1. Introduction
Climate is seen as an important factor that controls species distributions, at least across broad, spatial scales [
1,
2,
3]. Determining the relationship between species distributions and climate is an important goal in many ecological studies, particularly in forecasting potential impacts of climate change [
4,
5]. One tool that has been heavily relied on in making these forecasts is species distribution modeling, commonly used to evaluate extinction risk of species to assess conservation impacts [
6] and develop risk assessments for invasive species based on habitat suitability [
7].
The field of species distribution modeling has rapidly expanded over the last decade. Correlative species distribution models are developed by relating locations of organisms to environmental characteristics at observed sites to make predictions about suitable habitat across a geographic extent [
8]. These correlative methods, also known as ecological niche models, habitat suitability models, or environmental matching models, make several assumptions. One assumption is that the environmental predictors included in the model constrain the species’ distribution [
9]. In addition, when projecting the model in space or time, these models also assume that the correlation structure among the predictors is constant and that the niche is conserved [
10].
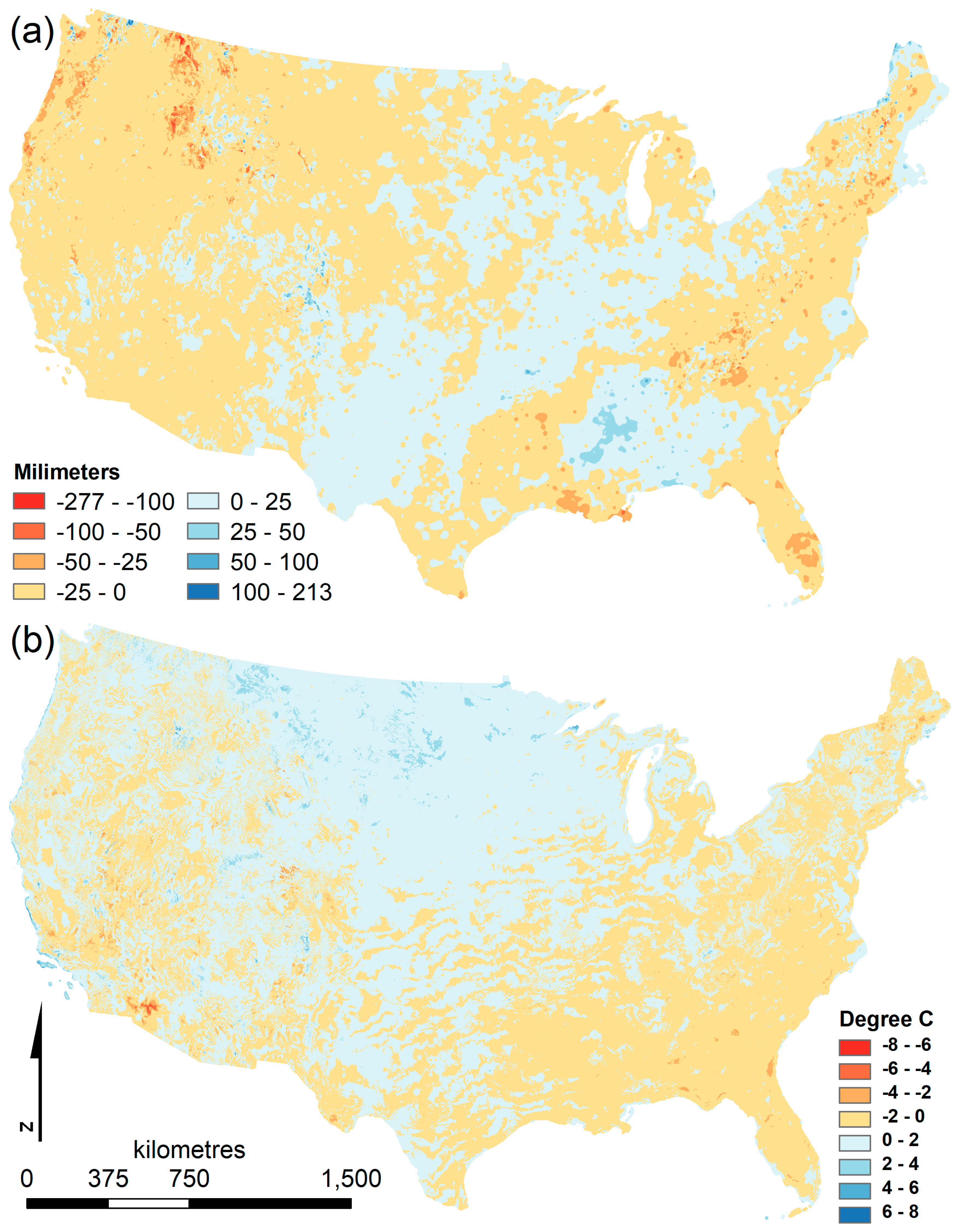
Climate averages over time are often described as climate “normals”, a standard mandated by the World Meteorological Organization, consisting of an arithmetic average of climate over a 30-year period and used as a reference point to compare to other time periods. The Parameter-elevation Regressions on Independent Slopes Model (PRISM) climate normals data set for the continental United States of America (USA) has been updated from monthly averages across 1971 to 2000 to include monthly averages across 1981 to 2010 [
11]. Despite the overlap of two-thirds of the years, there are substantial changes in the 30-year average precipitation, maximum temperature, and minimum temperature. For example, parts of the USA exhibited over 200 mm more precipitation during the driest month in the earlier period while other regions had less precipitation at a comparable magnitude (
Figure 1). Similar differences existed for minimum temperature of the coldest month, with spatial heterogeneity between areas that were colder or warmer between the two time periods.
In correlative models, model parameterization is dependent on the correlation structure between predictor variables and location data. Changes in correlation structure can be particularly important when extrapolating to novel climates [
