Application of Foundation Models for Colorectal Cancer Tissue Classification in Mass Spectrometry Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
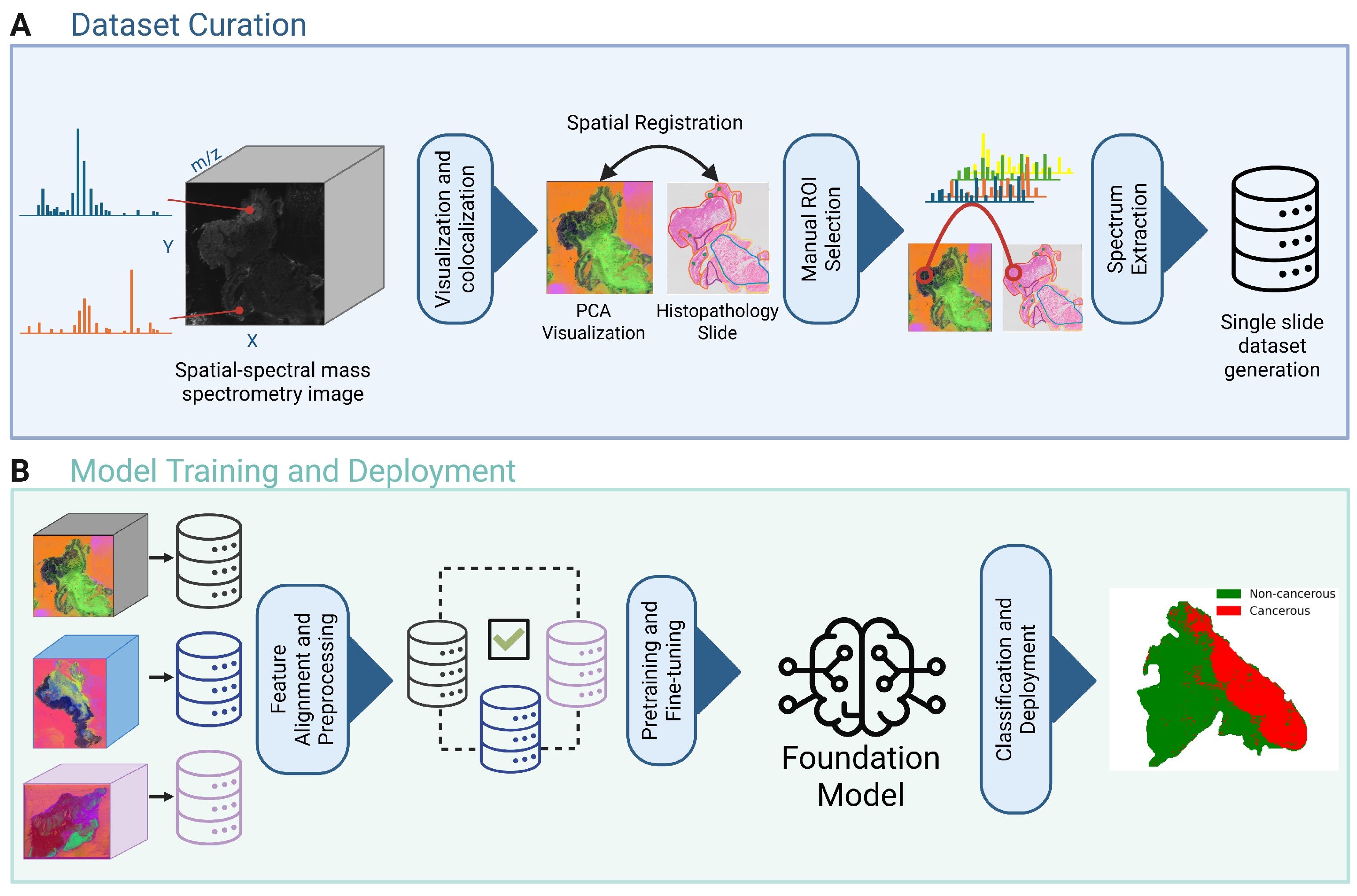
2.1.1. Dataset Curation
2.1.2. Preprocessing
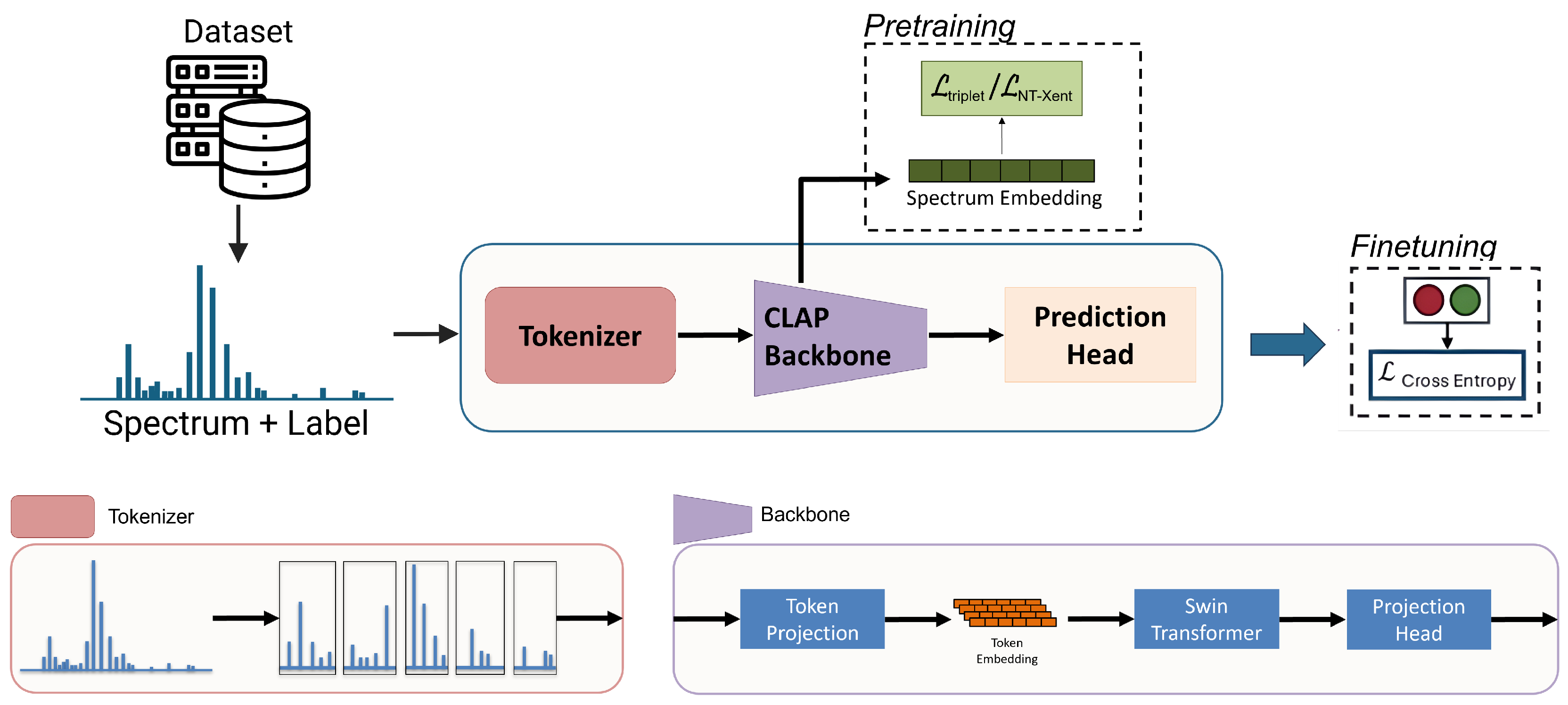
2.2. Models
2.3. Training
2.3.1. Pretraining
2.3.2. Finetuning
2.4. Experiments
