Dysregulated Gene Expression in Lymphoblasts from Parkinson’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Participant Cohort
2.2. Transcriptomic Cohort
2.3. Proteomic Cohort
2.4. Generation of Lymphoblastoid Cell Lines
2.5. Whole-Cell Protein Extraction and Mass Spectrometry
2.6. RNA Extraction and RNA-Seq
2.7. Software Programs and Statistical Analysis
3. Results
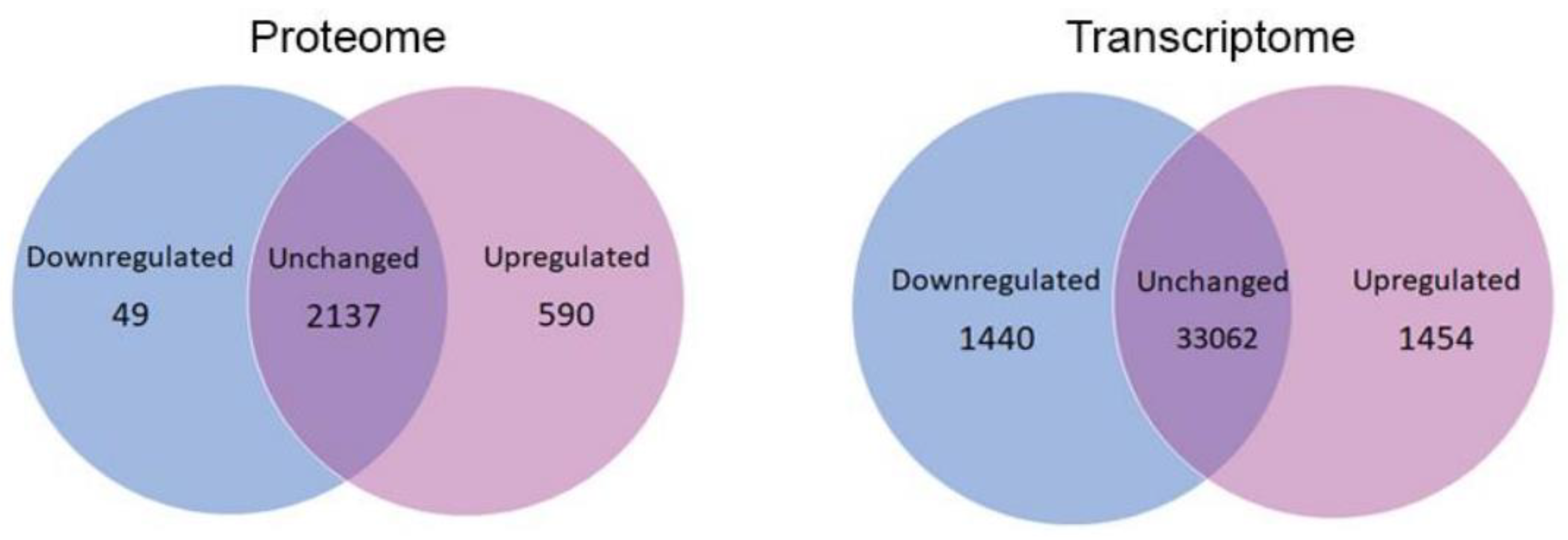
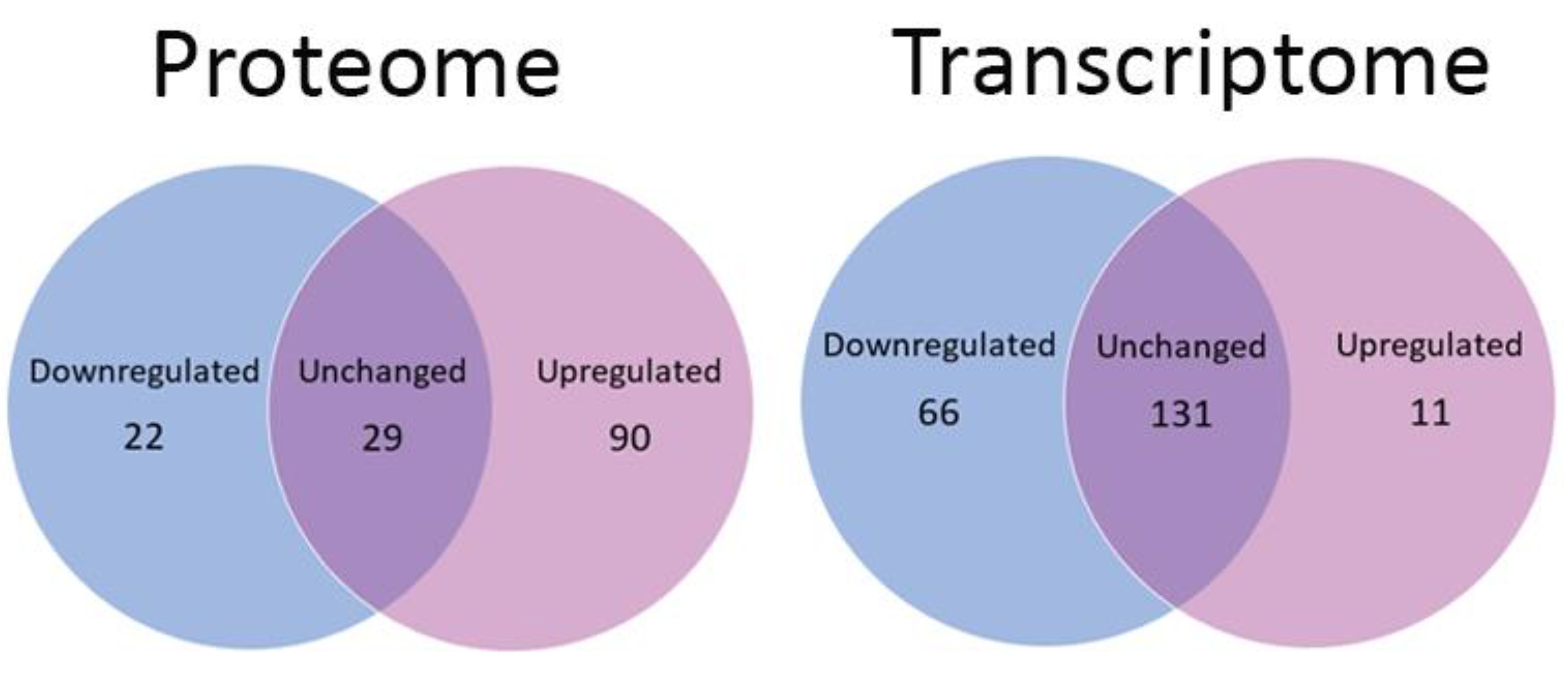
3.1. Global Changes in Parkinson Disease Proteomes and Transcriptomes
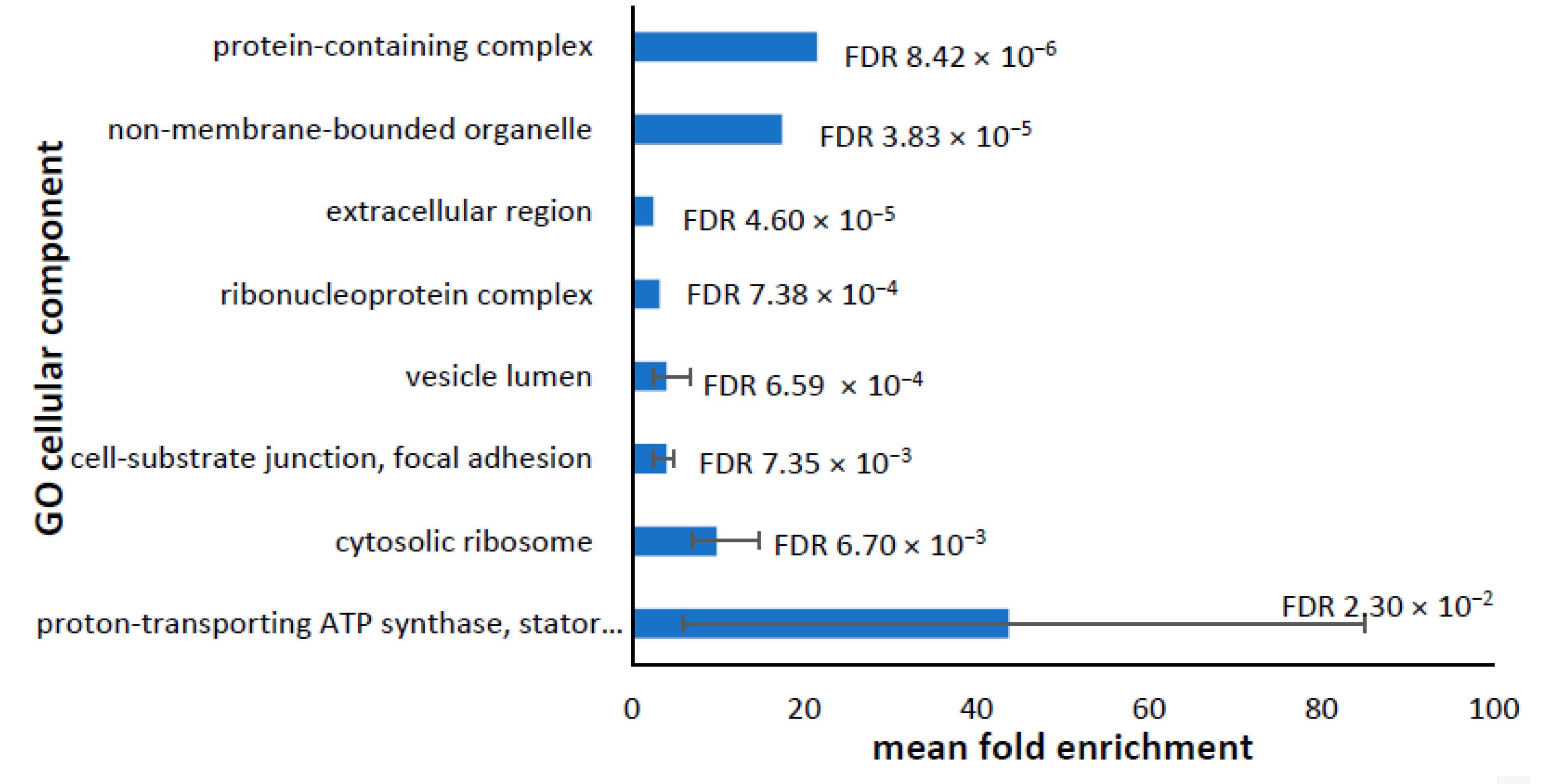
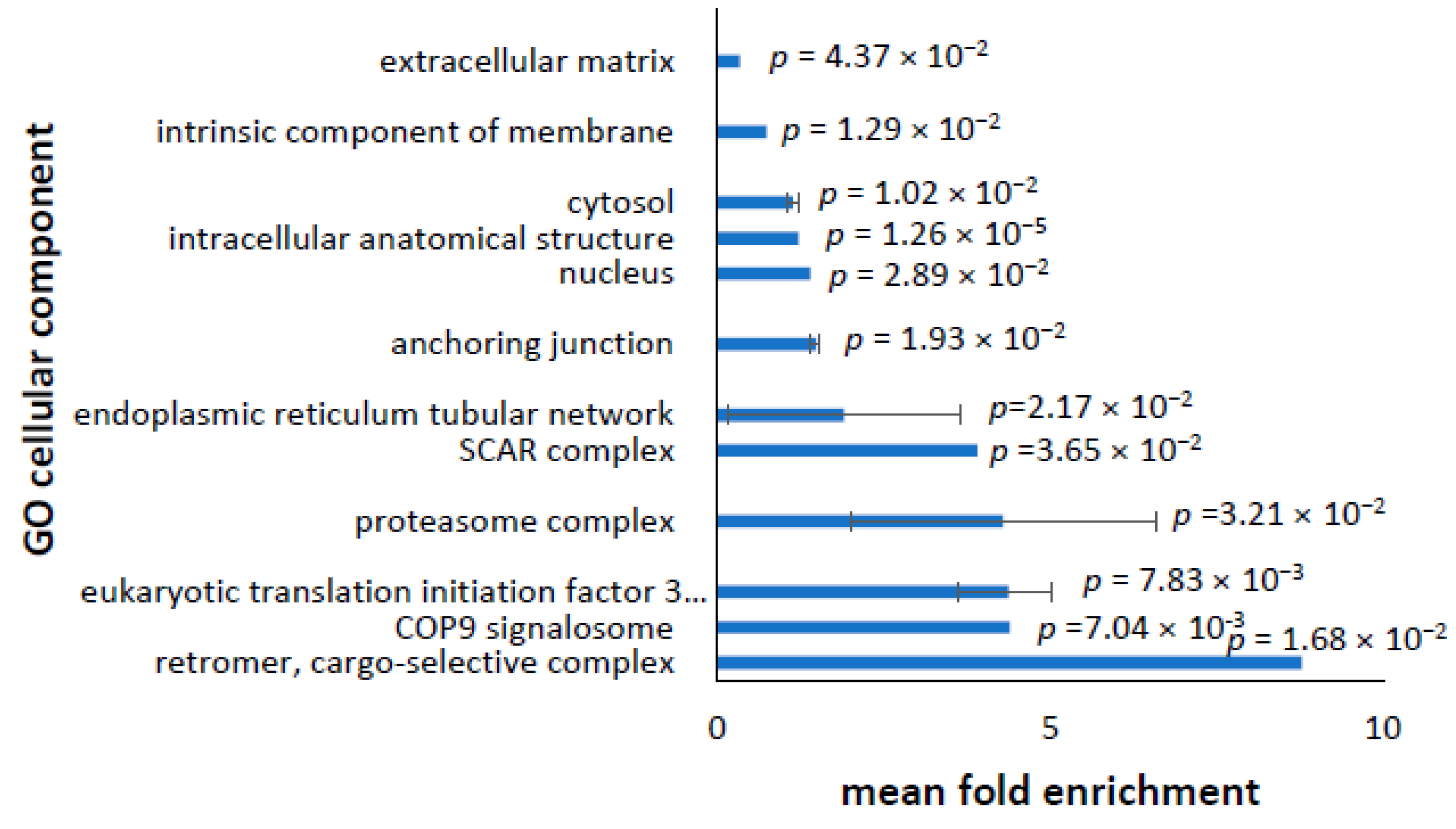
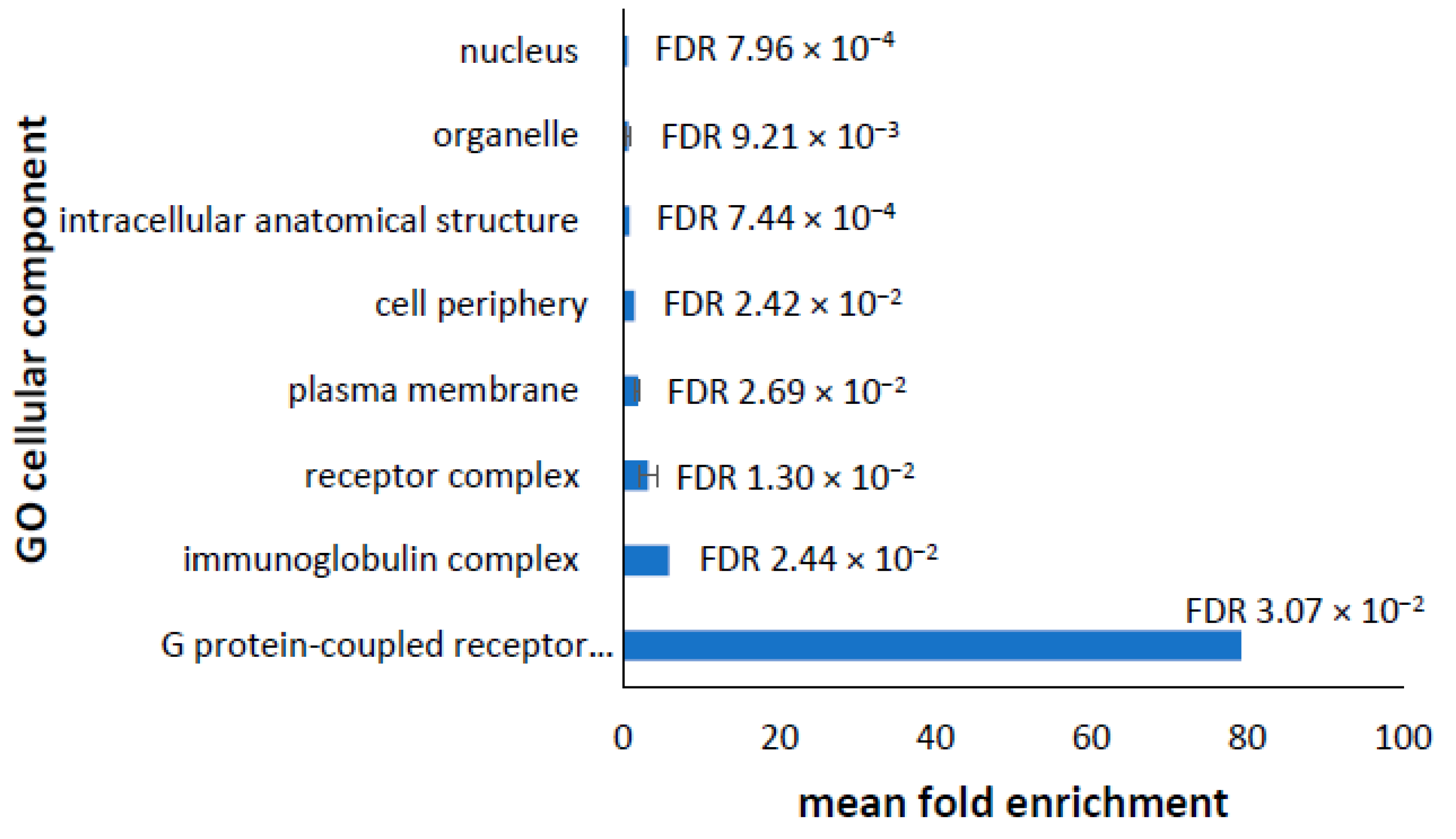
3.2. Cellular Component
3.3. Dysregulated Proteome
3.4. Dysregulated Transcriptome
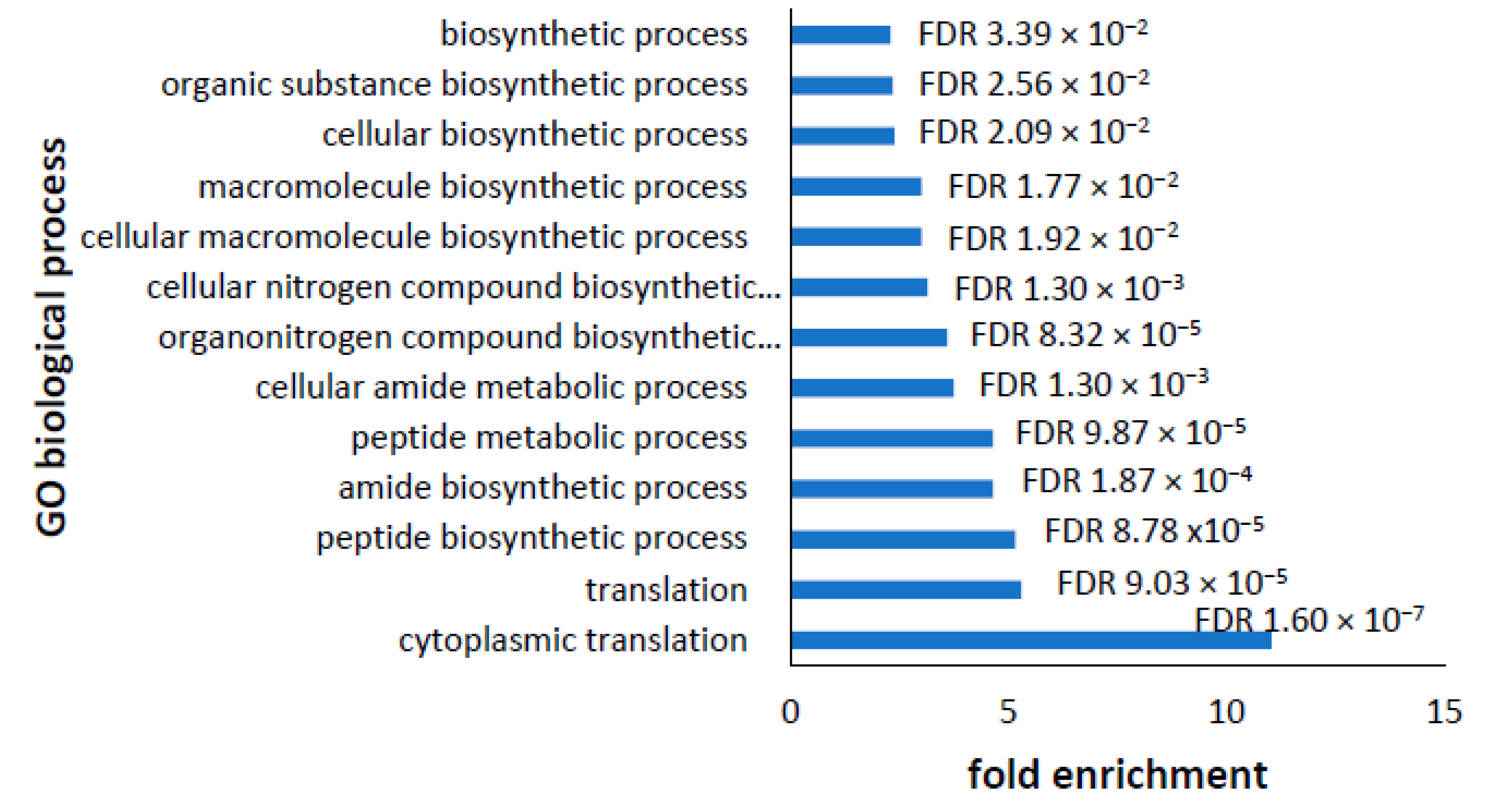
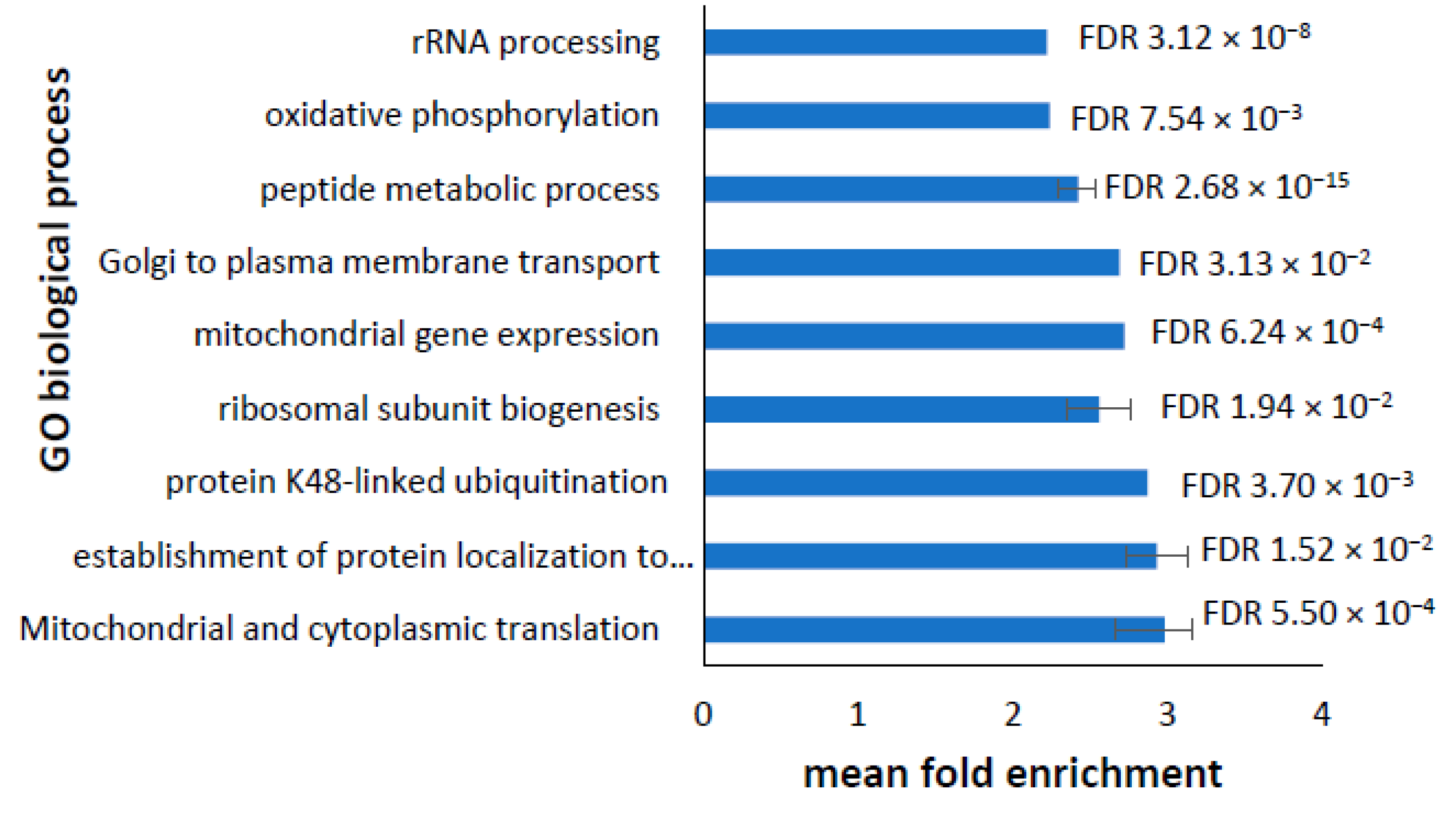
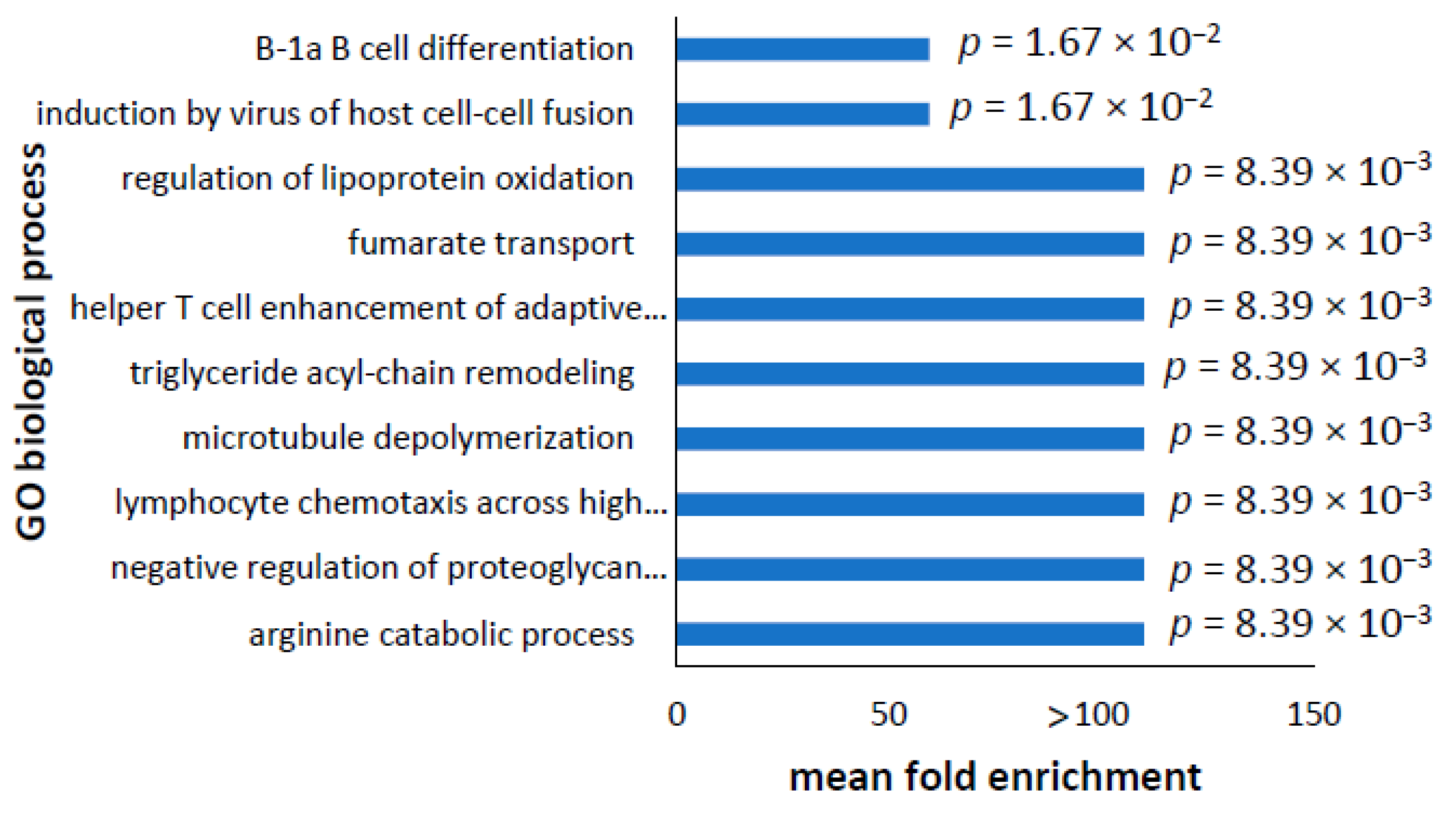
3.5. Analysis of Altered GO Biological Processes in Differentially Expressed Transcripts and Proteins in PD Cell Lines
3.6. Dysregulated Proteome
3.7. Dysregulated Transcriptome
3.8. Analysis of Mitochondrial Proteins
3.9. Dysregulated Mitochondrial Proteome
3.10. Dysregulated Transcriptome-Encoding Mitochondrial Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gammon, K. Neurodegenerative disease: Brain windfall. Nature 2014, 515, 299–300. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J. An Essay on the Shaking Palsy; Sherwood, Neely, and Jones: London, UK, 1817. [Google Scholar]
- Hardy, J. Genetic analysis of pathways to Parkinson disease. Neuron 2010, 68, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Giguere, N.; Burke Nanni, S.; Trudeau, L.E. On Cell Loss and Selective Vulnerability of Neuronal Populations in Parkinson’s Disease. Front. Neurol. 2018, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Trist, B.G.; Hare, D.J.; Double, K.L. Oxidative stress in the aging substantia nigra and the etiology of Parkinson’s disease. Aging Cell 2019, 18, e13031. [Google Scholar] [CrossRef]
- Papagiannakis, N.; Xilouri, M.; Koros, C.; Simitsi, A.M.; Stamelou, M.; Maniati, M.; Stefanis, L. Autophagy dysfunction in peripheral blood mononuclear cells of Parkinson’s disease patients. Neurosci. Lett. 2019, 704, 112–115. [Google Scholar] [CrossRef]
