Malaria Cell Image Classification Using Compact Deep Learning Architectures on Jetson TX2
Abstract
1. Introduction
2. Materials and Methods
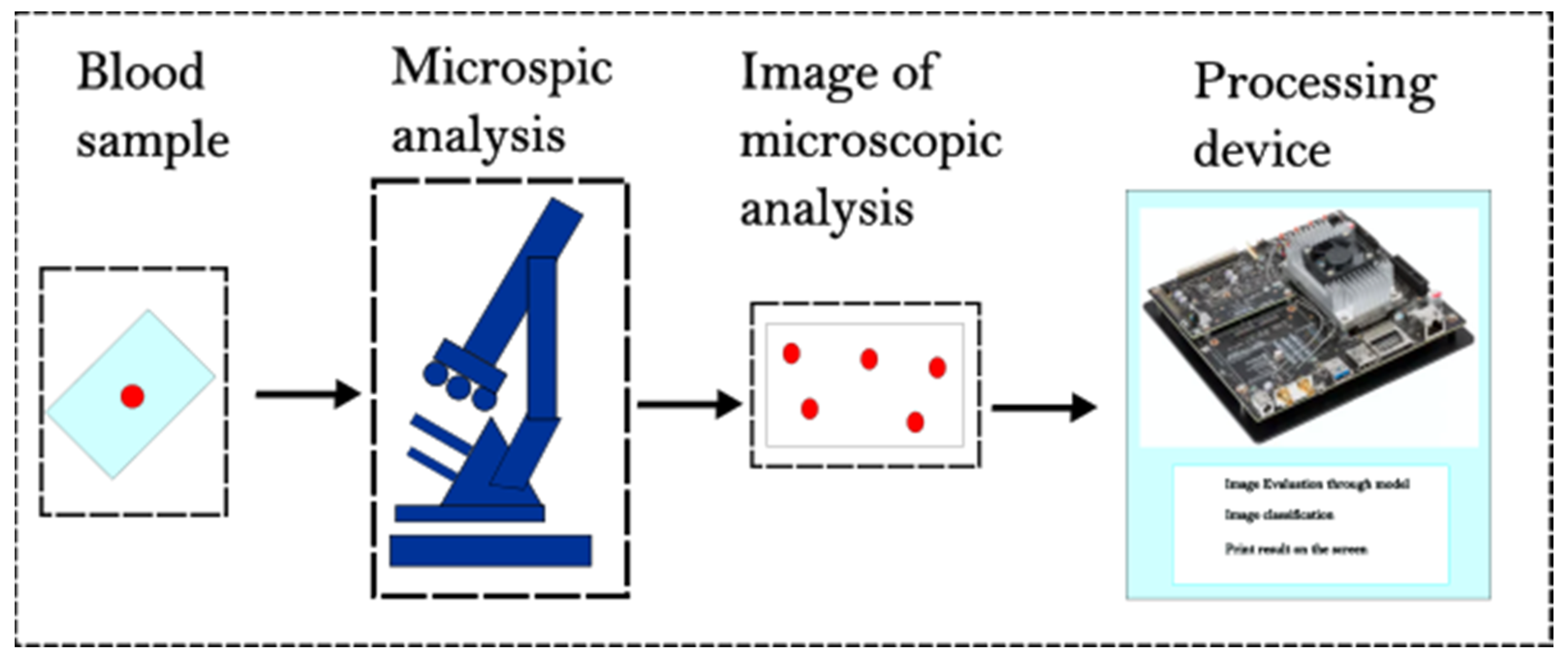
2.1. Dataset and Hardware
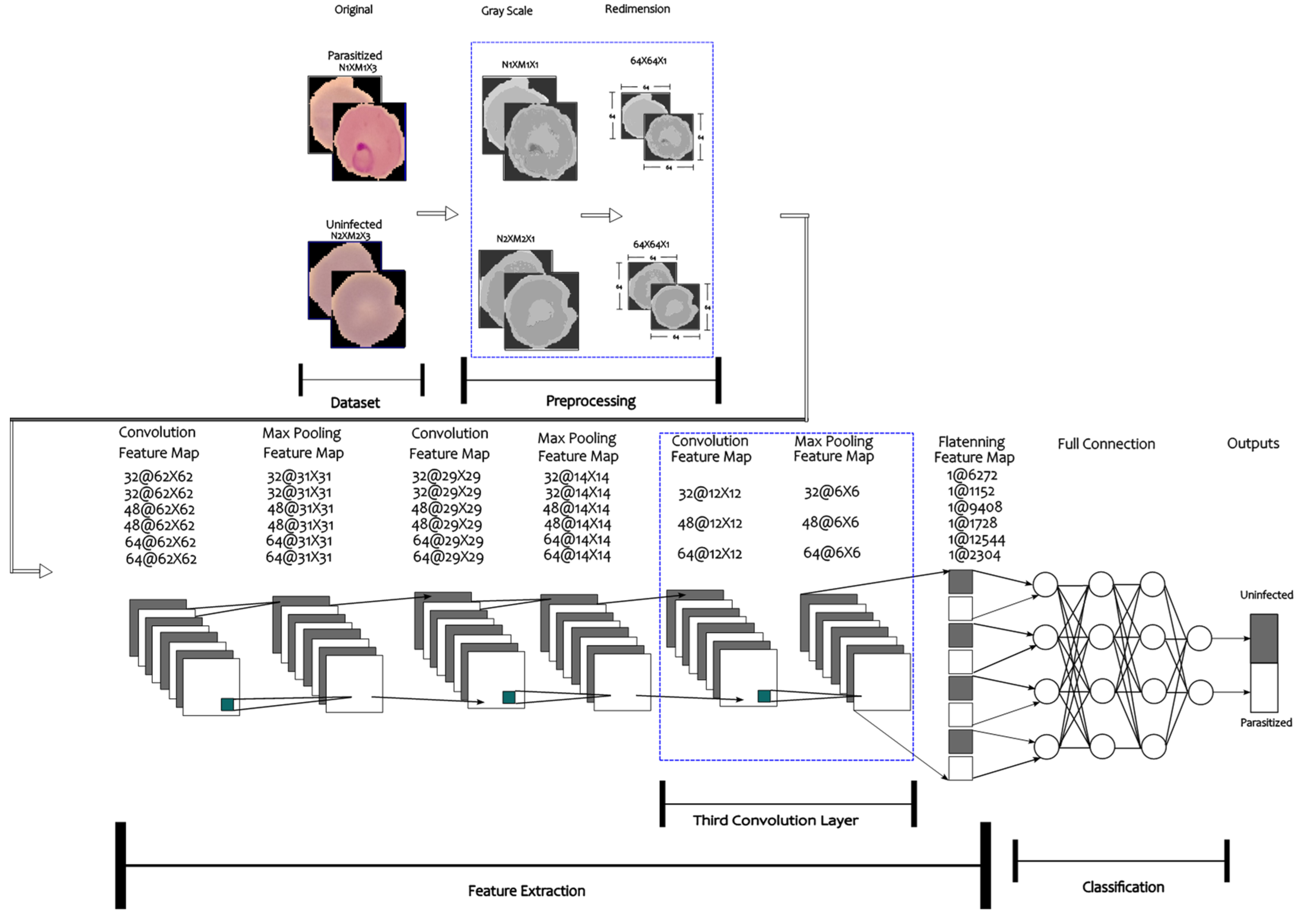
2.2. Convolutional Neural Network Architecture
2.3. Model Training
