Graph Learning and Deep Neural Network Ensemble for Supporting Cognitive Decline Assessment
Abstract
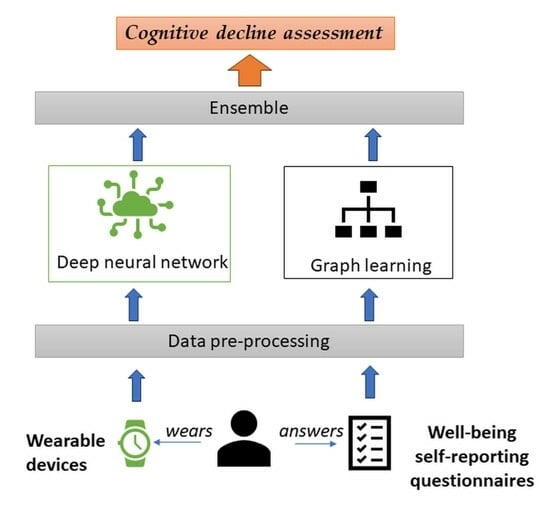
:1. Introduction
- a deep neural network technique that uses a CNN model to analyze objective activity data coming from wearable devices (fitness trackers, smart watches, smart patches, etc.), with a focus on daily activity fragmentation.
- a graph learning (GL) technique that captures structural insights from subjective self-reporting activities (through wellbeing assessment questionnaires) to offer a better understanding of symptoms associated with cognitive decline using Node2Vec embeddings and K-means clustering.
- an ensemble technique to aggregate the outputs of both models based on the Softmax function.
2. Related Work
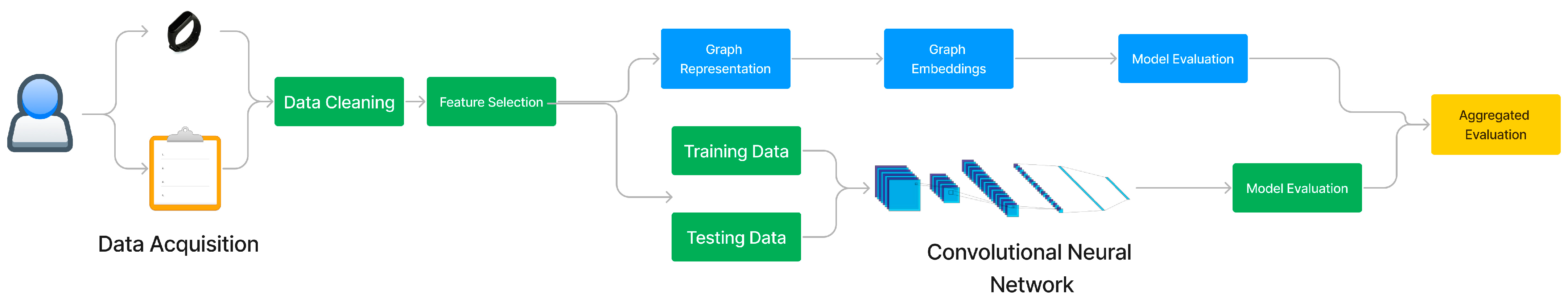
3. Materials and Methods
