USA 2015, 112, 2900–2905. [Google Scholar] [CrossRef]
- Kim, J.H. Multifaceted Chromatin Structure and Transcription Changes in Plant Stress Response. Int. J. Mol. Sci. 2021, 22, 2013. [Google Scholar] [CrossRef]
- Kim, J.M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef]
- Halder, K.; Chaudhuri, A.; Abdin, M.Z.; Majee, M.; Datta, A. Chromatin-Based Transcriptional Reprogramming in Plants under Abiotic Stresses. Plants 2022, 11, 1449. [Google Scholar] [CrossRef]
- Guarino, F.; Cicatelli, A.; Castiglione, S.; Agius, D.R.; Orhun, G.E.; Fragkostefanakis, S.; Leclercq, J.; Dobránszki, J.; Kaiserli, E.; Lieberman-Lazarovich, M.; et al. An Epigenetic Alphabet of Crop Adaptation to Climate Change. Front. Genet 2022, 13, 818727. [Google Scholar] [CrossRef]
- Ahuja, I.; de Vos, R.C.; Bones, A.M.; Hall, R.D. Plant Molecular Stress Responses Face Climate Change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef]
- Von Koskull-Döring, P.; Scharf, K.D.; Nover, L. The Diversity of Plant Heat Stress Transcription Factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar] [CrossRef]
- Pecinka, A.; Dinh, H.Q.; Baubec, T.; Rosa, M.; Lettner, N.; Scheid, O.M. Epigenetic Regulation of Repetitive Elements Is Attenuated by Prolonged Heat Stress in Arabidopsis. Plant Cell 2010, 22, 3118–3129. [Google Scholar] [CrossRef]
- Gan, L.; Wei, Z.; Yang, Z.; Li, F.; Wang, Z. Updated Mechanisms of GCN5—The Monkey King of the Plant Kingdom in Plant Development and Resistance to Abiotic Stresses. Cells 2021, 10, 979. [Google Scholar] [CrossRef]
- Hou, H.; Zhao, L.; Zheng, X.; Gautam, M.; Yue, M.; Hou, J.; Chen, Z.; Wang, P.; Li, L. Dynamic Changes in Histone Modification Are Associated with Upregulation of Hsf and rRNA Genes during Heat Stress in Maize Seedlings. Protoplasma 2019, 256, 1245–1256. [Google Scholar] [CrossRef]
- Wei, W.; Chen, J.Y.; Zeng, Z.X.; Kuang, J.F.; Lu, W.J.; Shan, W. The Ubiquitin E3 Ligase MaLUL2 Is Involved in High Temperature-Induced Green Ripening in Banana Fruit. Int. J. Mol. Sci. 2020, 21, 9386. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Wei, C.; Liu, X.; Wang, M.; Yu, F.; Xie, Q.; Tu, J. The RING Finger Ubiquitin E3 Ligase OsHTAS Enhances Heat Tolerance by Promoting H2O2-Induced Stomatal Closure in Rice. Plant Physiol. 2016, 170, 429–443. [Google Scholar] [CrossRef]
- Folsom, J.J.; Begcy, K.; Hao, X.; Wang, D.; Walia, H. Rice Fertilization-Independent Endosperm1 Regulates Seed Size under Heat Stress by Controlling Early Endosperm Development. Plant Physiol. 2014, 165, 238–248. [Google Scholar] [CrossRef]
- Weng, M.; Yang, Y.; Feng, H.; Pan, Z.; Shen, W.H.; Zhu, Y.; Dong, A. Histone Chaperone ASF1 Is Involved in Gene Transcription Activation in Response to Heat Stress in Arabidopsis thaliana. Plant Cell Environ. 2014, 37, 2128–2138. [Google Scholar] [CrossRef]
- Qiu, Y.; Li, M.; Jean, R.; Moore, C.M.; Chen, M. Daytime Temperature Is Sensed by Phytochrome B in Arabidopsis through a Transcriptional Activator HEMERA. Nat. Commun. 2019, 10, 140. [Google Scholar] [CrossRef]