- Vavilova, J.D.; Boyko, A.A.; Troyanova, N.I.; Ponomareva, N.V.; Fokin, V.F.; Fedotova, E.Y.; Streltsova, M.A.; Kust, S.A.; Grechikhina, M.V.; Shustova, O.A.; et al. Alterations in Proteostasis System Components in Peripheral Blood Mononuclear Cells in Parkinson Disease: Focusing on the HSP70 and p62 Levels. Biomolecules 2022, 12, 493. [Google Scholar] [CrossRef]
- Borrageiro, G.; Haylett, W.; Seedat, S.; Kuivaniemi, H.; Bardien, S. A review of genome-wide transcriptomics studies in Parkinson’s disease. Eur. J. Neurosci. 2018, 47, 1–16. [Google Scholar] [CrossRef]
- Dixit, A.; Mehta, R.; Singh, A.K. Proteomics in Human Parkinson’s Disease: Present Scenario and Future Directions. Cell Mol. Neurobiol. 2019, 39, 901–915. [Google Scholar] [CrossRef]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006, 27, 126–139. [Google Scholar] [CrossRef]
- Bolen, C.R.; Uduman, M.; Kleinstein, S.H. Cell subset prediction for blood genomic studies. BMC Bioinform. 2011, 12, 258. [Google Scholar] [CrossRef]
- Scherzer, C.R.; Offe, K.; Gearing, M.; Rees, H.D.; Fang, G.; Heilman, C.J.; Schaller, C.; Bujo, H.; Levey, A.I.; Lah, J.J. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch. Neurol. 2004, 61, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Annesley, S.J.; Lay, S.T.; De Piazza, S.W.; Sanislav, O.; Hammersley, E.; Allan, C.Y.; Francione, L.M.; Bui, M.Q.; Chen, Z.P.; Ngoei, K.R.; et al. Immortalized Parkinson’s disease lymphocytes have enhanced mitochondrial respiratory activity. DMM 2016, 9, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Hauser, D.N.; Hastings, T.G. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease and monogenic parkinsonism. Neurobiol. Dis. 2013, 51, 35–42. [Google Scholar] [CrossRef]
- Chen, S.; Annesley, S.J.; Jasim, R.A.F.; Musco, V.J.; Sanislav, O.; Fisher, P.R. The Parkinson’s disease-associated protein DJ-1 plays a positive nonmitochondrial role in endocytosis in Dictyostelium cells. Dis. Model Mech. 2017, 10, 1261–1271. [Google Scholar] [CrossRef]
- Haylett, W.S.C.; van der Westhuizen, F.; van Dyk, H.; van der Merwe, L.; van der Merwe, C.; Loos, B.; Carr, J.; Kinnear, C.; Bardien, S. Altered Mitochondrial Respiration and Other Features of Mitochondrial Function in Parkin-Mutant Fibroblasts from Parkinson’s Disease Patients. Parkinson’s Dis. 2016, 2016, 1819209. [Google Scholar]