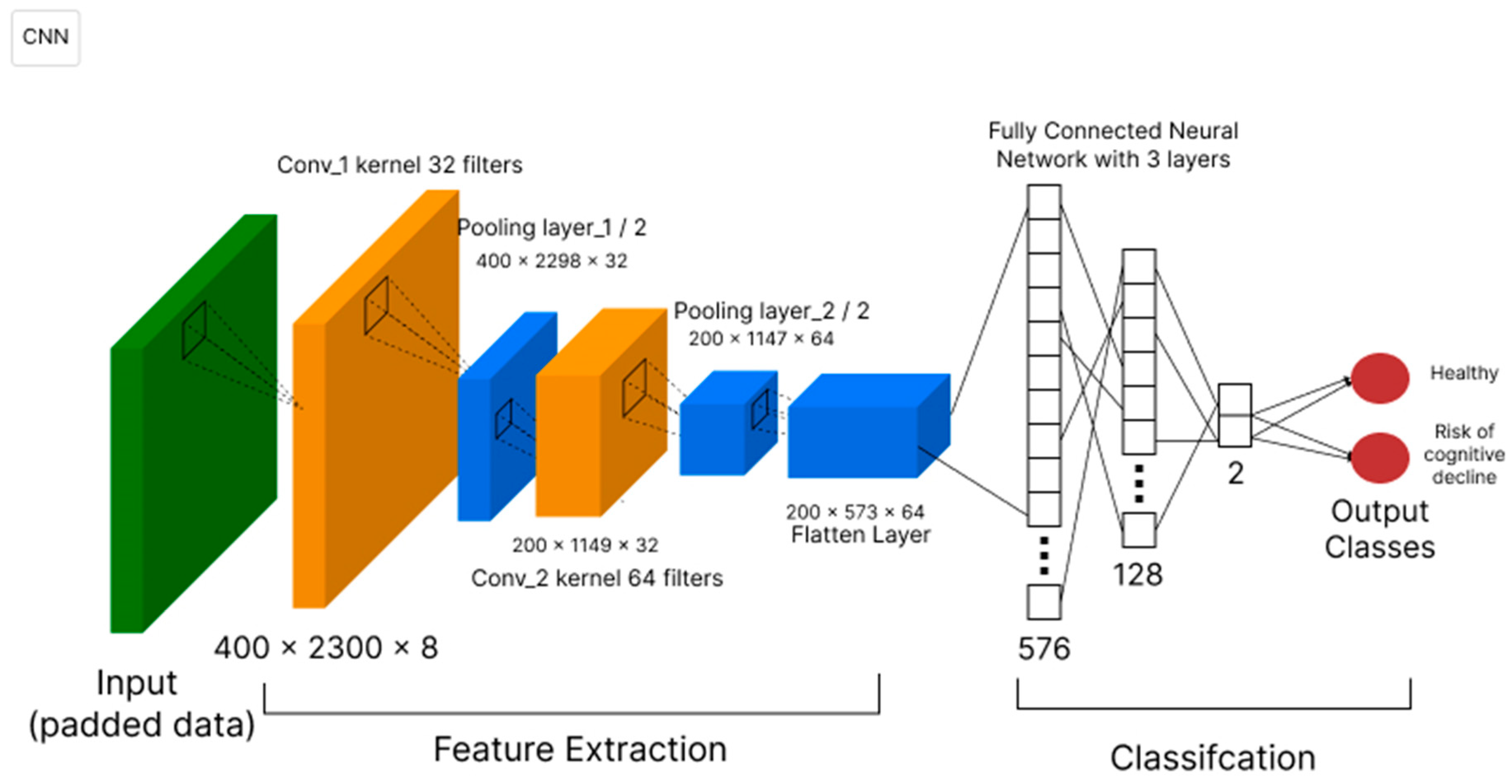
3.1. Deep Neural Network Technique
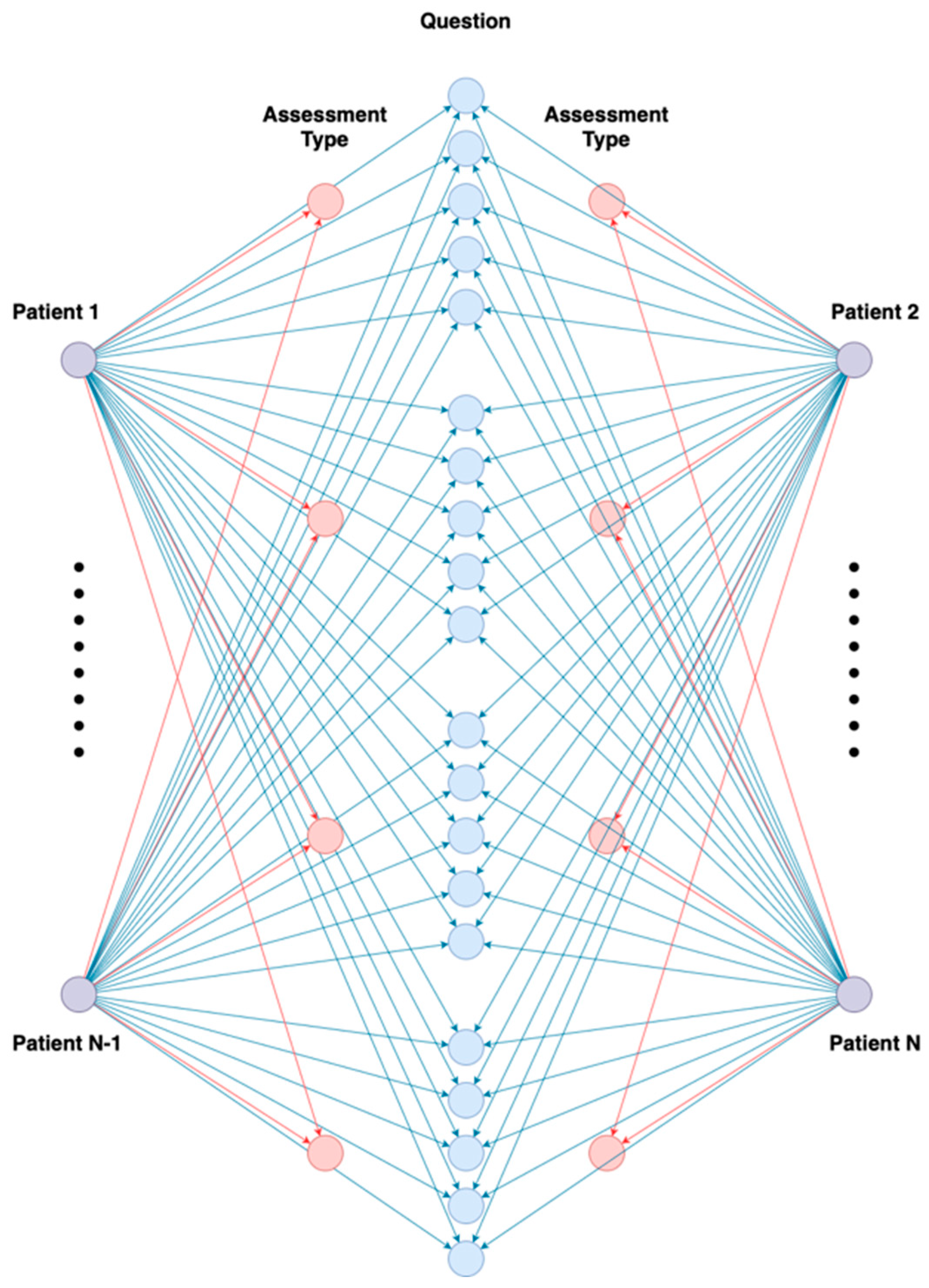
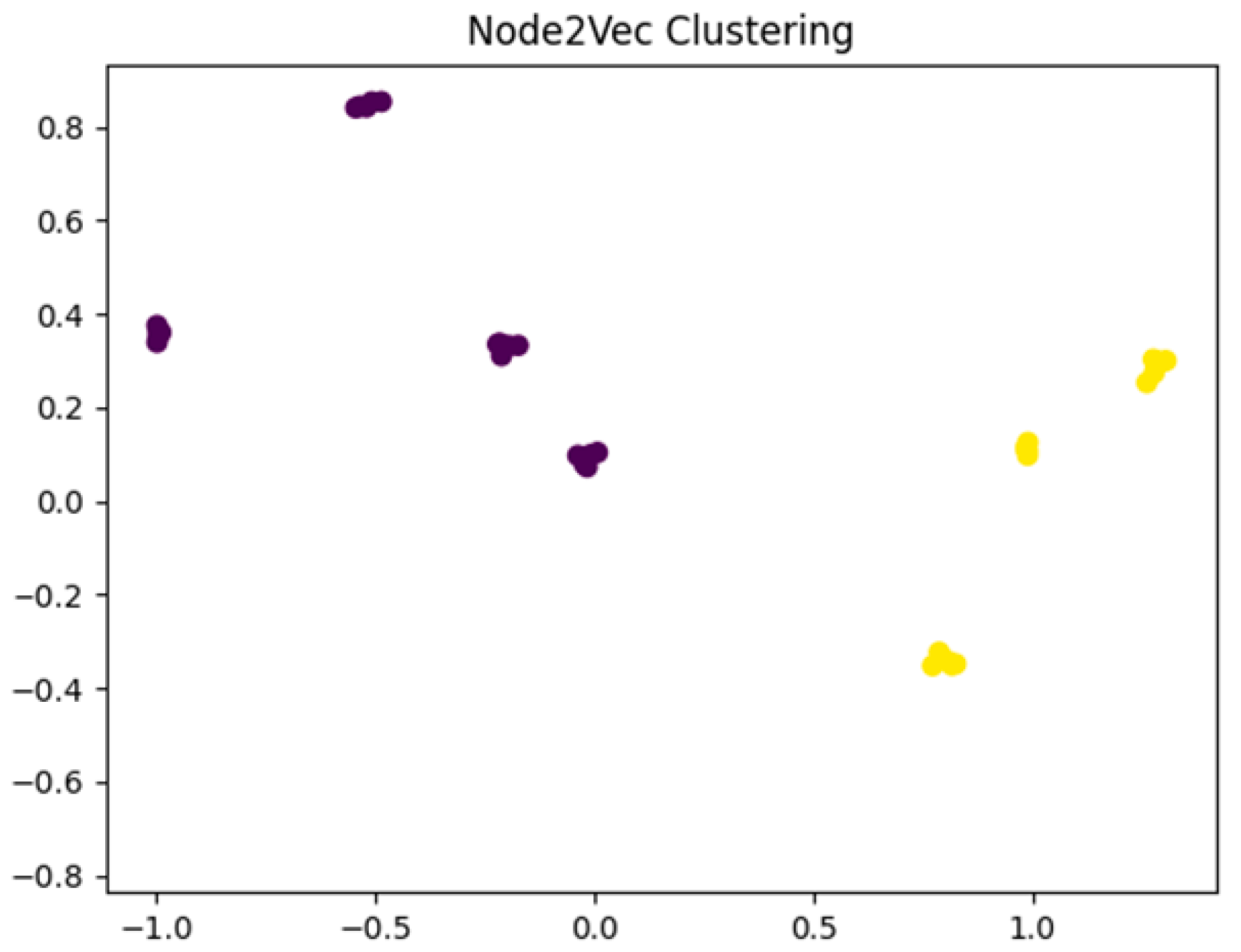
3.2. Graph Learning Technique
- Reducing the dimensionality: In certain scenarios, graphs can be large and complex, being difficult to evaluate. Graph embeddings solve this issue by reducing the dimensionality of the graph while keeping the essential features.
- Feature Leaning: Graph embeddings capture the inherent features and relationships between nodes and edges. This representation can be used for numerous downstream tasks, including node classification, link prediction, and graph clustering.
- Integration with ML Models: GL offers a way to use traditional ML algorithms on graphs. After the graph is represented as vectors, tasks such as classification, regression, and clustering can be easily achieved by using classic techniques.