- Loss function: binary cross entropy. Optimizer: Adam, with an initial learning rate of 0.001. Evaluation metrics: accuracy, specificity, recall, precision, and F1-score.
- Data split: The dataset was split into 80% for training and 20% for validation.
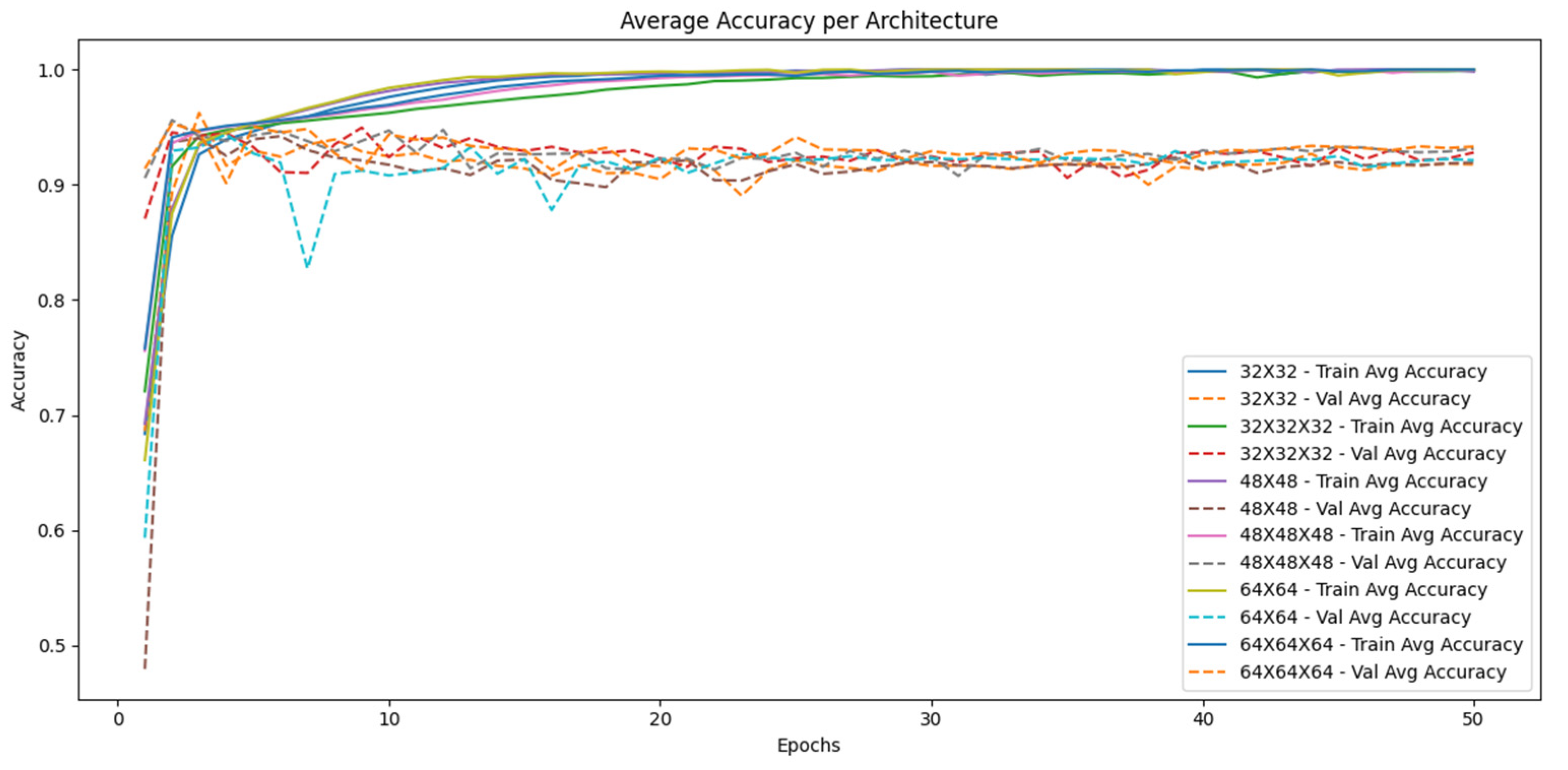
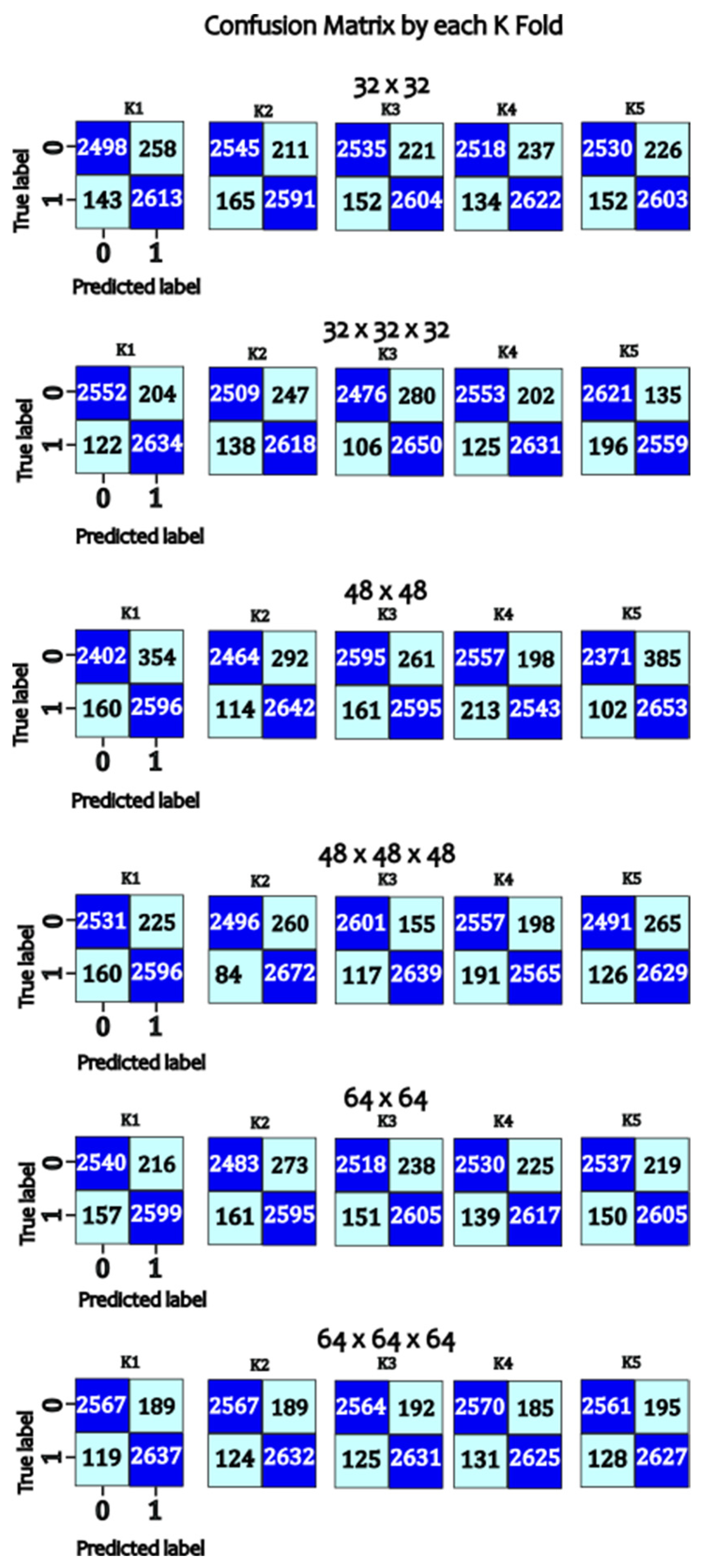
- Epochs: The model was trained for 50 epochs with a batch size of 32. Evaluation and Validation Model performance was evaluated using a separate test dataset not used during training. Performance metrics included overall accuracy, specificity, recall, precision, and F1-score. In addition, confusion matrices were generated to analyze false positives and false negatives. The codes are available in the repository Ref. [35].