- Ugalde, C.L.; Annesley, S.J.; Gordon, S.E.; Mroczek, K.; Perugini, M.A.; Lawson, V.A.; Fisher, P.R.; Finkelstein, D.I.; Hill, A.F. Misfolded alpha-synuclein causes hyperactive respiration without functional deficit in live neuroblastoma cells. DMM 2020, 13, dmm040899. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef]
- Thomas, P.D.; Kejariwal, A.; Guo, N.; Mi, H.; Campbell, M.J.; Muruganujan, A.; Lazareva-Ulitsky, B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006, 34, W645–W650. [Google Scholar] [CrossRef]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Dong, Q.; Muruganujan, A.; Gaudet, P.; Lewis, S.; Thomas, P.D. PANTHER version 7: Improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010, 38, D204–D210. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.D.; Selvaratnam, T.; Lee, J.C.T.; Chao, Y.X.; Tan, E.K. Molecular targets for modulating the protein translation vital to proteostasis and neuron degeneration in Parkinson’s disease. Transl. Neurodegener. 2019, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Hoozemans, J.J.; van Haastert, E.S.; Eikelenboom, P.; de Vos, R.A.; Rozemuller, J.M.; Scheper, W. Activation of the unfolded protein response in Parkinson’s disease. Biochem. Biophys. Res. Commun. 2007, 354, 707–711. [Google Scholar] [CrossRef]
- Mutez, E.; Nkiliza, A.; Belarbi, K.; de Broucker, A.; Vanbesien-Mailliot, C.; Bleuse, S.; Duflot, A.; Comptdaer, T.; Semaille, P.; Blervaque, R.; et al. Involvement of the immune system, endocytosis and EIF2 signaling in both genetically determined and sporadic forms of Parkinson’s disease. Neurobiol. Dis. 2014, 63, 165–170. [Google Scholar] [CrossRef]
- Sheikh, B.N.; Guhathakurta, S.; Akhtar, A. The non-specific lethal (NSL) complex at the crossroads of transcriptional control and cellular homeostasis. EMBO Rep. 2019, 20, e47630. [Google Scholar] [CrossRef]
- Deshpande, P.; Flinkman, D.; Hong, Y.; Goltseva, E.; Siino, V.; Sun, L.; Peltonen, S.; Elo, L.L.; Kaasinen, V.; James, P.; et al. Protein synthesis is suppressed in sporadic and familial Parkinson’s disease by LRRK2. FASEB J. 2020, 34, 14217–14233. [Google Scholar] [CrossRef]