- Random Walks Generation: generate random walks in the graph, a series of nodes.
- Skip-gram Model Training: generate meaningful node embeddings, predict neighboring nodes, and capture the local and global structures of the network.
- Embedding Extraction: extract node embeddings.
4. Evaluation Results
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pais, R.; Ruano, L.; Carvalho, O.P.; Barros, H. Global Cognitive Impairment Prevalence and Incidence in Community Dwelling Older Adults—A Systematic Review. Geriatrics 2020, 5, 84. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Association. 2020 Alzheimer’s Disease Facts and Figures; Wiley: New York, NY, USA, 2020. [Google Scholar]
- Garg, D.; Gupta, A.; Agarwal, A.; Mishra, B.; Srivastava, M.V.P.; Basheer, A.; Vishnu, V.Y. Latest Trends in Outcome Measures in Dementia and Mild Cognitive Impairment Trials. Brain Sci. 2022, 12, 922. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, M.N.; Boada, M.; Borson, S.; Chilukuri, M.; Doraiswamy, P.M.; Dubois, B.; Ingram, J.; Iwata, A.; Porsteinsson, A.P.; Possin, K.L.; et al. Rationale for Early Diagnosis of Mild Cognitive Impairment (MCI) supported by Emerging Digital Technologies. J. Prev. Alzheimer’s Dis. 2020, 7, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.; O’Neill, B.; Evans, J.J.; Coen, R.F.; Lawlor, B.A. A review of screening tests for cognitive impairment. J. Neurol. Neurosurg. Psychiatry 2007, 78, 790–799. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Mental Health Atlas. 2017. Available online: https://www.who.int/mental_health/evidence/atlas/mental_health_atlas_2017/en/ (accessed on 15 November 2023).