- Zhang, L.; Luo, P.; Bai, J.; Wu, L.; Di, D.W.; Liu, H.Q.; Li, J.J.; Liu, Y.L.; Khaskheli, A.J.; Zhao, C.M.; et al. Function of Histone H2B Monoubiquitination in Transcriptional Regulation of Auxin Biosynthesis in Arabidopsis. Commun. Biol. 2021, 4, 206. [Google Scholar] [CrossRef]
- Hammoudi, V.; Beerens, B.; Jonker, M.J.; Helderman, T.A.; Vlachakis, G.; Giesbers, M.; Kwaaitaal, M.; van den Burg, H.A. The Protein Modifier SUMO Is Critical for Integrity of the Arabidopsis Shoot Apex at Warm Ambient Temperatures. J. Exp. Bot. 2021, 72, 7531–7548. [Google Scholar] [CrossRef]
- Liu, X.H.; Lyu, Y.S.; Yang, W.; Yang, Z.T.; Lu, S.J.; Liu, J.X. A Membrane Associated NAC Transcription Factor OsNTL3 Is Involved in Thermotolerance in Rice. Plant Biotechnol. J. 2020, 18, 1317–1329. [Google Scholar] [CrossRef]
- Wang, J.; Meng, X.; Yuan, C.; Harrison, A.P.; Chen, M. The Roles of Cross-Talk Epigenetic Patterns in Arabidopsis thaliana. Briefings Funct. Genom. 2016, 15, 278–287. [Google Scholar] [CrossRef]
- Verma, N.; Giri, S.K.; Singh, G.; Gill, R.; Kumar, A. Epigenetic Regulation of Heat and Cold Stress Responses in Crop Plants. Plant Gene. 2022, 29, 100351. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Chong, K. Cold Signaling in Plants: Insights into Mechanisms and Regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef]
- Chan, Z.; Wang, Y.; Cao, M.; Gong, Y.; Mu, Z.; Wang, H.; Hu, Y.; Deng, X.; He, X.J.; Zhu, J.K. RDM4 Modulates Cold Stress Resistance in Arabidopsis Partially through the CBF-Mediated Pathway. N. Phytol. 2016, 209, 1527–1539. [Google Scholar] [CrossRef]
- Min, H.J.; Jung, Y.J.; Kang, B.G.; Kim, W.T. CaPUB1, a Hot Pepper U-Box E3 Ubiquitin Ligase, Confers Enhanced Cold Stress Tolerance and Decreased Drought Stress Tolerance in Transgenic Rice (Oryza sativa L.). Mol. Cells 2016, 39, 250–257. [Google Scholar]
- Zeng, Z.; Zhang, W.; Marand, A.P.; Zhu, B.; Buell, C.R.; Jiang, J. Cold Stress Induces Enhanced Chromatin Accessibility and Bivalent Histone Modifications H3K4me3 and H3K27me3 of Active Genes in Potato. Genome Biol. 2019, 20, 1–17. [Google Scholar] [CrossRef]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef]
- Yung, W.S.; Li, M.W.; Sze, C.C.; Wang, Q.; Lam, H.M. Histone Modifications and Chromatin Remodeling in Plants in Response to Salt Stress. Physiol. Plant. 2021, 173, 1495–1513. [Google Scholar] [CrossRef]
- Singroha, G.; Kumar, S.; Gupta, O.P.; Singh, G.P.; Sharma, P. Uncovering the Epigenetic Marks Involved in Mediating Salt Stress Tolerance in Plants. Front. Genet. 2022, 13, 811732. [Google Scholar] [CrossRef]
- Ramu, V.S.; Oh, S.; Lee, H.K.; Nandety, R.S.; Oh, Y.; Lee, S.; Nakashima, J.; Tang, Y.; Senthil-Kumar, M.; Mysore, K.S. A novel role of salt-and drought-induced RING 1 protein in modulating plant defense against hemibiotrophic and necrotrophic pathogens. Mol. Plant Microbe Interact. 2021, 34, 297–308. [Google Scholar] [CrossRef]
- Ma, J.T.; Yin, C.C.; Zhou, M.L.; Wang, Z.L.; Wu, Y.M. A Novel DREB Transcription Factor from Halimodendron halodendron Leads to Enhanced Drought and Salt Tolerance in Arabidopsis. Biol. Plant. 2015, 59, 74–82. [Google Scholar] [CrossRef]