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
- TP = True Positives;
- TN = True Negatives;
- FP = False Positives;
- FN = False Negatives.
References
- World Health Organization. Fact Sheet About Malaria. Available online: https://www.who.int/news-room/fact-sheets/detail/malaria (accessed on 19 September 2024).
- Landier, J.; Parker, D.M.; Thu, A.M.; Lwin, K.M.; Delmas, G.; Nosten, F.H. The role of early detection and treatment in malaria elimination. Malar. J. 2016, 15, 363. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, D.; Hunziker, P. Transmission-blocking strategies: The roadmap from laboratory bench to the community. Malar. J. 2016, 15, 95. [Google Scholar] [CrossRef] [PubMed]
- Shahbodaghi, S.; Rathjen, N. Malaria: Prevention, Diagnosis, and Treatment. Am. Fam. Physician 2022, 106, 270–278. [Google Scholar] [PubMed]
- Chima, J.S.; Shah, A.; Shah, K.; Ramesh, R. Malaria Cell Image Classification using Deep Learning. Int. J. Recent Technol. Eng. 2020, 8, 5553–5559. [Google Scholar] [CrossRef]
- Cai, Z.; Ma, C.; Li, J.; Liu, C. Hybrid Amplitude Ordinal Partition Networks for ECG Morphology Discrimination: An Application to PVC Recognition. IEEE Trans. Instrum. Meas 2024, 73, 4008113. [Google Scholar] [CrossRef]
- Ibrahim, E.; Zaghden, N.; Mejdoub, M. Semantic Analysis System to Recognize Moving Objects by Using a Deep Learning Model. IEEE Access 2024, 12, 80740–80753. [Google Scholar] [CrossRef]
- Malu, G.; Uday, N.; Sherly, E.; Abraham, A.; Bodhey, N.K. CirMNet: A Shape-based Hybrid Feature Extraction Technique using CNN and CMSMD for Alzheimer’s MRI Classification. IEEE Access 2024, 12, 80491–80504. [Google Scholar] [CrossRef]
- Tseng, C.H.; Chien, S.J.; Wang, P.S.; Lee, S.J.; Pu, B.; Zeng, X.J. Real-time Automatic M-mode Echocardiography Measurement with Panel Attention. IEEE J. Biomed. Health Inform. 2024, 28, 5383–5395. [Google Scholar] [CrossRef]
- Salah, S.; Chouchene, M.; Sayadi, F. FPGA implementation of a Convolutional Neural Network for Alzheimer’s disease classification. In Proceedings of the 2024 21st International Multi-Conference on Systems, Signals & Devices (SSD), Erbil, Iraq, 22–25 April 2024; pp. 193–198. [Google Scholar] [CrossRef]
- Gondkar, R.R.; Gondkar, S.R.; Kavitha, S.; RV, S.B. Hybrid Deep Learning Based GRU Model for Classifying the Lung Cancer from CT Scan Images. In Proceedings of the 2024 Third International Conference on Distributed Computing and Electrical Circuits and Electronics (ICDCECE), Ballari, India, 26–27 April 2024; pp. 1–8. [Google Scholar] [CrossRef]
- Preetha, R.; Priyadarsini, M.J.P.; Nisha, J.S. Automated Brain Tumor Detection from Magnetic Resonance Images Using Fine-Tuned EfficientNet-B4 Convolutional Neural Network. IEEE Access 2024, 12, 112181–112195. [Google Scholar] [CrossRef]
- NVIDIA. NVIDIA Jetson TX2: High Performance AI at the Edge. Available online: https://www.nvidia.com/en-us/autonomous-machines/embedded-systems/jetson-tx2/ (accessed on 26 November 2024).