- van Harten, A.C.; Mielke, M.M.; Swenson-Dravis, D.M.; Hagen, C.E.; Edwards, K.K.; Roberts, R.O.; Geda, Y.E.; Knopman, D.S.; Petersen, R.C. Subjective cognitive decline and risk of MCI: The Mayo Clinic Study of Aging. Neurology 2018, 91, e300–e312. [Google Scholar] [CrossRef] [PubMed]
- McCollum, L.; Karlawish, J. Cognitive Impairment Evaluation and Management. Med. Clin. N. Am. 2020, 104, 807–825. [Google Scholar] [CrossRef]
- Edmonds, E.C.; Delano-Wood, L.; Clark, L.R.; Jak, A.J.; Nation, D.A.; McDonald, C.R.; Libon, D.J.; Au, R.; Galasko, D.; Salmon, D.P.; et al. Susceptibility of the conventional criteria for mild cognitive impairment to false-positive diagnostic errors. Alzheimer’s Dement. 2015, 11, 415–424. [Google Scholar] [CrossRef]
- Lim, Y.Y.; Kong, J.; Maruff, P.; Jaeger, J.; Huang, E.; Ratti, E. Longitudinal Cognitive Decline in Patients with Mild Cognitive Impairment or Dementia Due to Alzheimer’s Disease. J. Prev. Alzheimer’s Dis. 2022, 9, 178–183. [Google Scholar] [CrossRef]
- Ahamed, F.; Shahrestani, S.; Cheung, H. Internet of Things and Machine Learning for Healthy Ageing: Identifying the Early Signs of Dementia. Sensors 2020, 20, 6031. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, J.; Yu, X.; Wang, S.; Zhang, Y. A Survey of Deep Learning for Alzheimer’s Disease. Mach. Learn. Knowl. Extr. 2023, 5, 611–668. [Google Scholar] [CrossRef]
- Moradi, E.; Pepe, A.; Gaser, C.; Huttunen, H.; Tohka, J.; Alzheimer’s Disease Neuroimaging Initiative. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. NeuroImage 2015, 104, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Alashwal, H.; El Halaby, M.; Crouse, J.J.; Abdalla, A.; Moustafa, A.A. The Application of Unsupervised Clustering Methods to Alzheimer’s Disease. Front. Comput. Neurosci. 2019, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Taherdoost, H. Deep Learning and Neural Networks: Decision-Making Implications. Symmetry 2023, 15, 1723. [Google Scholar] [CrossRef]
- Lee, G.; Nho, K.; Kang, B.; Sohn, K.A.; Kim, D.; Alzheimer’s Disease Neuroimaging Initiative. Predicting Alzheimer’s disease progression using multi-modal deep learning approach. Sci. Rep. 2019, 9, 1952. [Google Scholar] [CrossRef] [PubMed]
- Wanigatunga, A.; Liu, F.; Wang, H.; Urbanek, J.; An, Y.; Simonsick, E.; Resnick, S.; Schrack, J. Daily Physical Activity Patterns: A Window on Cognitive Decline in the Baltimore Longitudinal Study of Aging (BLSA). Innov. Aging 2021, 5 (Suppl. S1), 443–444. [Google Scholar] [CrossRef]
- Shukla, A.; Tiwari, R.; Tiwari, S. Review on Alzheimer Disease Detection Methods: Automatic Pipelines and Machine Learning Techniques. Sci 2023, 5, 13. [Google Scholar] [CrossRef]
- Muhammed Niyas, K.P.; Thiyagarajan, P. A systematic review on early prediction of Mild cognitive impairment to alzheimers using machine learning algorithms. Int. J. Intell. Netw. 2023, 4, 74–88. [Google Scholar] [CrossRef]
- Javed, A.R.; Saadia, A.; Mughal, H.; Gadekallu, T.R.; Rizwan, M.; Maddikunta, P.K.R.; Mahmud, M.; Liyanage, M.; Hussain, A. Artificial Intelligence for Cognitive Health Assessment: State-of-the-Art, Open Challenges and Future Directions. Cogn. Comput. 2023, 15, 1767–1812. [Google Scholar] [CrossRef]
- Gomes, K.B.; Pereira, R.G.; Braga, A.A.; Guimarães, H.C.; Resende, E.d.P.F.; Teixeira, A.L.; Barbosa, M.T.; Junior, W.M.; Carvalho, M.d.G.; Caramelli, P. Machine Learning-Based Routine Laboratory Tests Predict One-Year Cognitive and Functional Decline in a Population Aged 75+ Years. Brain Sci. 2023, 13, 690. [Google Scholar] [CrossRef]
