A Genomic Perspective on the Evolutionary Diversity of the Plant Cell Wall
Abstract
1. Introduction
2. Cell Wall Diversity and Plant Evolution
3. Recent Updates and Developments in the Databases of Plant Genomes and Cell Wall Genes
4. Comparative Plant Genome Analysis of Plants with a View toward Characterizing of Cell Wall Diversity
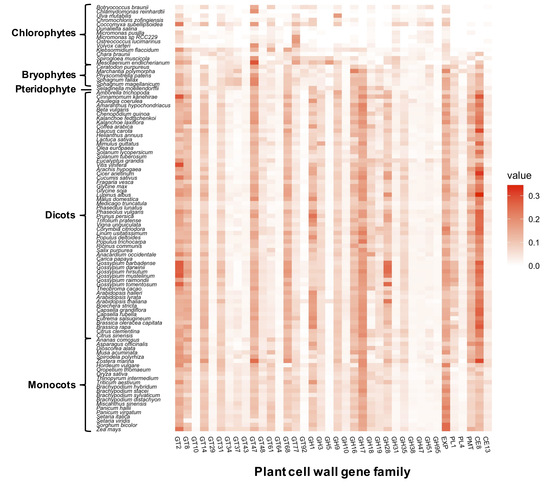
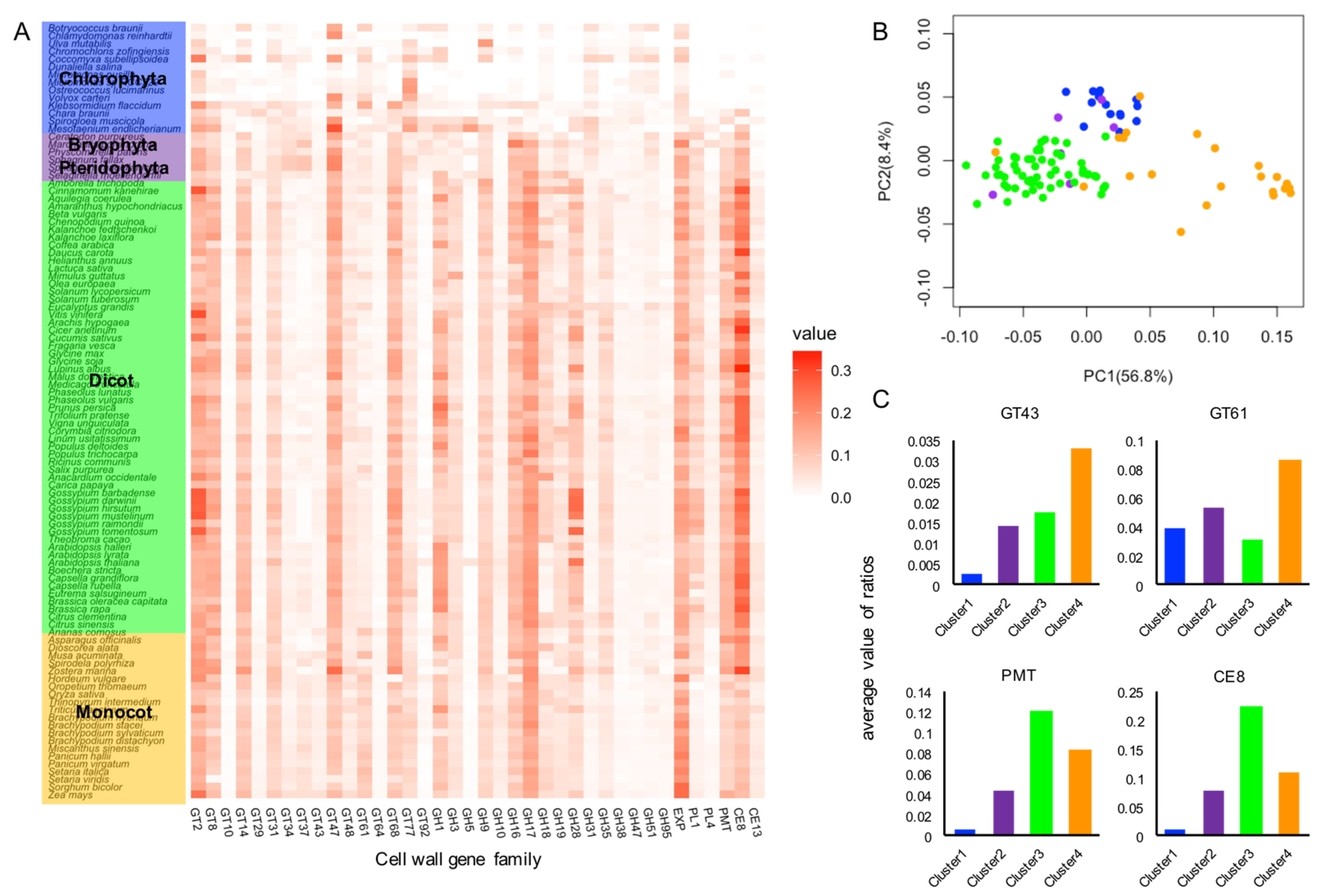
4.1. Comparative Analysis of the Cell Wall Gene Families among 100 Plant Species
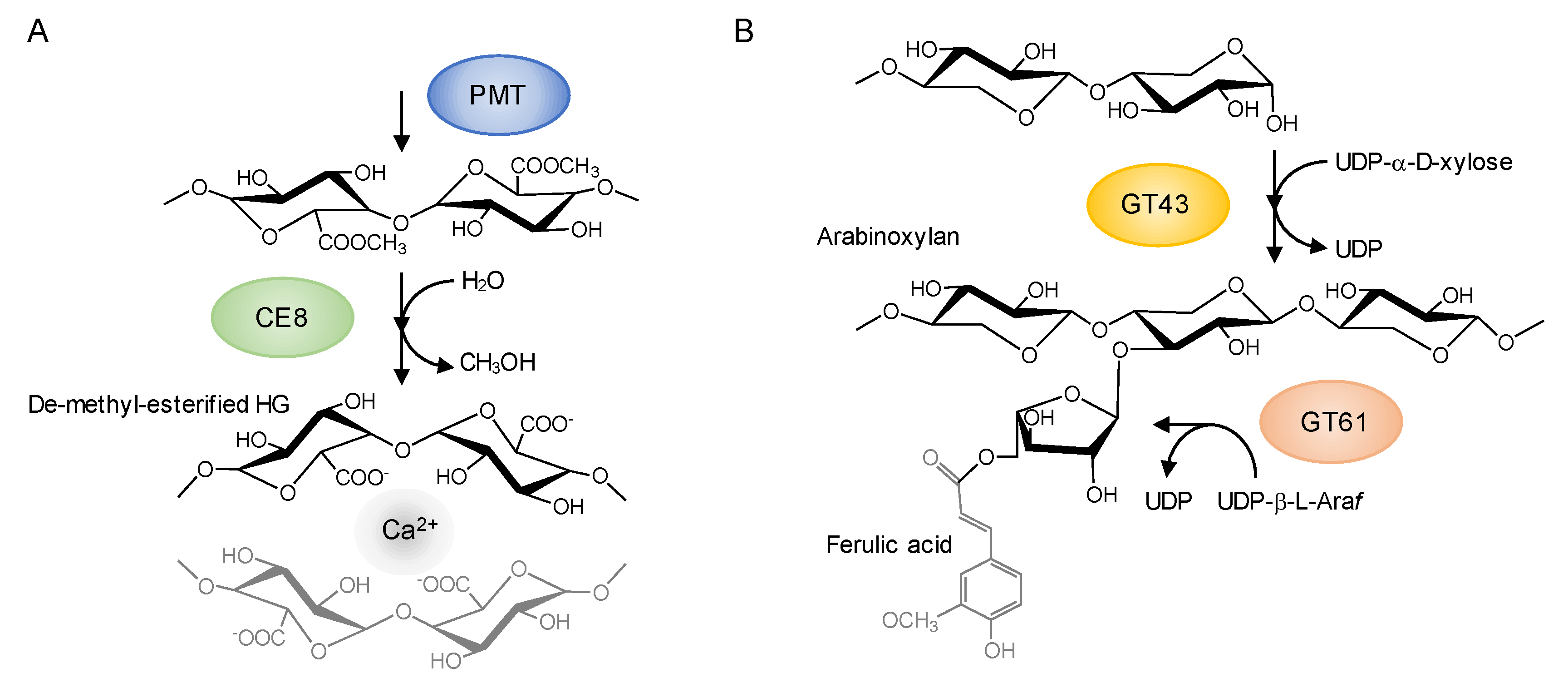
4.2. Identification of Cell Wall Gene Families that Contribute to the Diversity Patterns of Cell Wall Gene Family Members in Plant Linkages
5. Concluding Remarks and Perspectives
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a systems approach to understanding plant-cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- McNeil, M.; Darvill, A.G.; Fry, S.C.; Albersheim, P. Structure and function of the primary cell walls of plants. Annu. Rev. Biochem. 1984, 53, 625–663. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T. Xyloglucans in the primary cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 139–168. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, K. Construction and Restructuring of the cellulose-xyloglucan framework in the apoplast as mediated by the xyloglucan-related protein family—A hypothetical scheme. J. Plant Res. 1998, 111, 159–166. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Expansive growth of plant cell walls. Plant Physiol. Biochem. 2000, 38, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.C. Primary cell wall metabolism: Tracking the careers of wall polymers in living plant cells. New Phytol. 2005, 161, 641–675. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Re-constructing our models of cellulose and primary cell wall assembly. Curr. Opin. Plant Biol. 2014, 22, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 2012, 158, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015, 56, 180–194. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.C.; Bush, M.; Milioni, D.; Sado, P.; Stacey, N.J.; Catchpole, G.; Defernez, M.; Carpita, N.C.; Hofte, H.; Ulvskov, P.; et al. Approaches to understanding the functional architecture of the plant cell wall. Phytochemistry 2001, 57, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.; Keegstra, K. Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 2010, 13, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Cano-Delgado., A.; Lee, J.Y.; Demura, T. Regulatory mechanisms for specification and patterning of plant vascular tissues. Annu. Rev. Cell Dev. Biol. 2010, 26, 605–637. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Somssich, M.; Nakata, M.T.; Unda, F.; Atsuzawa, K.; Kaneko, Y.; Wang, T.; Bågman, A.-M.; Gaudinier, A.; Yoshida, K.; et al. Complete substitution of a secondary cell wall with a primary cell wall in Arabidopsis. Nat. Plants 2018, 4, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zheng, Y.; Cosgrove, D.J. Spatial organization of cellulose microfibrils and matrix polysaccharides in primary plant cell walls as imaged by multichannel atomic force microscopy. Plant J. 2016, 85, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Vavylonis, D.; Durachko, D.M.; Cosgrove, D.J. Nanoscale movements of cellulose microfibrils in primary cell walls. Nat. Plants 2017, 3, 17056. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Zhao, S.; Shen, W.; Collings, C.; Ding, S.-Y. Direct Measurement of Plant Cellulose Microfibril and Bundles in Native Cell Walls. Front. Plant Sci. 2020, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Simmons, T.J.; Mortimer, J.C.; Bernardinelli, O.D.; Pöppler, A.C.; Brown, S.P.; de Azevedo, E.R.; Dupree, R.; Dupree, P. Folding of xylan onto cellulose fibrils in plant cell walls revealed by solid-state NMR. Nat. Commun. 2016, 7, 13902. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Kirui, A.; Dickwella Widanage, M.C.; Mentink-Vigier, F.; Cosgrove, D.J.; Wang, T. Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid-state NMR. Nat. Commun. 2019, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Knox, J.P. Revealing the structural and functional diversity of plant cell walls. Curr. Opin. Plant Biol. 2018, 11, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Pattathil, S.; Avci, U.; Baldwin, D.; Swennes, A.G.; McGill, J.A.; Popper, Z.; Bootten, T.; Albert, A.; Davis, R.H.; Chennareddy, C.; et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 2010, 153, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, C.; Bartetzko, M.P.; Senf, D.; Dallabernadina, P.; Boos, I.; Andersen, M.C.F.; Kotake, T.; Knox, J.P.; Hahn, M.G.; Clausen, M.H.; et al. A synthetic glycan microarray enables epitope mapping of plant cell wall glycan-directed antibodies. Plant Physiol. 2017, 175, 1094–1104. [Google Scholar] [CrossRef] [PubMed]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Ordaz-Ortiz, J.J.; Knox, J.P. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 2009, 344, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, X.; Chen, Y.; Wagner, E.; Cosgrove, D.J. Xyloglucan in the primary cell wall: Assessment by FESEM, selective enzyme digestions and nanogold affinity tags. Plant J. 2018, 92, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.T.; Carroll, A.; Akhmetova, L.; Somerville, C. Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 2010, 152, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Anderson, C.T. Functional analysis of cellulose and xyloglucan in the walls of stomatal guard cells of Arabidopsis. Plant Physiol. 2016, 170, 1398–1419. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Kuki, H.; Kuroha, T.; Nishitani, K. Arabidopsis regenerating protoplast: A powerful model system for combining the proteomics of cell wall proteins and the visualization of cell wall dynamics. Proteomes 2016, 4, 34. [Google Scholar] [CrossRef] [PubMed]
- Kuki, H.; Higaki, T.; Yokoyama, R.; Kuroha, T.; Shinohara, N.; Hasezawa, S.; Nishitani, K. Quantitative confocal imaging method for analyzing cellulose dynamics during cell wall regeneration in Arabidopsis mesophyll protoplasts. Plant Direct 2017, 1, e00021. [Google Scholar] [CrossRef] [PubMed]
- Kuki, H.; Yokoyama, R.; Kuroha, T.; Nishitani, K. Xyloglucan is not essential for the formation and integrity of the cellulose network in the primary cell wall regenerated from Arabidopsis protoplast. Plants 2020, 9, 629. [Google Scholar] [CrossRef] [PubMed]
- Macfie, S.; Welbourn, P.M. The cell wall as a barrier to uptake of metal ions in the unicellular green alga Chlamydomonas reinhardtii (Chlorophyceae). Arch. Environ. Contam. Toxicol. 2000, 39, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Mao, F.L.; Yin, Y.B.; Zhou, F.F.; Chou, W.C.; Zhou, C.; Chen, H.; Xu, Y. pDAWG: An integrated database for plant cell wall genes. Bioenerg. Res. 2009, 2, 209–216. [Google Scholar] [CrossRef]
- Ekstrom, A.; Taujale, R.; McGinn, N.; Yin, Y. PlantCAZyme: A database for plant carbohydrate-active enzymes. Database (Oxford) 2014, 2014, bau079. [Google Scholar] [CrossRef]
- Blanc, G.; Agarkova, I.; Grimwood, J.; Kuo, A.; Brueggeman, A.; Dunigan, D.D.; Gurnon, J.; Ladunga, I.; Lindquist, E.; Lucas, S.; et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012, 13, R39. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Harholt, J.; Ulvskov, P.; Johansen, I.E.; Fangel, J.U.; Doblin, M.S.; Bacic, A.; Willats, W.G.T. Evidence for land plant cell wall biosynthetic mechanisms in charophyte green algae. Ann. Bot. 2014, 114, 1217–1236. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, I.; Pettolino, F.A.; Bacic, A.; Ralph, J.; Lu, F.; O’Neill, M.A.; Fei, Z.; Rose, J.K.C.; Domozych, D.S.; Willats, W.G.T. The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 2011, 68, 201–211. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, C.; Gregson, T.; Murray, L.; Sadler, I.H.; Fry, S.C. Sugar composition of the pectic polysaccharides of charophytes, the closest algal relatives of land-plants: Presence of 3-O-methyl-d-galactose residues. Ann. Bot. 2015, 116, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Wachananawat, B.; Kuroha, T.; Takenaka, Y.; Kajiura, H.; Naramoto, S.; Yokoyama, R.; Ishizaki, K.; Nishitani, K.; Ishimizu, T. Diversity of pectin rhamnogalacturonan I rhamnosyltransferases in glycosyltransferase family 106. Front. Plant Sci. 2020, 11, 997. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Ishii, T.; Matsumoto, S.; Higuchi, M.; Darvill, A.; Albersheim, P.; O’Neill, M.A. Occurrence of the primary cell wall polysaccharide rhamnogalacturonan II in pteridophytes, lycophytes, and bryophytes: Implications for the evolution of vascular plants. Plant Physiol. 2004, 134, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Herburger, K.; Xin, A.; Holzinger, A. Homogalacturonan Accumulation in Cell Walls of the Green Alga Zygnema sp. (Charophyta) Increases Desiccation Resistance. Front. Plant Sci. 2019, 10, 540. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 445–476. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 2008, 11, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Fincher, G.B. Revolutionary times in our understanding of cell wall biosynthesis and re-modelling in the grasses. Plant Physiol. 2009, 149, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Buanafina, M.M.dO. Feruloylation in grasses: Current and future perspectives. Mol. Plant 2009, 2, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.J.; Trethewey, J.A.K. The distribution of ester-linked ferulic acid in the cell walls of angiosperms. Phytochem. Rev. 2010, 9, 19–33. [Google Scholar] [CrossRef]
- Ishii, T. Structure and functions of feruloylated polysaccharides. Plant Sci. 1997, 127, 111–127. [Google Scholar] [CrossRef]
- Burr, S.J.; Fry, S.C. Extracellular cross-linking of maize arabinoxylans by oxidation of feruloyl esters to form oligoferuloyl esters and ether-like bonds. Plant J. 2009, 58, 554–567. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.A. Evolutionary history of the grasses. Plant Physiol. 2001, 125, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Popper, Z.A.; Michel, G.; Herve, C.; Domozych, D.S.; Willats, W.G.T.; Tuohy, M.G.; Kloareg, B.; Stengel, D.B. Evolution and diversity of plant cell walls: From algae to flowering plants. Annu. Rev. Plant Biol. 2011, 62, 567–590. [Google Scholar] [CrossRef] [PubMed]