- Ori, A.; Toyama, B.H.; Harris, M.S.; Bock, T.; Iskar, M.; Bork, P.; Ingolia, N.T.; Hetzer, M.W.; Beck, M. Integrated Transcriptome and Proteome Analyses Reveal Organ-Specific Proteome Deterioration in Old Rats. Cell Syst. 2015, 1, 224–237. [Google Scholar] [CrossRef]
- Wei, Y.N.; Hu, H.Y.; Xie, G.C.; Fu, N.; Ning, Z.B.; Zeng, R.; Khaitovich, P. Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging. Genome Biol. 2015, 16, 41. [Google Scholar] [CrossRef]
- Jan, A.; Jansonius, B.; Delaidelli, A.; Bhanshali, F.; An, Y.A.; Ferreira, N.; Smits, L.M.; Negri, G.L.; Schwamborn, J.C.; Jensen, P.H.; et al. Activity of translation regulator eukaryotic elongation factor-2 kinase is increased in Parkinson disease brain and its inhibition reduces alpha synuclein toxicity. Acta Neuropathol. Commun. 2018, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.M.; Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 2009, 10, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Mu, Z.; Wang, L.; Deng, W.; Wang, J.; Wu, G. Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane. Cell Discov. 2017, 3, 17049. [Google Scholar] [CrossRef] [PubMed]
- Inoki, K.; Zhu, T.; Guan, K.L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003, 115, 577–590. [Google Scholar] [CrossRef]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.K.; Chao, Y.X.; West, A.; Chan, L.L.; Poewe, W.; Jankovic, J. Parkinson disease and the immune system-associations, mechanisms and therapeutics. Nat. Rev. Neurol. 2020, 16, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, T.M.; Chang, Y.S.; Chen, W.S.; Sheu, J.J.; Chen, Y.H.; Chen, J.H. Autoimmune rheumatic diseases and the risk of Parkinson disease: A nationwide population-based cohort study in Taiwan. Ann. Med. 2018, 50, 83–90. [Google Scholar] [CrossRef]
- Witoelar, A.; Jansen, I.E.; Wang, Y.; Desikan, R.S.; Gibbs, J.R.; Blauwendraat, C.; Thompson, W.K.; Hernandez, D.G.; Djurovic, S.; Schork, A.J.; et al. Genome-wide Pleiotropy Between Parkinson Disease and Autoimmune Diseases. JAMA Neurol. 2017, 74, 780–792. [Google Scholar] [CrossRef]
- Li, X.; Sundquist, J.; Sundquist, K. Subsequent risks of Parkinson disease in patients with autoimmune and related disorders: A nationwide epidemiological study from Sweden. Neurodegener. Dis. 2012, 10, 277–284. [Google Scholar] [CrossRef]