- Chiu, S.-I.; Fan, L.-Y.; Lin, C.-H.; Chen, T.-F.; Lim, W.S.; Jang, J.-S.R.; Chiu, M.-J. Machine Learning-Based Classification of Subjective Cognitive Decline, Mild Cognitive Impairment, and Alzheimer’s Dementia Using Neuroimage and Plasma Biomarkers. ACS Chem. Neurosci. 2022, 13, 3263–3270. [Google Scholar] [CrossRef]
- Zhou, T.; Thung, K.H.; Liu, M.; Shi, F.; Zhang, C.; Shen, D. Multi-modal latent space inducing ensemble SVM classifier for early dementia diagnosis with neuroimaging data. Med. Image Anal. 2020, 60, 101630. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, S.; Cai, W.; Che, H.; Pujol, S.; Kikinis, R.; Feng, D.; Fulham, M.J. Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s disease. IEEE Trans. Biomed. Eng. 2015, 62, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Sheng, C.; Jin, S.; Zhang, Q.; Zhang, S.; Zhang, L.; Ding, C.; Wang, L.; Wang, L.; Han, Y.; et al. Generative adversarial network constrained multiple loss autoencoder: A deep learning-based individual atrophy detection for Alzheimer’s disease and mild cognitive impairment. Hum. Brain Mapp. 2023, 44, 1129–1146. [Google Scholar] [CrossRef]
- Taye, M.M. Understanding of Machine Learning with Deep Learning: Architectures, Workflow, Applications and Future Directions. Computers 2023, 12, 91. [Google Scholar] [CrossRef]
- Sarraf, S.; DeSouza, D.D.; Anderson, J.; Tofighi, G.; Alzheimer’s Disease Neuroimaging Initiative. DeepAD: Alzheimer Disease Classification via Deep Convolutional Neural Networks using MRI and fMRI. bioRxiv 2017. [Google Scholar] [CrossRef]
- Stalin Babu, G.; Rao, S.N.T.; Rao, R.R. Alzheimer’s Disease Prediction via Optimized Deep Learning Framework. In Proceedings of the Second International Conference on Advances in Computer Engineering and Communication Systems, Singapore, 23 January 2022. [Google Scholar]
- Diaz, V.; Rodriguez, G. Machine Learning for Detection of Cognitive Impairment. Acta Polytech. Hung. 2022, 19, 195–213. [Google Scholar] [CrossRef]
- Jeon, Y.; Kang, J.; Kim, B.C.; Lee, K.H.; Song, J.-I.; Gwak, J. Early Alzheimer’s Disease Diagnosis Using Wearable Sensors and Multilevel Gait Assessment: A Machine Learning Ensemble Approach. IEEE Sens. J. 2023, 23, 10041–10053. [Google Scholar] [CrossRef]
- Lin, F.; Han, J.; Xue, T.; Lin, J.; Chen, S.; Zhu, C.; Lin, H.; Chen, X.; Lin, W.; Huang, H. Predicting cognitive impairment in outpatients with epilepsy using machine learning techniques. Sci. Rep. 2021, 11, 20002. [Google Scholar] [CrossRef]
- Lin, L.; Xiong, M.; Zhang, G.; Kang, W.; Sun, S.; Wu, S.; Initiative Alzheimer’s Disease Neuroimaging. A Convolutional Neural Network and Graph Convolutional Network Based Framework for AD Classification. Sensors 2023, 23, 1914. [Google Scholar] [CrossRef]
- Tang, H.; Guo, L.; Dennis, E.; Thompson, P.M.; Huang, H.; Ajilore, O.; Leow, A.D.; Zhan, L. Classifying Stages of Mild Cognitive Impairment via Augmented Graph Embedding. In Multimodal Brain Image Analysis and Mathematical Foundations of Computational Anatomy; Zhu, D., Yan, J., Huang, H., Shen, L., Thompson, P.M., Westin, C.-F., Pennec, X., Joshi, S., Nielsen, M., Fletcher, T., et al., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 30–38. [Google Scholar]
- Xu, M.; Sanz, D.L.; Garces, P.; Maestu, F.; Li, Q.; Pantazis, D. A Graph Gaussian Embedding Method for Predicting Alzheimer’s Disease Progression with MEG Brain Networks. IEEE Trans. Biomed. Eng. 2021, 68, 1579–1588. [Google Scholar] [CrossRef]
- Lei, B.; Zhu, Y.; Yu, S.; Hu, H.; Xu, Y.; Yue, G.; Wang, T.; Zhao, C.; Chen, S.; Yang, P.; et al. Multi-scale enhanced graph convolutional network for mild cognitive impairment detection. Pattern Recognit. 2023, 134, 109106. [Google Scholar] [CrossRef]