- Alonso-Ramírez, A.A.; Mwata-Velu, T.; García-Capulín, C.H.; Rostro-González, H.; Prado-Olivarez, J.; Gutiérrez-López, M.; Barranco-Gutiérrez, A.I. Classifying Parasitized and Uninfected Malaria Red Blood Cells Using Convolutional-Recurrent Neural Networks”. IEEE Access 2022, 10, 97348–97359. [Google Scholar] [CrossRef]
- Arunagiri, V.; Rajesh, B. Deep Learning Approach to Detect Malaria from Microscopic Images. Multimed. Tools Appl. 2020, 79, 15297–15317. [Google Scholar] [CrossRef]
- Yebasse, M.; Cheoi, K.; Ko, J. Malaria Disease Cell Classification with Highlighting Small Infected Regions. IEEE Access 2023, 11, 15945–15953. [Google Scholar] [CrossRef]
- Suraksha, S.; Santhosh, C.; Vishwa, B. Classification of Malaria Cell Images Using Deep Learning Approach. In Proceedings of the 2023 Third International Conference on Advances in Electrical, Computing, Communication and Sustainable Technologies (ICAECT), Bhilai, India, 5–6 January 2023; pp. 1–5. [Google Scholar] [CrossRef]
- Pan, W.D.; Dong, Y.; Wu, D. Classification of Malaria-Infected Cells Using Deep Convolutional Neural Networks. In Machine Learning; Farhadi, H., Ed.; IntechOpen: Rijeka, Croatia, 2018; Chapter 8. [Google Scholar] [CrossRef]
- Pattanaik, P.; Mittal, M.; Khan, M. Unsupervised Deep Learning CAD Scheme for the Detection of Malaria in Blood Smear Microscopic Images. IEEE Access 2020, 8, 94936–94946. [Google Scholar] [CrossRef]
- Molina-Borrás, A.; Rojas, C.; del Río, J.; Bermejo, J.; Gutiérrez, J. Automatic Identification of Malaria and Other Red Blood Cell Inclusions Using Convolutional Neural Networks. Comput. Biol. Med. 2021, 136, 104680. [Google Scholar] [CrossRef]
- Siłka, W.; Sobczak, J.; Duda, J.; Wieczorek, M. Malaria Detection Using Advanced Deep Learning Architecture. Sensors 2023, 23, 1501. [Google Scholar] [CrossRef]
- Mittal, S. A Survey on Optimized Implementation of Deep Learning Models on the NVIDIA Jetson Platform. J. Syst. Archit. 2019, 97, 428–442. [Google Scholar] [CrossRef]
- Saypadith, S.; Aramvith, S. Real-Time Multiple Face Recognition using Deep Learning on Embedded GPU System. In Proceedings of the APSIPA Annual Summit and Conference, Honolulu, HI, USA, 12–15 November 2018; pp. 1318–1324. [Google Scholar] [CrossRef]
- Jung, S.; Kim, Y.; Lee, H.; Jang, J.; Hwang, J. Perception, Guidance, and Navigation for Indoor Autonomous Drone Racing Using Deep Learning. IEEE Robot. Autom. Lett. 2018, 3, 2539–2544. [Google Scholar] [CrossRef]
- Amert, T.; Otterness, N.; Yang, M.; Anderson, J.H.; Smith, F.D. GPU Scheduling on the NVIDIA TX2: Hidden Details Revealed. In Proceedings of the 2017 IEEE Real-Time Systems Symposium (RTSS), Paris, France, 5–8 December 2017; pp. 104–115. [Google Scholar] [CrossRef]
- Mohan, H.M.; Singh, D.; Sadiq, M.; Dey, P.; Maji, S.; Pati, S.K. Edge Artificial Intelligence: Real-Time Noninvasive Technique for Vital Signs of Myocardial Infarction Recognition Using Jetson Nano. Adv. Hum.-Comput. Interact. 2021, 2021, 6483003. [Google Scholar] [CrossRef]
- Lou, L.; Liang, H.; Wang, Z. Deep-Learning-Based COVID-19 Diagnosis and Implementation in Embedded Edge-Computing Device. Diagnostics 2023, 13, 1329. [Google Scholar] [CrossRef]
- Shihadeh, J.; Ansari, A.; Ozunfunmi, T. Deep Learning Based Image Classification for Remote Medical Diagnosis. In Proceedings of the 2018 IEEE Global Humanitarian Technology Conference (GHTC), San Jose, CA, USA, 18–21 October 2018; pp. 1–8. [Google Scholar] [CrossRef]
- Liu, B.; Chen, C.; Wan, S.; Qiao, P.; Pei, Q. An Edge Traffic Flow Detection Scheme Based on Deep Learning in an Intelligent Transportation System. IEEE Trans. Intell. Transp. Syst. 2020, 22, 1840–1852. [Google Scholar] [CrossRef]
- Choe, C.; Choe, M.; Jung, S. Run Your 3D Object Detector on NVIDIA Jetson Platforms:A Benchmark Analysis. Sensors 2023, 23, 4005. [Google Scholar] [CrossRef] [PubMed]
- Beyaz, A.; Saripinar, Z. Sugar Beet Seed Classification for Production Quality Improvement by Using YOLO and NVIDIA Artificial Intelligence Boards. Sugar Tech 2024. [Google Scholar] [CrossRef]
- Rajaraman, S.; Antani, S.K.; Poostchi, M.; Silamut, K.; Hossain, M.A.; Maude, R.J.; Jaeger, S.; Thoma, G.R. Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images. PeerJ 2018, 6, e4568. [Google Scholar] [CrossRef] [PubMed]
- U.S. National Library of Medicine. Malaria Datasheet. Available online: https://lhncbc.nlm.nih.gov/LHC-research/LHC-projects/image-processing/malaria-datasheet.html (accessed on 27 September 2024).
- Chollet, F. Deep Learning with Python, 1st ed.; Manning Publications Co.: Shelter Island, NY, USA, 2017; ISBN 1617294438. [Google Scholar]
- Alonso-Ramírez, A.-A.; Barranco-Gutiérrez, A.-I.; Méndez-Gurrola, I.-I.; Gutiérrez-López, M.; Prado-Olivarez, J.; Pérez-Pinal, F.-J.; Villegas-Saucillo, J.J.; García-Muñoz, J.-A.; García-Capulín, C.-H. MalariaClassification_JetsonTX2. Available online: https://github.com/adanantonio07A/MalariaClassification_JetsonTX2 (accessed on 26 November 2024).