- Gerhard, A.; Pavese, N.; Hotton, G.; Turkheimer, F.; Es, M.; Hammers, A.; Eggert, K.; Oertel, W.; Banati, R.B.; Brooks, D.J. In vivo imaging of microglial activation with [11C](R)-PK11195 PET in idiopathic Parkinson’s disease. Neurobiol. Dis. 2006, 21, 404–412. [Google Scholar] [CrossRef]
- Lin, C.H.; Chen, C.C.; Chiang, H.L.; Liou, J.M.; Chang, C.M.; Lu, T.P.; Chuang, E.Y.; Tai, Y.C.; Cheng, C.; Lin, H.Y.; et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J. Neuroinflamm. 2019, 16, 129. [Google Scholar] [CrossRef] [PubMed]
- Brodacki, B.; Staszewski, J.; Toczylowska, B.; Kozlowska, E.; Drela, N.; Chalimoniuk, M.; Stepien, A. Serum interleukin (IL-2, IL-10, IL-6, IL-4), TNFalpha, and INFgamma concentrations are elevated in patients with atypical and idiopathic parkinsonism. Neurosci. Lett. 2008, 441, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Mogi, M.; Harada, M.; Narabayashi, H.; Inagaki, H.; Minami, M.; Nagatsu, T. Interleukin (IL)-1 beta, IL-2, IL-4, IL-6 and transforming growth factor-alpha levels are elevated in ventricular cerebrospinal fluid in juvenile parkinsonism and Parkinson’s disease. Neurosci. Lett. 1996, 211, 13–16. [Google Scholar] [CrossRef]
- Qiu, X.; Xiao, Y.; Wu, J.; Gan, L.; Huang, Y.; Wang, J. C-Reactive Protein and Risk of Parkinson’s Disease: A Systematic Review and Meta-Analysis. Front. Neurol. 2019, 10, 384. [Google Scholar] [CrossRef]
- Planken, A.; Kurvits, L.; Reimann, E.; Kadastik-Eerme, L.; Kingo, K.; Koks, S.; Taba, P. Looking beyond the brain to improve the pathogenic understanding of Parkinson’s disease: Implications of whole transcriptome profiling of Patients’ skin. BMC Neurol. 2017, 17, 6. [Google Scholar] [CrossRef]
- Merrill, A.H., Jr.; Schmelz, E.M.; Wang, E.; Dillehay, D.L.; Rice, L.G.; Meredith, F.; Riley, R.T. Importance of sphingolipids and inhibitors of sphingolipid metabolism as components of animal diets. J. Nutr. 1997, 127, 830S–833S. [Google Scholar] [CrossRef]
- Merrill, A.H., Jr.; Sullards, M.C.; Wang, E.; Voss, K.A.; Riley, R.T. Sphingolipid metabolism: Roles in signal transduction and disruption by fumonisins. Environ. Health Perspect. 2001, 109 (Suppl. S2), 283–289. [Google Scholar] [CrossRef]