12], so understanding how correlations vary in space and time is important. If there is a temporal disconnect between location data collection and predictor variable coverage, and the predictor variable’s correlation structure is not constant through time, this could potentially have a large effect on model results.
The environmental data used to produce correlative species distribution models, including climate, are often based on what data are readily available rather than decisions made based on the research question or species of interest [
8]. For example, the global WorldClim data set, which only provides climate normals (1950 and 2000 [
13] or 1970 to 2000), was cited in 1950 of 6380 articles published between 2013 to 2017 (google scholar search including climate AND [“species distribution model” OR “niche model” OR “habitat suitability model” OR “environmental matching model”]). These results indicate that a high percentage of species distribution models used the readily available WorldClim data set rather than a data set with customizable time periods. The PRISM data set is commonly mentioned in papers focused in the USA (293 of 2720 articles included PRISM). Unlike WorldClim, this dataset is downloadable for two periods of climate normals and is available as monthly data that could be used to develop project-specific climate normals, albeit at a coarser spatial resolution. As readily available climate normals datasets continue to be used in species distribution models, it is important to assess the potential impact of the temporal period of climate data on model results.
Our objectives were to evaluate the sensitivity of correlative species distribution models to the choice of partially overlapping climate normals period, including variable importance, variable response curves, and prediction agreement, while also examining the consistency of the correlation structure between the two baseline data sets. We did this using readily available data and standard best practices for creating correlative models to mimic common applications of these models. We evaluated whether model results were more sensitive to a time period or model algorithm choice. We hypothesized that the effect of climate normals data would be less for a long-lived species (e.g., tree) than for a short-lived species (e.g., insect). In addition, we hypothesized that simpler model algorithms would be less sensitive to the choice of climate normals data time period. We investigated the sensitivity of species distribution models to climate time period used by creating models with species data for a variety of taxa with commonly used climate data that matched the 30-year time span recommended by the Intergovernmental Panel on Climate Change. We compared the uncertainty from baseline climate data to uncertainty from the model technique, one of the greatest sources of uncertainty from those quantified [
14], and assessed the consistency of predictions.