- Ramakrishnan, A.B.; Sridevi, M.; Vasudevan, S.K.; Manikandan, R.; Gandomi, A.H. Optimizing brain tumor classification with hybrid CNN architecture: Balancing accuracy and efficiency through oneAPI optimization. Inform. Med. Unlocked 2024, 44, 101436. [Google Scholar] [CrossRef]
- Lv, E.; Kang, X.; Wen, P.; Tian, J.; Zhang, M. A novel benign and malignant classification model for lung nodules based on multi-scale interleaved fusion integrated network. Sci. Rep. 2024, 14, 27506. [Google Scholar] [CrossRef]

| Parasitized | 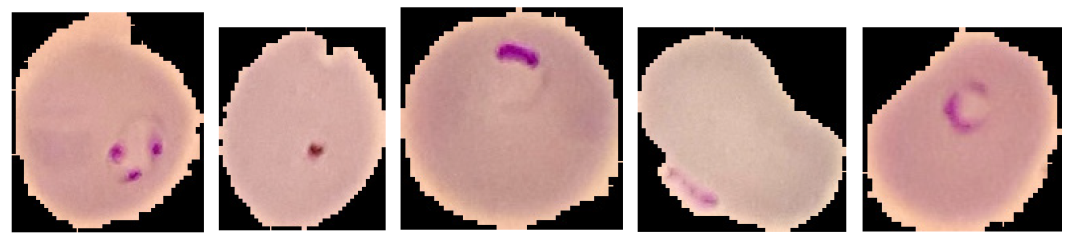 |
| Uninfected |  |
| PR: Parasitized (Resized [64 × 64]) |  |
| PU: Uninfected (Resized [64 × 64]) |  |
| PR (Grayscale) |  |
| PU (Grayscale) |  |
| Model | K-Fold | Accuracy | Specificity | Recall | Precision | F1-Score |
|---|---|---|---|---|---|---|
| 32 × 32 | 1 | 97.27 | 98.64 | 95.98 | 98.68 | 97.31 |
| 2 | 97.32 | 98.73 | 95.99 | 98.77 | 97.36 | |
| 3 | 97.44 | 98.72 | 96.22 | 98.75 | 97.47 | |
| 4 | 97.11 | 98.89 | 95.46 | 98.93 | 97.16 | |
| 5 | 97.28 | 98.84 | 95.82 | 98.88 | 97.32 | |
| 32 × 32 × 32 | 1 | 97.12 | 99.03 | 95.36 | 99.06 | 97.18 |
| 2 | 97.64 | 98.97 | 96.39 | 99 | 97.68 | |
| 3 | 97.71 | 99.04 | 96.46 | 99.06 | 97.74 | |
| 4 | 97.59 | 98.52 | 96.7 | 98.55 | 97.61 | |
| 5 | 97.7 | 98.93 | 96.53 | 98.95 | 97.73 | |
| 48 × 48 | 1 | 97.27 | 98.83 | 95.8 | 98.87 | 97.31 |
| 2 | 97.23 | 98.79 | 95.77 | 98.83 | 97.27 | |
| 3 | 97.46 | 98.73 | 96.26 | 98.77 | 97.5 | |
| 4 | 97.19 | 98.83 | 95.65 | 98.87 | 97.23 | |
| 5 | 97.05 | 98.88 | 95.35 | 98.93 | 97.1 | |