2.4.1. Model Evaluation
2.4.2. Ablation Studies
2.5. Implementation Details
3. Results
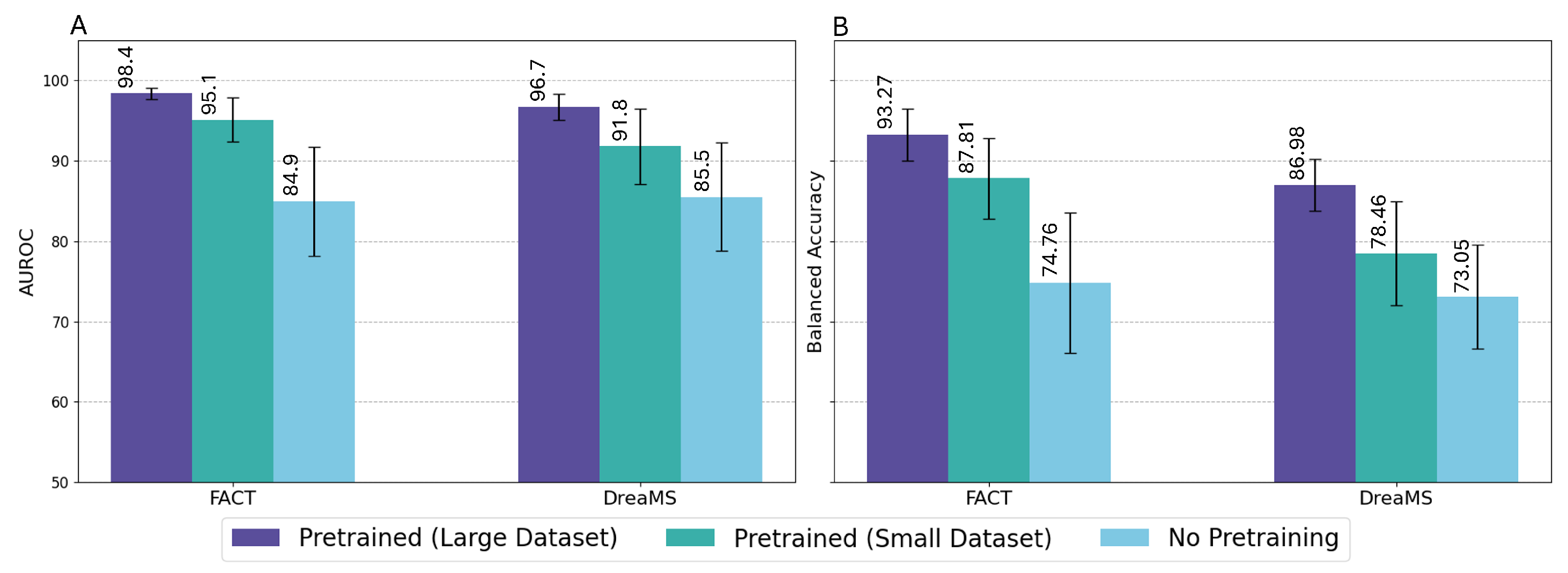
3.1. Baseline Comparison
3.2. Ablation Studies
3.3. Qualitative Comparison
4. Discussion
4.1. Ablation Studies
4.2. Clinical Relevance and Use Cases
4.3. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Colorectal Cancer Fact Sheet. Available online: https://www.who.int/news-room/fact-sheets/detail/colorectal-cancer (accessed on 14 November 2024).
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef] [PubMed]
- World Cancer Research Fund. Colorectal Cancer Statistics. Available online: https://www.wcrf.org/preventing-cancer/cancer-statistics/colorectal-cancer-statistics/ (accessed on 10 March 2025).