3. Results
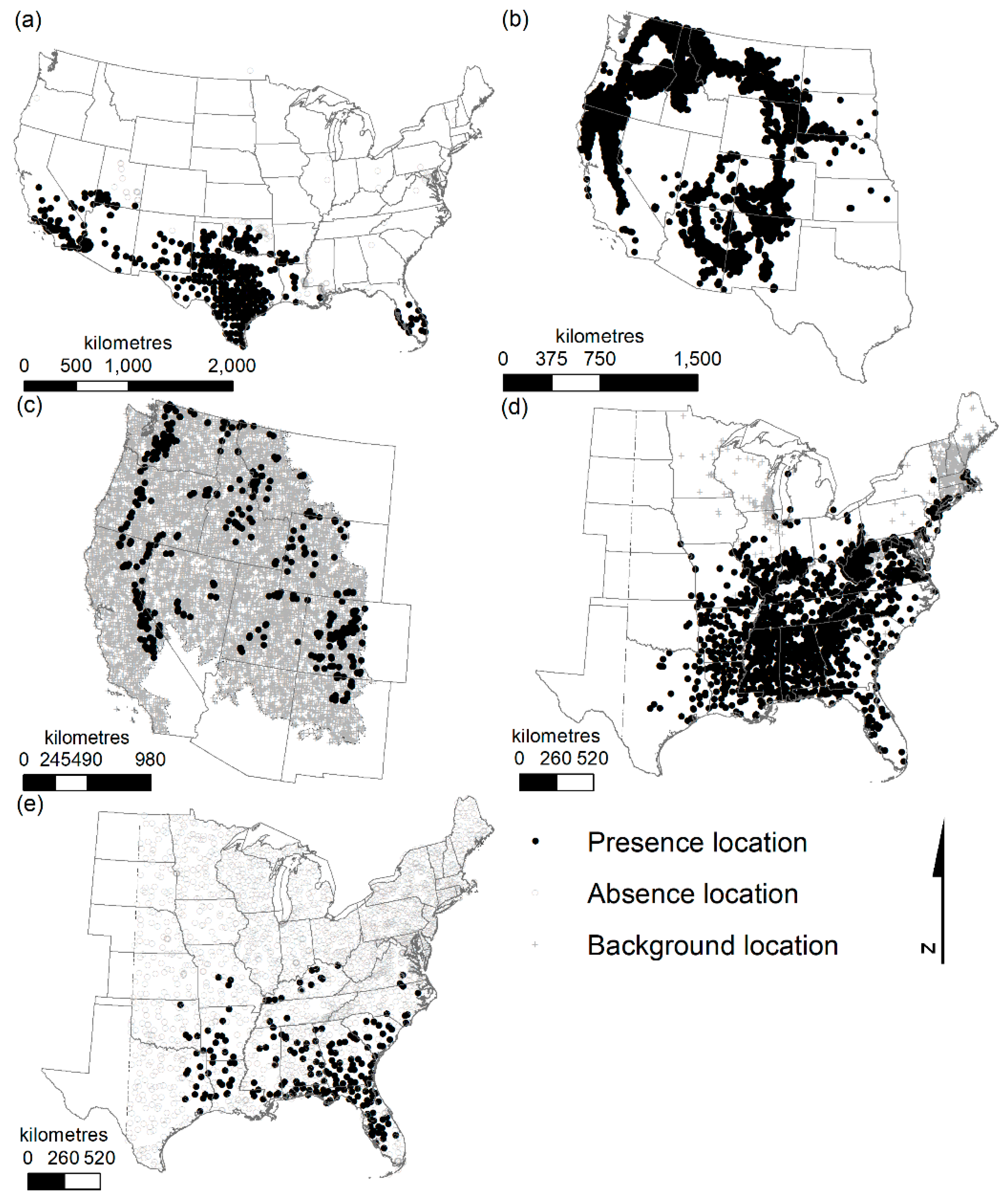
All models performed well, with high cross-validation discrimination capacity (e.g., most independent split AUC values >0.9, all >0.8). Calibration plots also indicated good agreement between predicted probabilities of occurrence and observed occupancy, with a few exceptions (e.g., American pika and kudzu, which had background rather than absence data). However, for these analyses we were primarily concerned with discrimination ability.