| 48 × 48 × 48 | 1 | 96.67 | 98.66 | 94.83 | 98.72 | 96.73 |
| 2 | 97.48 | 98.97 | 96.08 | 99 | 97.52 | |
| 3 | 97.8 | 99 | 96.66 | 99.02 | 97.83 | |
| 4 | 97.67 | 99.04 | 96.37 | 99.06 | 97.7 | |
| 5 | 97.75 | 98.87 | 96.67 | 98.9 | 97.77 | |
| 64 × 64 | 1 | 97.26 | 98.86 | 95.77 | 98.9 | 97.31 |
| 2 | 97.25 | 98.79 | 95.8 | 98.82 | 97.29 | |
| 3 | 97.4 | 98.68 | 96.19 | 98.72 | 97.44 | |
| 4 | 97.5 | 98.89 | 96.19 | 98.92 | 97.53 | |
| 5 | 97.32 | 98.82 | 95.91 | 98.86 | 97.36 | |
| 64 × 64 × 64 | 1 | 97.67 | 99.07 | 96.36 | 99.09 | 97.71 |
| 2 | 97.66 | 98.93 | 96.46 | 98.96 | 97.69 | |
| 3 | 97.88 | 99.06 | 96.75 | 99.08 | 97.9 | |
| 4 | 97.72 | 99.05 | 96.47 | 99.07 | 97.76 | |
| 5 | 97.67 | 98.96 | 96.44 | 98.99 | 97.7 |
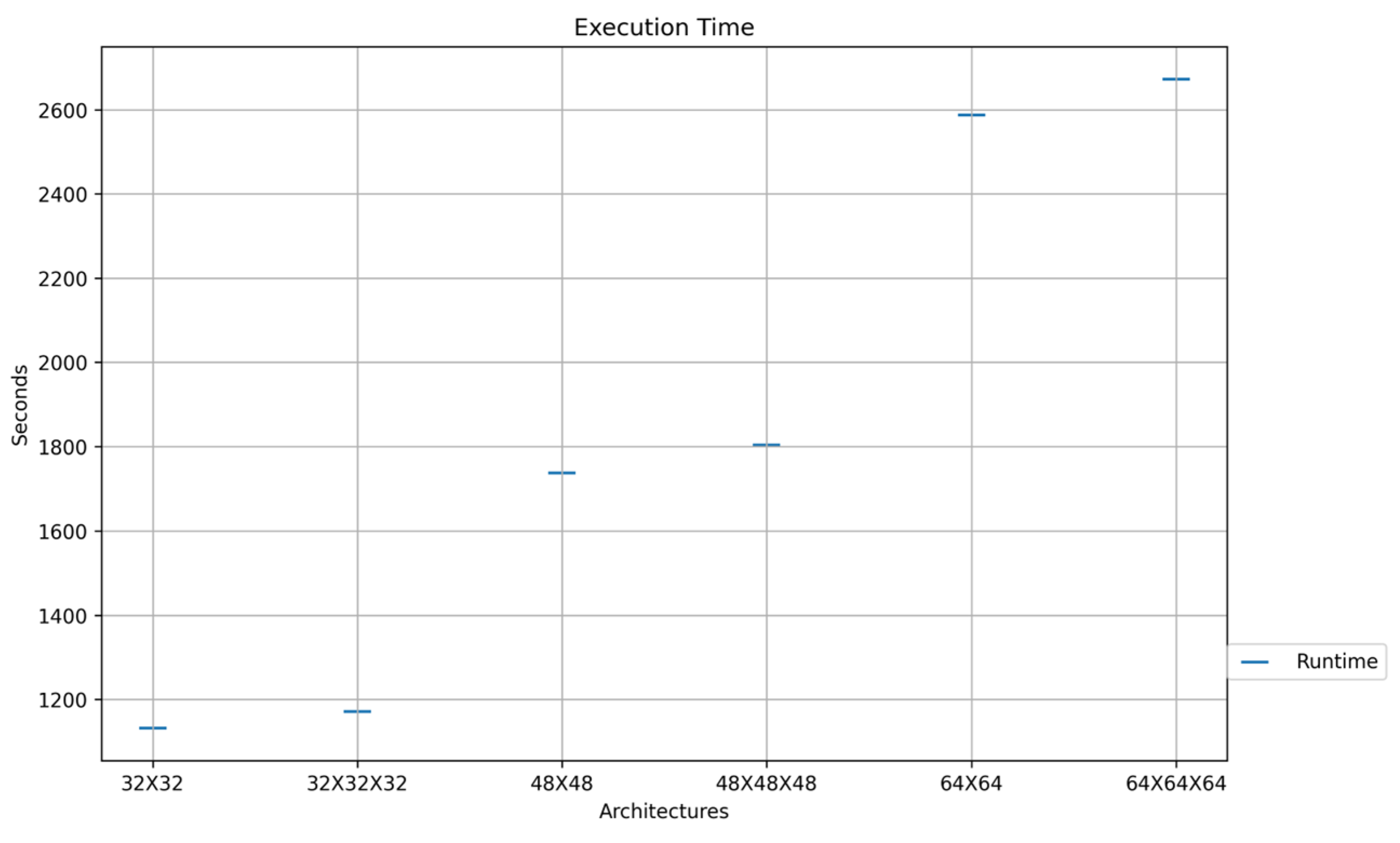
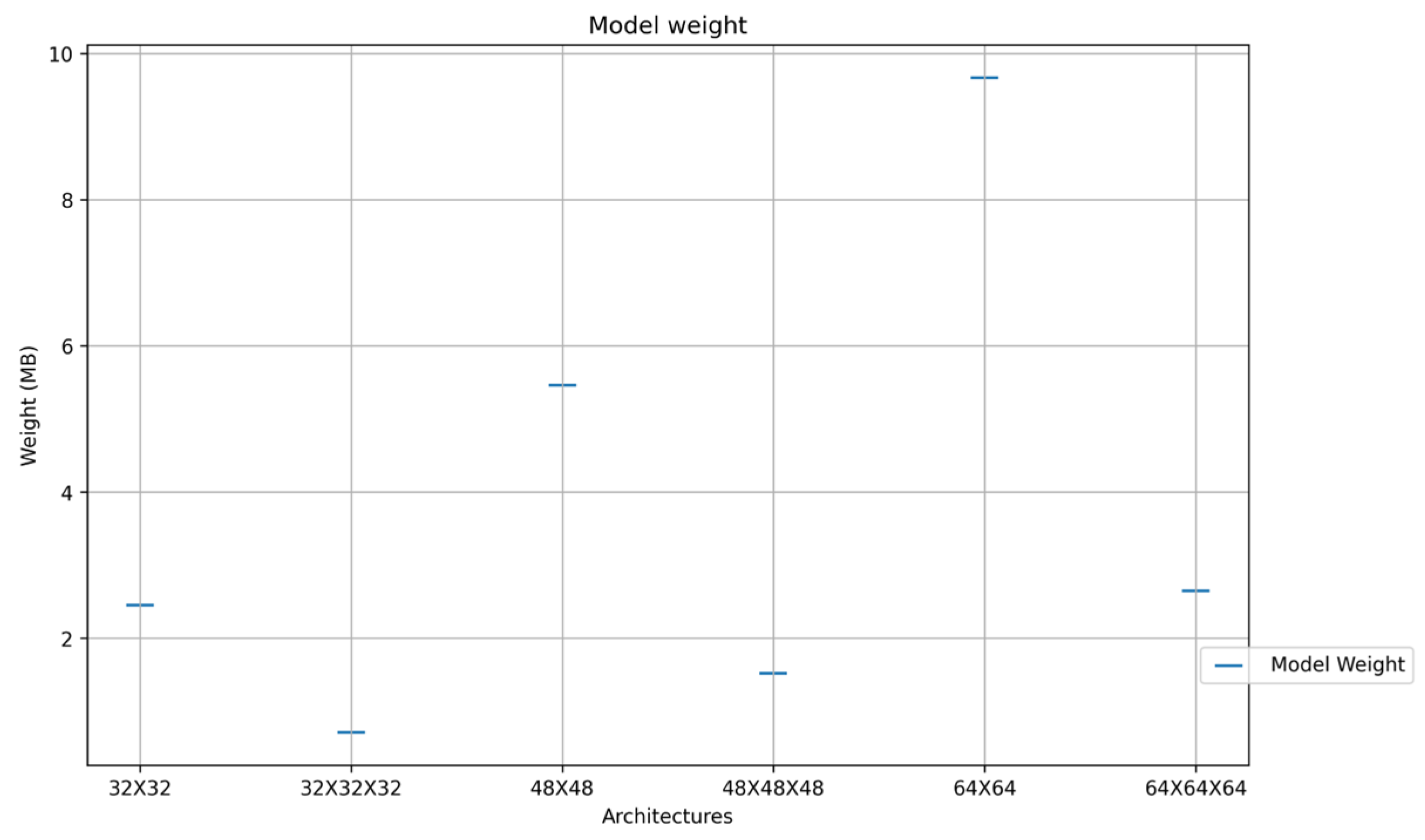
| Model | Accuracy | Classification Execution(s) |
|---|---|---|
| 32 × 32 | 97.28 | 0.0014876 |
| 48 × 48 | 97.55 | 0.0015972 |
| 64 × 64 | 97.24 | 0.0023 |
| 32 × 32 × 32 | 97.47 | 0.0025032 |
| 48 × 48 × 48 | 97.35 | 0.0034522 |
| 64 × 64 × 64 | 97.72 | 0.0038254 |
| Reference | Accuracy | Lowest Classification Execution Time |
|---|---|---|
| Alonso-Ramirez A. A. et al. (2022) Ref. [14] first approach | 99.89% | 0.125 s |
| Alonso-Ramirez A. A. et al. (2022) Ref. [14] second approach | 99.89% | 0.130 s |
| Alonso-Ramirez A. A. et al. (2024) minimal arch | 97.28% | 0.0014876 s |