- Ahmedt-Aristizabal, D.; Armin, M.A.; Denman, S.; Fookes, C.; Petersson, L. Graph-Based Deep Learning for Medical Diagnosis and Analysis: Past, Present and Future. Sensors 2021, 21, 4758. [Google Scholar] [CrossRef] [PubMed]
- Holm, A.N.; Wright, D.; Augenstein, I. Revisiting Softmax for Uncertainty Approximation in Text Classification. Information 2023, 14, 420. [Google Scholar] [CrossRef]
- Appenzeller, A.; Leitner, M.; Philipp, P.; Krempel, E.; Beyerer, J. Privacy and Utility of Private Synthetic Data for Medical Data Analyses. Appl. Sci. 2022, 12, 12320. [Google Scholar] [CrossRef]
- Chen, R.J.; Lu, M.Y.; Chen, T.Y.; Williamson, D.F.K.; Mahmood, F. Synthetic data in machine learning for medicine and healthcare. Nat. Biomed. Eng. 2021, 5, 493–497. [Google Scholar] [CrossRef]
- Grover, A.; Leskovec, J. Node2vec: Scalable Feature Learning for Networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ‘16), San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 855–864. [Google Scholar]

| Feature | Description |
|---|---|
| Daily Activity Count | Total number of activities performed daily. It helps in understanding the general activity level, which can be indicative of changes in physical and cognitive health. |
| Multi-dimensional Activity Encoding | Encoded array with activity type, start and end timestamps, and duration. This provides a detailed view of the patient’s daily routine, allowing the chance to identify any deviations or irregularities that might signal cognitive decline. |
| Total Active Hours | Cumulative duration of moderate to high energy expenditure activities in a day. It is an important metric for assessing the physical aspect of cognitive health, as changes in active hours can reflect alterations in energy levels and overall wellness. |
| Average Daily Activity Count | The mean activity count over the monitoring period, offering insights into the subject’s routine consistency, which is often disrupted in cognitive decline. |
| Daily Activity Duration Variability | The standard deviation of active daily activity durations. High variability can indicate irregular activity patterns, a potential marker of cognitive issues. |
| Activity Fragmentation | Reciprocal of the mean bout length, indicating the rhythm and continuity of activity bouts. Disrupted activity fragmentation is often associated with cognitive decline, as it reflects challenges in maintaining regular, sustained activities. |
| Average Fragmentation | Mean activity fragmentation over the entire monitoring span |
| Metric | Healthy | Risk of Cognitive Decline |
|---|---|---|
| Precision | 100% | 80% |
| Recall | 100% | 90% |
| F1-Score | 100% | 81% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonesi, G.; Rancea, A.; Cioara, T.; Anghel, I. Graph Learning and Deep Neural Network Ensemble for Supporting Cognitive Decline Assessment. Technologies 2024, 12, 3. https://doi.org/10.3390/technologies12010003
Antonesi G, Rancea A, Cioara T, Anghel I. Graph Learning and Deep Neural Network Ensemble for Supporting Cognitive Decline Assessment. Technologies. 2024; 12(1):3. https://doi.org/10.3390/technologies12010003
Chicago/Turabian StyleAntonesi, Gabriel, Alexandru Rancea, Tudor Cioara, and Ionut Anghel. 2024. "Graph Learning and Deep Neural Network Ensemble for Supporting Cognitive Decline Assessment" Technologies 12, no. 1: 3. https://doi.org/10.3390/technologies12010003
APA StyleAntonesi, G., Rancea, A., Cioara, T., & Anghel, I. (2024). Graph Learning and Deep Neural Network Ensemble for Supporting Cognitive Decline Assessment. Technologies, 12(1), 3. https://doi.org/10.3390/technologies12010003