3.1. Variable Correlation
Despite regional heterogeneity visible in the maps of differences in climate between the two climate normals periods (
Figure 1), correlations between predictor variable values from 1971 to 2000 compared to the same predictor variable in 1981 to 2010 were similar (
Table 1 and
Table S1 diagonals). For all variables except mean temperature of the wettest quarter [bio8] in the eastern USA (
Table S1b), correlations were |r| > 0.9. Mean temperature of the wettest quarter consistently had the lowest correlation, perhaps due to changes in the consecutive three-month period that comprised the quarter, as maps of this predictor are spatially disjointed.
Correlations between predictor variables within a single climate normals period also remained consistent when compared across climate normals periods (
Table 1 mirror image above and below the diagonal). Differences in correlation coefficients for predictor variable pairs between the two time periods were commonly <0.05.
3.2. Variable Importance
Variable retention and importance ranking generally changed with a change in climate normals periods regardless of species or model algorithm (
Table 2). While this was not the case for every pair of species and model type, each species and model type had at least one difference in variable retention or ranking. For kudzu, however, the only difference among models was that the 1971 to 2000 climate normals GLM model retained mean diurnal range, though with a very low relative importance value (0.1%;
Table 2d). This predictor was dropped by the 1981 to 2010 climate normals GLM model. The other four variables were consistent between the GLM models in their order of importance. Africanized honey bee models also showed consistency in variable importance ranking among models with only one difference, where the third and fourth most important variables switched places in the RF model (
Table 2a). Across model types maintaining a consistent climate period, each pairing had a difference in variable importance ranking or variable retention for three of five species.
Most differences among model pairings with a change in climate normals period involved a switch in the order of variable importance. In some cases variables were dropped during the model fitting process for one climate normals period while they were retained in the model fitting process using the other climate normals period. For example, the top two predictors in BRT models of Bachman’s sparrow were the same, but mean temperature of the wettest quarter was retained as a third predictor in the model using 1981 to 2010 climate normals data, but dropped from the model using 1971 to 2000 climate data (
Table 2e). However, this predictor contributed relatively little to the model (6.1%).
Ponderosa pine and American pika, both modeled in the western USA, exhibited the greatest differences in variable importance between paired models from the two different time periods. These two species also had the greatest number of predictors retained after the variable selection correlation step (nine and eight, respectively, compared to five in the other species’ models).
3.3. Prediction Agreement
The choice of model algorithm generally caused greater differences in predictions between models than the choice of climate normals period (
Table 3). The magnitude of differences in predictions between species, however, differed. Ponderosa pine had the largest difference in model algorithm choice (average of 16%), and American pika had the smallest (average of 3%). Africanized honey bees had the greatest difference related to selection of climate normals data (12.1% for the MARS models), and American pika, again, had the smallest difference (1.6% for both the GLM and RF models). However, American pika are relatively rare on the landscape. If we only consider areas predicted as suitable by at least one model, we have a much higher level of disagreement (1.6% to 4.9% compared to 14.0% to 40.7%).