- Dasgupta, S.; Ray, S.K. Diverse Biological Functions of Sphingolipids in the CNS: Ceramide and Sphingosine Regulate Myelination in Developing Brain but Stimulate Demyelination during Pathogenesis of Multiple Sclerosis. J. Neurol. Psychol. 2017, 5, 7. [Google Scholar] [CrossRef]
- Lee, Y.J.; Wang, S.; Slone, S.R.; Yacoubian, T.A.; Witt, S.N. Defects in very long chain fatty acid synthesis enhance alpha-synuclein toxicity in a yeast model of Parkinson’s disease. PLoS ONE 2011, 6, e15946. [Google Scholar] [CrossRef]
- Sidransky, E.; Nalls, M.A.; Aasly, J.O.; Aharon-Peretz, J.; Annesi, G.; Barbosa, E.R.; Bar-Shira, A.; Berg, D.; Bras, J.; Brice, A.; et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl. J. Med. 2009, 361, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Abbott, S.K.; Li, H.; Munoz, S.S.; Knoch, B.; Batterham, M.; Murphy, K.E.; Halliday, G.M.; Garner, B. Altered ceramide acyl chain length and ceramide synthase gene expression in Parkinson’s disease. Mov. Disord. 2014, 29, 518–526. [Google Scholar] [CrossRef]
- Hu, L.; Dong, M.X.; Huang, Y.L.; Lu, C.Q.; Qian, Q.; Zhang, C.C.; Xu, X.M.; Liu, Y.; Chen, G.H.; Wei, Y.D. Integrated Metabolomics and Proteomics Analysis Reveals Plasma Lipid Metabolic Disturbance in Patients with Parkinson’s Disease. Front. Mol. Neurosci. 2020, 13, 80. [Google Scholar] [CrossRef]
- Altamura, N.; Capitanio, N.; Bonnefoy, N.; Papa, S.; Dujardin, G. The Saccharomyces cerevisiae OXA1 gene is required for the correct assembly of cytochrome c oxidase and oligomycin-sensitive ATP synthase. FEBS Lett. 1996, 382, 111–115. [Google Scholar] [CrossRef]
- Garcia-Aguilar, A.; Cuezva, J.M. A Review of the Inhibition of the Mitochondrial ATP Synthase by IF1 in vivo: Reprogramming Energy Metabolism and Inducing Mitohormesis. Front. Physiol. 2018, 9, 1322. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bermudez, J.; Cuezva, J.M. The ATPase Inhibitory Factor 1 (IF1): A master regulator of energy metabolism and of cell survival. Biochim. Biophys. Acta 2016, 1857, 1167–1182. [Google Scholar] [CrossRef]
- De Rasmo, D.; Micelli, L.; Santeramo, A.; Signorile, A.; Lattanzio, P.; Papa, S. cAMP regulates the functional activity, coupling efficiency and structural organization of mammalian FOF1 ATP synthase. Biochim. Biophys. Acta 2016, 1857, 350–358. [Google Scholar] [CrossRef]