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Duan, B.; Zhao, Y.; Bai, J.; Wang, B.J.; Duan, X.; Luo, X.; Zhang, R.; Pu, Y.; Kou, M.; Lei, J.; et al. Colorectal Cancer: An Overview. In Gastrointestinal Cancers; Morgado-Diaz, J.A., Ed.; Exon Publications: Brisbane City, QLD, Australia, 2022; Chapter 1. [Google Scholar] [CrossRef]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.U.; Karuniawati, H.; Jairoun, A.A.; Urbi, Z.; Ooi, D.J.; John, A.; Lim, Y.C.; Kibria, K.M.K.; Mohiuddin, A.K.M.; Ming, L.C.; et al. Colorectal cancer: A review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers 2022, 14, 1732. [Google Scholar] [CrossRef]
- Zhou, D.; Tian, F.; Tian, X.; Sun, L.; Huang, X.; Zhao, F.; Zhou, N.; Chen, Z.; Zhang, Q.; Yang, M.; et al. Diagnostic evaluation of a deep learning model for optical diagnosis of colorectal cancer. Nat. Commun. 2020, 11, 2961. [Google Scholar] [CrossRef]
- Duncan, K.D.; Pětrošová, H.; Lum, J.J.; Goodlett, D.R. Mass spectrometry imaging methods for visualizing tumor heterogeneity. Curr. Opin. Biotechnol. 2024, 86, 103068. [Google Scholar] [CrossRef]
- Liu, K.; Wu, T.; Chen, P.; Tsai, Y.; Roth, H.; Wu, M.; Liao, W.; Wang, W. Deep learning to distinguish pancreatic cancer tissue from non-cancerous pancreatic tissue: A retrospective study with cross-racial external validation. Lancet Digit. Health 2020, 2, e303–e313. [Google Scholar] [CrossRef]
- Gao, Y.; Zeng, S.; Xu, X.; Li, H.; Yao, S. Deep learning-enabled pelvic ultrasound images for accurate diagnosis of ovarian cancer in China: A retrospective, multicentre, diagnostic study. Lancet Digit. Health 2022, 4, e179–e187. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Han, B.; Kim, K.; Han, K.; Nam, H.; Lee, E.; Kim, E.; Chang, J. Changes in cancer detection and false-positive recall in mammography using artificial intelligence: A retrospective, multireader study. Lancet Digit. Health 2020, 2, e138–e148. [Google Scholar] [CrossRef]
- Yao, L.; Li, S.; Tao, Q.; Mao, Y.; Dong, J.; Lu, C.; Han, C.; Qiu, B.; Huang, Y.; Huang, X.; et al. Deep learning for colorectal cancer detection in contrast-enhanced CT without bowel preparation: A retrospective, multicentre study. EBioMedicine 2024, 104, 105183. [Google Scholar] [CrossRef]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef]
- Zhao, T.; Gu, Y.; Yang, J.; Usuyama, N.; Lee, H.H.; Kiblawi, S.; Naumann, T.; Gao, J.; Crabtree, A.; Abel, J.; et al. A foundation model for joint segmentation, detection and recognition of biomedical objects across nine modalities. Nat. Methods 2025, 22, 166–176. [Google Scholar] [CrossRef]
- Wilson, P.F.R.; To, M.N.N.; Jamzad, A.; Gilany, M.; Harmanani, M.; Elghareb, T.; Fooladgar, F.; Wodlinger, B.; Abolmaesumi, P.; Mousavi, P. ProstNFound: Integrating Foundation Models with Ultrasound Domain Knowledge and Clinical Context for Robust Prostate Cancer Detection. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2024, Marrakesh, Morocco, 6–10 October 2024; Springer Nature: Cham, Switzerland, 2024; Volume LNCS 15006. [Google Scholar]