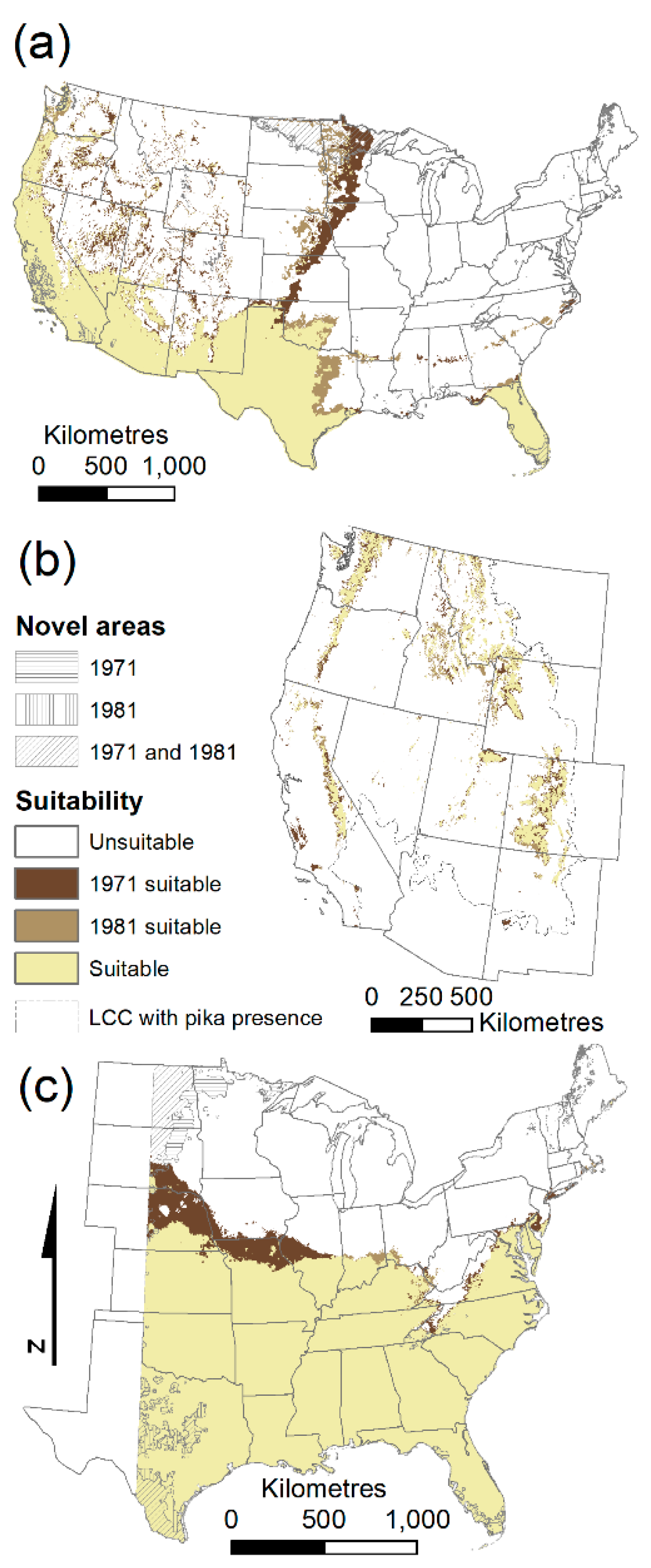
Most differences between model predictions, either from algorithm choice or climate normals period choice, occurred at the geographic edges of ranges, rather than within the interior of the predicted suitable habitat (
Figure 3;
Supplementary material Figure S1). For example, most of the disagreement between the choice of climate normals for kudzu was along the northern edge of the predicted suitable habitat (
Figure 3c).