- Demine, S.; Renard, P.; Arnould, T. Mitochondrial Uncoupling: A Key Controller of Biological Processes in Physiology and Diseases. Cells 2019, 8, 795. [Google Scholar] [CrossRef]
- Kunji, E.R.S.; King, M.S.; Ruprecht, J.J.; Thangaratnarajah, C. The SLC25 Carrier Family: Important Transport Proteins in Mitochondrial Physiology and Pathology. Physiology 2020, 35, 302–327. [Google Scholar] [CrossRef]
- Rupprecht, A.; Brauer, A.U.; Smorodchenko, A.; Goyn, J.; Hilse, K.E.; Shabalina, I.G.; Infante-Duarte, C.; Pohl, E.E. Quantification of uncoupling protein 2 reveals its main expression in immune cells and selective up-regulation during T-cell proliferation. PLoS ONE 2012, 7, e41406. [Google Scholar] [CrossRef]
- Kumar, R.T.A.; Singothu, S.; Singh, S.B.; Bhandari, V. Uncoupling proteins as a therapeutic target for the development of new era drugs against neurodegenerative disorder. Biomed. Pharmacother. 2022, 147, 112656. [Google Scholar] [CrossRef] [PubMed]
- Stanczak-Mrozek, K.I.; Kolanowska, M.; Grieb, P. Application of Western blotting for the detection of uncoupling protein-2 (UCP-2) in mitochondria from smokers and non-smokers. Adv. Respir. Med. 2009, 77, 118–122. [Google Scholar]
- Jezek, P.; Holendova, B.; Garlid, K.D.; Jaburek, M. Mitochondrial Uncoupling Proteins: Subtle Regulators of Cellular Redox Signaling. Antioxid. Redox Signal. 2018, 29, 667–714. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.; Covian, R.; Aponte, A.M.; Glancy, B.; Taylor, J.F.; Chess, D.; Balaban, R.S. Regulation of oxidative phosphorylation complex activity: Effects of tissue-specific metabolic stress within an allometric series and acute changes in workload. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R1034–R1048. [Google Scholar] [CrossRef]
- Willers, I.M.; Martinez-Reyes, I.; Martinez-Diez, M.; Cuezva, J.M. miR-127-5p targets the 3′UTR of human beta-F1-ATPase mRNA and inhibits its translation. Biochim. Biophys. Acta 2012, 1817, 838–848. [Google Scholar] [CrossRef] [PubMed]


| Complex | Proteins Upregulated (Average Fold-Change) | Proteins Downregulated (Average Fold-Change) | Proportion Test (Significance p-Value) | t-Test (Significance p-Value) |