- Swinburne, N.C.; Jackson, C.B.; Pagano, A.M.; Stember, J.N.; Schefflein, J.; Marinelli, B.; Panyam, P.K.; Autz, A.; Chopra, M.S.; Holodny, A.I.; et al. Foundational Segmentation Models and Clinical Data Mining Enable Accurate Computer Vision for Lung Cancer. J. Imaging Inform. Med. 2024, 38, 1552–1562. [Google Scholar] [CrossRef]
- Ben Hamida, A.; Devanne, M.; Weber, J.; Truntzer, C.; Derangère, V.; Ghiringhelli, F.; Forestier, G.; Wemmert, C. Deep learning for colon cancer histopathological images analysis. Comput. Biol. Med. 2021, 136, 104730. [Google Scholar] [CrossRef]
- Connolly, L.; Fooladgar, F.; Jamzad, A.; Kaufmann, M.; Syeda, A.; Ren, K.; Abolmaesumi, P.; Rudan, J.F.; McKay, D.; Fichtinger, G.; et al. ImSpect: Image-driven self-supervised learning for surgical margin evaluation with mass spectrometry. Int. J. Comput. Assist. Radiol. Surg. 2024, 19, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Bushuiev, R.; Bushuiev, A.; Samusevich, R.; Brungs, C.; Sivic, J.; Pluskal, T. Emergence of molecular structures from repository-scale self-supervised learning on tandem mass spectra. ChemRxiv 2024. [Google Scholar] [CrossRef]
- Farahmand, M.; Jamzad, A.; Fooladgar, F.; Connolly, L.; Kaufmann, M.; Ren, K.Y.M.; Rudan, J.; McKay, D.; Fichtinger, G.; Mousavi, P. FACT: Foundation Model for Assessing Cancer Tissue Margins with Mass Spectrometry. Int. J. Comput. Assist. Radiol. Surg. 2025, 20, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xie, C.; Miao, R.; Xu, J.; Xu, X.; Fang, J.; Wang, X.; Liu, W.; Liao, X.; Wang, J.; et al. DeepION: A Deep Learning-Based Low-Dimensional Representation Model of Ion Images for Mass Spectrometry Imaging. Anal. Chem. 2024, 96, 3829–3836. [Google Scholar] [CrossRef]
- Abdelmoula, W.M.; Stopka, S.A.; Randall, E.C.; Regan, M.; Agar, J.N.; Sarkaria, J.N.; Wells, W.M.; Kapur, T.; Agar, N.Y.R. massNet: Integrated processing and classification of spatially resolved mass spectrometry data using deep learning for rapid tumor delineation. Bioinformatics 2022, 38, 2015–2021. [Google Scholar] [CrossRef]
- Li, Z.; Sun, Y.; An, F.; Chen, H.; Liao, J. Self-supervised clustering analysis of colorectal cancer biomarkers based on multi-scale whole slides image and mass spectrometry imaging fused images. Talanta 2023, 263, 124727. [Google Scholar] [CrossRef]
- Kaufmann, M.; Iaboni, N.; Jamzad, A.; Hurlbut, D.; Ren, K.Y.M.; Rudan, J.F.; Mousavi, P.; Fichtinger, G.; Varma, S.; Caycedo-Marulanda, A.; et al. Metabolically Active Zones Involving Fatty Acid Elongation Delineated by DESI-MSI Correlate with Pathological and Prognostic Features of Colorectal Cancer. Metabolites 2023, 13, 508. [Google Scholar] [CrossRef]
- Jamzad, A.; Warren, J.; Syeda, A.; Kaufmann, M.; Iaboni, N.; Nicol, C.J.B.; Rudan, J.; Ren, K.Y.M.; Hurlbut, D.; Varma, S.; et al. MassVision: An Open-Source End-to-End Platform for AI-Driven Mass Spectrometry Imaging Analysis. Anal. Chem. 2025, ASAP. [Google Scholar] [CrossRef]