BRT models for Africanized honey bees for both time periods selected the same subset of predictors with similar levels of variable importance. However, the geographic predictions of suitability were still quite different (8.1% difference;
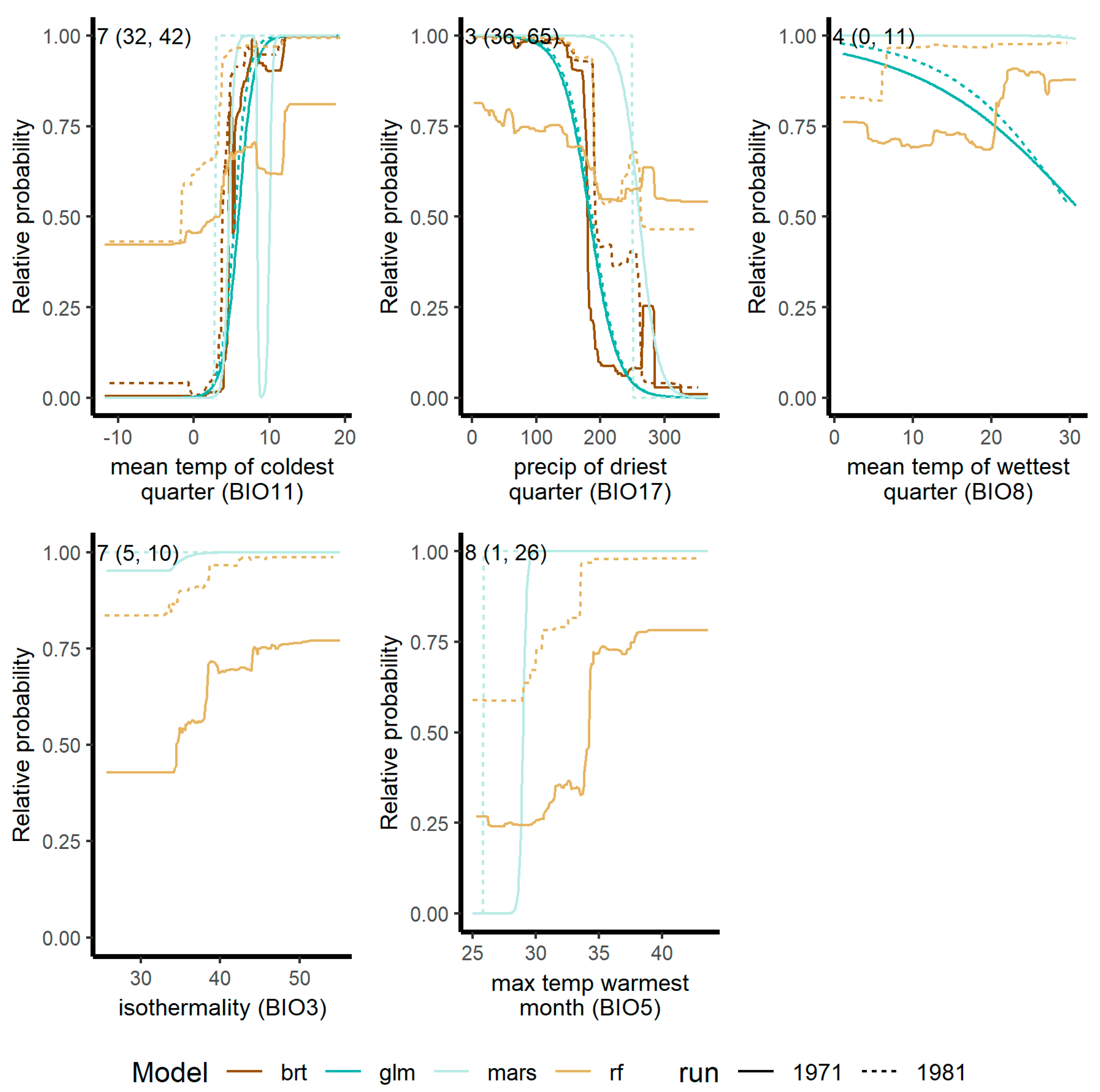
Figure 3a) because the response curve shapes differed (
Figure 4). All species had some differences in response curves (
Supplemental information Figure S2).
4. Discussion
We found the choice of model algorithm had a larger effect on prediction results among models than did the choice of climate normals period, although the climate normals time period did affect model results. Our findings that choice of model algorithm contributed considerable uncertainty to model results are consistent with others [
14,
36]. This underscores the importance of evaluating multiple model algorithms when developing habitat suitability models [
37]. There are several potential sources of uncertainty in species distribution modeling that have received attention in the literature [
9,
38,
39,
40,
41,
42,
43], often focusing on quantifiable uncertainty. Some researchers have compared the relative contribution of different components of uncertainty to models [
14,
36]. These components range from quality of location data (e.g., misidentification or inaccurate coordinates) to variance arising from different modeling algorithms, finding that model algorithm choice contributed the greatest amount of uncertainty among the factors they examined. We also found that these differences were consistent across taxa, despite the differences in life history and occurrence information (e.g., availability of absence data) among species we tested.