|---|---|---|---|---|
| Complex V | 2 (1.65) | 10 (0.85) | 0.04 | 0.45 |
| Complex IV | 7 (1.70) | 3 (0.86) | 0.18 | 0.021 |
| Complex III | 6 (2.24) | 3 (0.78) | 0.35 | 0.11 |
| Complex II | 2 (1.26) | 0 | 0.32 | 0.054 |
| Complex I | 19 (2.63) | 7 (0.79) | 0.0023 | 7.86 × 10−4 |
| TCA | 17 (1.8) | 7 (0.92) | 0.0094 | 0.0168 |
| Mitochondrial transport (TIMM/TOMM complex) | 7 (1.99) | 2 (0.54) | 0.059 | 0.0509 |
| SLC25 transporters | 6 (5.02) | 5 (0.91) | 1 | 0.103 |
| Others | LAMTOR (1.88), PRKAA1 (2.4), OXA1L (0.69) (Assembly of CV & CIV), IF1 which inhibits ATP activity (4.0) | |||
| Transcripts Upregulated (Average Fold-Change) | Transcripts Downregulated (Average Fold-Change) | Proportion Test (Significance p-Value) | t-Test (Significance p-Value) | |
|---|---|---|---|---|
| Complex I | 10 (1.04) | 34 (0.91) | 9.41 × 10−7 | 3.94 × 10−6 |
| Complex II | 2 (1.02) | 6 (0.90) | 0.13 | 9.01 × 10−3 |
| Complex III | 1 (1.10) | 10 (0.89) | 6.47 × 10−4 | 3.96 ×10−3 |
| Complex IV | 10 (1.07) | 25 (0.90) | 8.18 × 10−4 | 5.41 × 10−3 |
| Complex V | 3 (1.08) | 16 (0.90) | 9.89 × 10−5 | 8.41 × 10−4 |
| TCA | 7 (1.08) | 11 (0.94) | 0.32 | 0.40 |
| SLC25 transporters | 18 (1.34) | 34 (0.91) | 0.0033 | 0.83 |
| TIMM/TOMM | 4 (1.05) | 19 (0.91) | 7.11 × 10−5 | 1.50 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Annesley, S.J.; Allan, C.Y.; Sanislav, O.; Evans, A.; Fisher, P.R. Dysregulated Gene Expression in Lymphoblasts from Parkinson’s Disease. Proteomes 2022, 10, 20. https://doi.org/10.3390/proteomes10020020
Annesley SJ, Allan CY, Sanislav O, Evans A, Fisher PR. Dysregulated Gene Expression in Lymphoblasts from Parkinson’s Disease. Proteomes. 2022; 10(2):20. https://doi.org/10.3390/proteomes10020020
Chicago/Turabian StyleAnnesley, Sarah Jane, Claire Yvonne Allan, Oana Sanislav, Andrew Evans, and Paul Robert Fisher. 2022. "Dysregulated Gene Expression in Lymphoblasts from Parkinson’s Disease" Proteomes 10, no. 2: 20. https://doi.org/10.3390/proteomes10020020
APA StyleAnnesley, S. J., Allan, C. Y., Sanislav, O., Evans, A., & Fisher, P. R. (2022). Dysregulated Gene Expression in Lymphoblasts from Parkinson’s Disease. Proteomes, 10(2), 20. https://doi.org/10.3390/proteomes10020020