- Patanun, O.; Ueda, M.; Itouga, M.; Kato, Y.; Utsumi, Y.; Matsui, A.; Tanaka, M.; Utsumi, C.; Sakakibara, H.; Yoshida, M.; et al. The Histone Deacetylase Inhibitor Suberoylanilide Hydroxamic Acid Alleviates Salinity Stress in Cassava. Front. Plant Sci. 2017, 7, 2039–3389. [Google Scholar] [CrossRef]
- Mehdi, S.; Derkacheva, M.; Ramström, M.; Kralemann, L.; Bergquist, J.; Hennig, L. The WD40 Domain Protein MSI1 Functions in a Histone Deacetylase Complex to Fine-Tune Abscisic Acid Signaling. Plant Cell 2016, 28, 42. [Google Scholar] [CrossRef]
- Dong, W.; Gao, T.; Wang, Q.; Chen, J.; Lv, J.; Song, Y. Salinity Stress Induces Epigenetic Alterations to the Promoter of MsMYB4 Encoding a Salt-Induced MYB Transcription Factor. Plant Physiol. Biochem. 2020, 155, 709–715. [Google Scholar] [CrossRef]
- Ullah, F.; Xu, Q.; Zhao, Y.; Zhou, D.X. Histone Deacetylase HDA710 Controls Salt Tolerance by Regulating ABA Signaling in Rice. J. Integr. Plant Biol. 2021, 63, 451–467. [Google Scholar] [CrossRef]
- Zheng, M.; Lin, J.; Liu, X.; Chu, W.; Li, J.; Gao, Y.; An, K.; Song, W.; Xin, M.; Yao, Y.; et al. Histone Acetyltransferase TaHAG1 Acts as a Crucial Regulator to Strengthen Salt Tolerance of Hexaploid Wheat. Plant Physiol. 2021, 186, 1951–1969. [Google Scholar] [CrossRef]
- Barber, V.A.; Juday, G.P.; Finney, B.P. Reduced Growth of Alaskan White Spruce in the Twentieth Century from Temperature-Induced Drought Stress. Nat. Cell Biol. 2000, 405, 668–673. [Google Scholar] [CrossRef]
- Lim, S.D.; Lee, C.; Jang, C.S. The Rice RING E3 Ligase, OsCTR1, Inhibits Trafficking to the Chloroplasts of OsCP12 and OsRP1, and Its Overexpression Confers Drought Tolerance in Arabidopsis. Plant Cell Environ. 2014, 37, 1097–1113. [Google Scholar] [CrossRef]
- Wang, W.S.; Pan, Y.J.; Zhao, X.Q.; Dwivedi, D.; Zhu, L.H.; Ali, J.; Fu, B.Y.; Li, Z.K. Drought-Induced Site-Specific DNA Methylation and Its Association with Drought Tolerance in Rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 1951–1960. [Google Scholar] [CrossRef]
- Bao, Y.; Song, W.M.; Wang, P.; Yu, X.; Li, B.; Jiang, C.; Shiu, S.H.; Zhang, H.; Bassham, D.C. COST1 Regulates Autophagy to Control Plant Drought Tolerance. Proc. Natl. Acad. Sci. USA 2020, 117, 7482–7493. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, F.; Wu, Y.; Lou, L.; Liu, L.; Tian, M.; Ning, Y.; Shu, K.; Tang, S.; Xie, Q. The RING Finger Ubiquitin E3 Ligase SDIR1 Targets SDIR1-INTERACTING PROTEIN1 for Degradation to Modulate the Salt Stress Response and ABA Signaling in Arabidopsis. Plant Cell 2015, 27, 214–227. [Google Scholar] [CrossRef]
- Gao, T.; Wu, Y.; Zhang, Y.; Liu, L.; Ning, Y.; Wang, D.; Tong, H.; Chen, S.; Chu, C.; Xie, Q. OsSDIR1 Overexpression Greatly Improves Drought Tolerance in Transgenic Rice. Plant Mol. Biol. 2011, 76, 145–156. [Google Scholar] [CrossRef]
- Cui, L.H.; Min, H.J.; Byun, M.Y.; Oh, H.G.; Kim, W.T. OsDIRP1, a Putative RING E3 Ligase, Plays an Opposite Role in Drought and Cold Stress Responses as a Negative and Positive Factor, Respectively, in Rice (Oryza sativa L.). Front. Plant Sci. 2018, 9, 1797. [Google Scholar] [CrossRef]