Our results also complement those of others looking more specifically at climate data. Braunisch et al. [
39] examined uncertainty introduced through the correlation filtering process to deal with collinearity between variables. Differences in predictions arose depending on the identity of the climate variables retained, similar to our findings of differences due to time period. In a similar study to this one, Roubicek et al. [
44] tested the importance of a temporal mismatch between occurrence data and climate data, using different baseline climate data with simulated location data for 18 different 10-year time spans, one 30-year time span, and one 50-year time span. They highlighted the importance of a temporal match between species location data and climate time period. However, our study differs from previous work in that we use a larger spatial extent (US compared to East coast of Australia) and real applications, rather than simulated species, including a diverse group of taxa with differing lifespans and data qualities. In addition, Roubicek et al. [
44] performed all modeling with Maxent, which is a presence–background technique, while we evaluate four commonly used techniques for presence–absence/pseudo–absence (but exclude Maxent as most modeled species had absence data). In this study we had temporal overlap between the two periods, with our location data generally spanning the climate normals periods covered. Our finding of model sensitivity to time period of climate normals is one aspect of uncertainty that was previously relatively unexplored.
Differences in variable importance and response curves due to the choice of climate normals time period resulted in measurable differences in predicted habitat suitability for the five species, even when only one-third of the data were different between the two periods. These differences arose despite the fact that values extracted from the two time periods were highly correlated (e.g.,
Table 1). If a subset of predictors was more highly correlated between the time periods, we could have tested if using the more correlated subset decreased differences between fitted models.
One use of correlative species distribution models istrying to predict shifting distributions due to climate change or range expansion of species [
45,
46,
47,
48]. As discussed elsewhere [
49], correlative models have their limitations for these types of assessments. While model type produced greater variation than climate normals time period, the choice of time period did affect all species and all model algorithms tested. These impacts occurred regardless of species taxa, spatial extent (entire US or a subset), niche breadth (generalist such as ponderosa to specialist such as pika), and nativity, all of which can affect model performance. Our hypothesis that long-lived species would be less impacted did not hold up; in fact, Africanized honey bees had less change in predictor importance than ponderosa. These differences may only be exacerbated when applied to novel locations or time periods [
50], as seen in other evaluations examining uncertainty [
39].
5. Conclusions
We showed that even with species-tuned model parameters, rather than generic model parameterization that is often applied to large suites of species, results are highly sensitive to both baseline climate period and model method. While the latter is well recognized as an important uncertainty and is often accounted for through ensemble modeling [
51], the impact of the former is not commonly assessed. We hypothesized that one particular modeling algorithm might consistently exhibit less impact, such as GLM because it is a simpler model with smoother response curves, but our results indicate that, instead, we have uncovered yet another source of uncertainty for model developers to evaluate on a case by case basis. Luckily, the development of somewhat automated software such as ModEco [
52], BIOMOD [
53] and VisTrails:SAHM [
26] facilitate running multiple modeling algorithms several times to test the sensitivity of a model to a particular decision (e.g., choice of model algorithm, threshold rules, predictor data sets, background selection, etc.). Another possibility is that as the availability of climate data and computational processing capabilities increase, climate normals relative to time of sampling could be used (e.g., 30-year average prior to sampling year, or even a longer climate average spanning the range of sampling dates such as 1971–2010 in our case). Our results further highlight the uncertainty around using these types of correlative models for forecasting future distributions of species to inform conservation decisions. Evaluating uncertainty in current models is a necessary first step before determining if it is wise to proceed with projections.