- Connolly, L.; Jamzad, A.; Kaufmann, M.; Farquharson, C.E.; Ren, K.; Rudan, J.F.; Fichtinger, G.; Mousavi, P. Combined Mass Spectrometry and Histopathology Imaging for Perioperative Tissue Assessment in Cancer Surgery. J. Imaging 2021, 7, 203. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, K.; Zhang, T.; Hui, Y.; Nezhurina, M.; Berg-Kirkpatrick, T.; Dubnov, S. Large-scale Contrastive Language-Audio Pretraining with Feature Fusion and Keyword-to-Caption Augmentation. arXiv 2024, arXiv:2211.06687. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. arXiv 2021, arXiv:2103.14030. [Google Scholar] [CrossRef]
- Chechik, G.; Sharma, V.; Shalit, U.; Bengio, S. Large scale online learning of image similarity through ranking. J. Mach. Learn. Res. 2010, 11, 1109–1135. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A simple framework for contrastive learning of visual representations. In Proceedings of the 37th International Conference on Machine Learning, Virtual, 13–18 July 2020; Volume 119, pp. 1597–1607. [Google Scholar]
- Fooladgar, F.; Jamzad, A.; Connolly, L.; Santilli, A.M.L.; Kaufmann, M.; Ren, K.Y.M.; Abolmaesumi, P.; Rudan, J.F.; McKay, D.; Fichtinger, G.; et al. Uncertainty estimation for margin detection in cancer surgery using mass spectrometry. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 2305–2313. [Google Scholar] [CrossRef]
- Alexandrov, T. Spatial Metabolomics and Imaging Mass Spectrometry in the Age of Artificial Intelligence. Annu. Rev. Biomed. Data Sci. 2020, 3, 61–87. [Google Scholar] [CrossRef]
- Yurekten, O.; Payne, T.; Tejera, N.; Amaladoss, F.X.; Martin, C.; Williams, M.; O’Donovan, C. MetaboLights: Open data repository for metabolomics. Nucleic Acids Res. 2023, 52, D640–D646. [Google Scholar] [CrossRef]



| Model | Balanced Accuracy | Sensitivity | Specificity | AUROC |
|---|---|---|---|---|
| PCA-LDA | ||||
| CLAP | ||||
| CLAP+SimCLR | ||||
| DreaMS | ||||
| FACT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabriel, A.; Jamzad, A.; Farahmand, M.; Kaufmann, M.; Iaboni, N.; Hurlbut, D.; Ren, K.Y.M.; Nicol, C.J.B.; Rudan, J.F.; Varma, S.; et al. Application of Foundation Models for Colorectal Cancer Tissue Classification in Mass Spectrometry Imaging. Technologies 2025, 13, 434. https://doi.org/10.3390/technologies13100434
Gabriel A, Jamzad A, Farahmand M, Kaufmann M, Iaboni N, Hurlbut D, Ren KYM, Nicol CJB, Rudan JF, Varma S, et al. Application of Foundation Models for Colorectal Cancer Tissue Classification in Mass Spectrometry Imaging. Technologies. 2025; 13(10):434. https://doi.org/10.3390/technologies13100434
Chicago/Turabian StyleGabriel, Alon, Amoon Jamzad, Mohammad Farahmand, Martin Kaufmann, Natasha Iaboni, David Hurlbut, Kevin Yi Mi Ren, Christopher J. B. Nicol, John F. Rudan, Sonal Varma, and et al. 2025. "Application of Foundation Models for Colorectal Cancer Tissue Classification in Mass Spectrometry Imaging" Technologies 13, no. 10: 434. https://doi.org/10.3390/technologies13100434
APA StyleGabriel, A., Jamzad, A., Farahmand, M., Kaufmann, M., Iaboni, N., Hurlbut, D., Ren, K. Y. M., Nicol, C. J. B., Rudan, J. F., Varma, S., Fichtinger, G., & Mousavi, P. (2025). Application of Foundation Models for Colorectal Cancer Tissue Classification in Mass Spectrometry Imaging. Technologies, 13(10), 434. https://doi.org/10.3390/technologies13100434