- Godwin, J.; Farrona, S. Plant Epigenetic Stress Memory Induced by Drought: A Physiological and Molecular Perspective. Plant Epigenetics Epigenomics 2020, 2093, 243–259. [Google Scholar]
- Müller-Xing, R.; Xing, Q.; Goodrich, J. Footprints of the Sun: Memory of UV and Light Stress in Plants. Front. Plant Sci. 2014, 5, 474. [Google Scholar]
- Jenkins, G.I. Signal Transduction in Responses to UV-B Radiation. Annu. Rev. Plant Biol. 2009, 60, 407–431. [Google Scholar] [CrossRef]
- Hideg, E.; Jansen, M.A.; Strid, A. UV-B Exposure, ROS, and Stress: Inseparable Companions or Loosely Linked Associates? Trends Plant Sci. 2013, 18, 107–115. [Google Scholar] [CrossRef]
- Li, J.; Yang, L.; Jin, D.; Nezames, C.D.; Terzaghi, W.; Deng, X.W. UV-B-Induced Photomorphogenesis in Arabidopsis. Protein Cell 2013, 4, 485–492. [Google Scholar] [CrossRef]
- Boyko, A.; Blevins, T.; Yao, Y.; Golubov, A.; Bilichak, A.; Ilnytskyy, Y.; Hollander, J.; Meins, F.J.; Kovalchuk, I. Transgenerational Adaptation of Arabidopsis to Stress Requires DNA Methylation and the Function of Dicer-Like Proteins. PLoS ONE 2010, 5, e9514. [Google Scholar] [CrossRef]
- Lang-Mladek, C.; Popova, O.; Kiok, K.; Berlinger, M.; Rakic, B.; Aufsatz, W.; Jonak, C.; Hauser, M.T.; Luschnig, C. Transgenerational Inheritance and Resetting of Stress-Induced Loss of Epigenetic Gene Silencing in Arabidopsis. Mol. Plant. 2010, 3, 594–602. [Google Scholar] [CrossRef]
- Sokolova, D.; Vengzhen, G.; Kravets, A. The Effect of DNA Methylation Modification Polymorphism of Corn Seeds on Their Germination Rate, Seedling Resistance and Adaptive Capacity under UV-C Exposure. Am. J. Plant Biol. 2014, 1, 1–14. [Google Scholar]
- Kim, J.S.; Lim, J.Y.; Shin, H.; Kim, B.G.; Yoo, S.D.; Kim, W.T.; Huh, J.H. ROS1-Dependent DNA Demethylation Is Required for ABA-Inducible NIC3 Expression. Plant Physiol. 2019, 179, 1810–1821. [Google Scholar] [CrossRef]
- Jung, I.J.; Ahn, J.W.; Jung, S.; Hwang, J.E.; Hong, M.J.; Choi, H.I.; Kim, J.B. Overexpression of Rice Jacalin-Related Mannose-Binding Lectin (OsJAC1) Enhances Resistance to Ionizing Radiation in Arabidopsis. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar] [CrossRef]
- Cruz, C.; Della Rosa, M.; Krueger, C.; Gao, Q.; Horkai, D.; King, M.; Field, L.; Houseley, J. Tri-Methylation of Histone H3 Lysine 4 Facilitates Gene Expression in Ageing Cells. Elife 2018, 7, e34081. [Google Scholar] [CrossRef]
- Nielsen, M.; Ard, R.; Leng, X.; Ivanov, M.; Kindgren, P.; Pelechano, V.; Marquardt, S. Transcription-Driven Chromatin Repression of Intragenic Transcription Start Sites. PLoS Genet. 2019, 15, e1007969. [Google Scholar] [CrossRef]
- Yadav, R.K.; Jablonowski, C.M.; Fernandez, A.G.; Lowe, B.R.; Henry, R.A.; Finkelstein, D.; Barnum, K.J.; Pidoux, A.L.; Kuo, Y.M.; Huang, J.; et al. Histone H3G34R Mutation Causes Replication Stress, Homologous Recombination Defects and Genomic Instability in S. pombe. Elife 2017, 6, e27406. [Google Scholar] [CrossRef]
- Guidotti, N. Development of Chemical Biology Tools to Reveal the Function and Organization of the Silenced Chromatin State; LCBM, Hamana Press: New York, NY, USA.