| Alonso-Ramirez A. A. et al. (2024) maximum arch | 97.72% | 0.0038254 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alonso-Ramírez, A.-A.; Barranco-Gutiérrez, A.-I.; Méndez-Gurrola, I.-I.; Gutiérrez-López, M.; Prado-Olivarez, J.; Pérez-Pinal, F.-J.; Villegas-Saucillo, J.J.; García-Muñoz, J.-A.; García-Capulín, C.-H. Malaria Cell Image Classification Using Compact Deep Learning Architectures on Jetson TX2. Technologies 2024, 12, 247. https://doi.org/10.3390/technologies12120247
Alonso-Ramírez A-A, Barranco-Gutiérrez A-I, Méndez-Gurrola I-I, Gutiérrez-López M, Prado-Olivarez J, Pérez-Pinal F-J, Villegas-Saucillo JJ, García-Muñoz J-A, García-Capulín C-H. Malaria Cell Image Classification Using Compact Deep Learning Architectures on Jetson TX2. Technologies. 2024; 12(12):247. https://doi.org/10.3390/technologies12120247
Chicago/Turabian StyleAlonso-Ramírez, Adán-Antonio, Alejandro-Israel Barranco-Gutiérrez, Iris-Iddaly Méndez-Gurrola, Marcos Gutiérrez-López, Juan Prado-Olivarez, Francisco-Javier Pérez-Pinal, J. Jesús Villegas-Saucillo, Jorge-Alberto García-Muñoz, and Carlos-Hugo García-Capulín. 2024. "Malaria Cell Image Classification Using Compact Deep Learning Architectures on Jetson TX2" Technologies 12, no. 12: 247. https://doi.org/10.3390/technologies12120247
APA StyleAlonso-Ramírez, A.-A., Barranco-Gutiérrez, A.-I., Méndez-Gurrola, I.-I., Gutiérrez-López, M., Prado-Olivarez, J., Pérez-Pinal, F.-J., Villegas-Saucillo, J. J., García-Muñoz, J.-A., & García-Capulín, C.-H. (2024). Malaria Cell Image Classification Using Compact Deep Learning Architectures on Jetson TX2. Technologies, 12(12), 247. https://doi.org/10.3390/technologies12120247