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef]
- Rang, F.J.; de Luca, K.L.; de Vries, S.S.; Valdes-Quezada, C.; Boele, E.; Nguyen, P.D.; Guerreiro, I.; Sato, Y.; Kimura, H.; Bakkers, J.; et al. Single-cell profiling of transcriptome and histone modifications with EpiDamID. Mol. Cell 2022, 82, 1956–1970. [Google Scholar] [CrossRef]
- Choudhary, M.; Singh, A.; Rakshit, S. Coping with low moisture stress: Remembering and responding. Physiol. Plant. 2021, 172, 1162–1169. [Google Scholar] [CrossRef]

| Modification Type | Regulator Name | Crop | Stress Type | References |
|---|---|---|---|---|
| Acetylation Acetyltransferase | GCN5, AtHAC1 | Arabidopsis and Poplar | Heat, salinity, and drought (Chimeric dCas9 HAT) | [35,36,37] |
| Acetylation Acetyltransferase | HAT, AREB | Poplar | Drought | [36] |
| Deacetylation (Deactylase) | HDAC, IDS1 | Rice | Salinity | [38] |
| Deactylation (Deactylase) | HDAC, MYB96 | Arabidopsis | Drought | [39] |
| Deactylation Deacetylase | HDA9, HDA15, HDA705, BdHD1, HD2C | Arabidopsis, Rice, and Brachypodium | Drought, salinity cold, and heat | [28,39,40,41,42] |
| H3K9 acetylation | HAT, GCN5, ZmEXPANSIN-B2 | Maize | Salinity | [43] |
| H3 hyperacetylation | HAT genes, OsHAT genes | Rice | Drought | [44,45] |
| Deactylation (Deactylase) | HDA9, CYP707A1, CYP707A2 | Arabidopsis | Drought | [46] |
| Deactylation (Deactylase) | BdHD1, WRKY24 | Purple False Brome or Stiff Brome | Drought | [47] |
| Acetylation | AtHAC1 | Arabidopsis | Heat | [48] |
| Acetylation | MYST, ELP3, GCN5 | Barley | Drought | [49] |
| Acetylation | OsHAC703, OsHAG703, OsHAF701, OsHAM70 | Rice | Drought | [50] |
| Deacetylation (Deactylase) | 84KHDA903 | Tobacco | Drought | [51] |
| Deacetylation (Deactylase) | HD2C, HSFA3, HSFC1, HSP10 | Arabidopsis | Heat | [42] |
| Acetylation | GCN5, PtrNAC006, | Black Cottonwood Tree | Drought | [52] |
| Recruiter | MYB96, IDS1, AREB1 | Arabidopsis, Rice, and Poplar | Drought and salinity | [38,39,52] |
| Methylation Methyltransferase | ATX1, ATX4/5 | Arabidopsis | Drought | [53,54] |
| Demethylation Demethylase | JMJ17 | Arabidopsis | Drought | [55] |
| Trimethylation | HMT | Arabidopsis | Gamma irradiation | [56] |
| Ubiquitination Ubiquitinase | HUB1/2, AtHUB2, OsHUB2 | Arabidopsis, Cotton, and Rice | Salinity and drought | [57,58,59,60,61] |
| Phosphorylation Kinase | MLK1/2 | Arabidopsis | Drought and salinity | [62,63,64] |
| Ubiquitinase and deubiquitinase | H2B | Rice | Drought | [61] |
| Sumoylation | SUMO E3 ligase (AtSIZ1, OsSIZ1) | Arabidopsis and Rice | Heat | [65,66,67] |
| Ubiquitination | SNAC1 gene | Wheat | Salt and drought | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, M.; Sidhu, A.K.; Samota, M.K.; Gupta, M.; Koli, P.; Choudhary, M. Post-Translational Modifications in Histones and Their Role in Abiotic Stress Tolerance in Plants. Proteomes 2023, 11, 38. https://doi.org/10.3390/proteomes11040038
Sharma M, Sidhu AK, Samota MK, Gupta M, Koli P, Choudhary M. Post-Translational Modifications in Histones and Their Role in Abiotic Stress Tolerance in Plants. Proteomes. 2023; 11(4):38. https://doi.org/10.3390/proteomes11040038
Chicago/Turabian StyleSharma, Madhvi, Amanpreet K. Sidhu, Mahesh Kumar Samota, Mamta Gupta, Pushpendra Koli, and Mukesh Choudhary. 2023. "Post-Translational Modifications in Histones and Their Role in Abiotic Stress Tolerance in Plants" Proteomes 11, no. 4: 38. https://doi.org/10.3390/proteomes11040038
APA StyleSharma, M., Sidhu, A. K., Samota, M. K., Gupta, M., Koli, P., & Choudhary, M. (2023). Post-Translational Modifications in Histones and Their Role in Abiotic Stress Tolerance in Plants. Proteomes, 11(4), 38. https://doi.org/10.3390/proteomes11040038