- Fangel, J.U.; Ulvskov, P.; Knox, J.P.; Mikkelsen, M.D.; Harholt, J.; Popper, Z.A.; Willats, W.G.T. Cell wall evolution and diversity. Front. Plant Sci. 2012, 3, 152. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Melkonian, M.; Smith, S.A.; Brockington, S.; Archibald, J.M.; Delaux, P.M.; Li, F.W.; Melkonian, B.; Mavrodiev, E.V.; Sun, W.; et al. 10KP: A phylodiverse genome sequencing plan. Gigascience 2018, 7, giy013. [Google Scholar] [CrossRef] [PubMed]
- Little, A.; Schwerdt, J.G.; Shirley, N.J.; Khor, S.F.; Neumann, K.; O’Donovan, L.A.; Lahnstein, J.; Collins, H.M.; Henderson, M.; Fincher, G.B.; et al. Revised phylogeny of the Cellulose Synthase gene superfamily: Insights into cell wall evolution. Plant Physiol. 2018, 177, 1124–1141. [Google Scholar] [CrossRef] [PubMed]
- Bulone, V.; Schwerdt, J.G.; Fincher, G.B. Co-evolution of enzymes involved in plant cell wall metabolism in the grasses. Front. Plant Sci. 2019, 10, 1009. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Helbert, W.; Poulet, L.; Drouillard, S.; Mathieu, S.; Loiodice, M.; Couturier, M.; Lombard, V.; Terrapon, N.; Turchetto, J.; Vincentelli, R.; et al. Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc. Natl. Acad. Sci. USA 2019, 116, 6063–6068. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, Y.; Kato, K.; Ogawa-Ohnishi, M.; Tsuruhama, K.; Kajiura, H.; Yagyu, K.; Takeda, A.; Takeda, Y.; Kunieda, T.; Hara-Nishimura, I.; et al. Pectin RG-I rhamnosyltransferases represent a novel plant-specific glycosyltransferase family. Nat. Plants 2018, 4, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Nishitani, K. Genomic basis for cell—wall diversity in plants: A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol. 2004, 45, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 2017, 171, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Sterck, L.; Billiau, K.; Abeel, T.; Rouzé, P.; de Peer, Y.V. ORCAE: Online resource for community annotation of eukaryotes. Nat. Methods 2012, 9, 1041. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Xian, W.; Fu, Y.; Marin, B.; Keller, J.; Wu, T.; Sun, W.; Li, X.; Xu, Y.; Zhang, Y.; et al. Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 2019, 179, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Maruyama, F.; Fujisawa, T.; Togashi, T.; Yamamoto, N.; Seo, M.; Sato, S.; Yamada, T.; Mori, H.; Tajima, N.; et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014, 5, 3978. [Google Scholar] [CrossRef] [PubMed]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Philippe, F.; Pelloux, P.; Rayon, C. Plant pectin acetylesterase structure and function: New insights from bioinformatic analysis. BMC Genom. 2017, 18, 456. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, T.W.; Der, J.P.; Honaas, L.A.; de Pamphilis, C.W.; Anderson, C.T. Phylogenetic analysis of pectin-related gene families in Physcomitrella patens and nine other plant species yields evolutionary insights into cell walls. BMC Plant Biol. 2014, 14, 79. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Braybrook, S.; Höfte, H. Cell wall mechanics and growth control in plants: The role of pectins revisited. Front. Plant Sci. 2012, 3, 121. [Google Scholar] [CrossRef] [PubMed]
- Domozych, D.S. Penium margaritaceum: A unicellular model organism for studying plant cell wall architecture and dynamics. Plants 2014, 3, 543–558. [Google Scholar] [CrossRef]
- Domozych, D.S.; Sørensen, I.; Popper, Z.A.; Ochs, J.; Andreas, A.; Fangel, J.U.; Pielach, A.; Sachs, C.; Brechka, H.; Ruisi-Besares, P.; et al. Pectin metabolism and assembly in the cell wall of the charophyte green alga Penium margaritaceum. Plant Physiol. 2014, 165, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Sørensen, I.; Sun, X.; Sun, H.; Behar, H.; Alseekh, S.; Philippe, G.; Lopez, K.P.; Sun, L.; Reed, R.; et al. The Penium margaritaceum genome: Hallmarks of the origins of land plants. Cell 2020, 181, 1097–1111. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Sakayama, H.; de Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara genome: Secondary complexity and implications for plant terrestrialization. Cell 2018, 174, 448–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, L.; Li, H.; Sahu, S.K.; Wang, H.; Xu, Y.; Xian, W.; Song, B.; Liang, H.; Cheng, S.; et al. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants 2020, 6, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Mouille, G.; Pelloux, J. Homogalacturonan methyl-esterification and plant development. Mol. Plant 2009, 2, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Braybrook, S.A.; Le Guillou, L.; Bron, E.; Kuhlemeier, C.; Höfte, H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 2011, 21, 1720–1726. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Greiner, S. Growth control by cell wall pectins. Protoplasma 2012, 249, S169–S175. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.R.; Lou, H.; Aubert, M.K.; Wilkinson, L.G.; Little, A.; Houston, K.; Pinto, S.C.; Shirley, N.J. Exploring the role of cell wall-related genes and polysaccharides during plant development. Plants 2018, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Huang, J.; Tatsumi, Y.; Abe, M.; Sugano, S.S.; Kojima, M.; Takebayashi, Y.; Kiba, T.; Yokoyama, R.; Nishitani, K.; et al. Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy. Nat. Commun. 2018, 9, 5290. [Google Scholar] [CrossRef] [PubMed]
- Levesque-Tremblay, G.; Pelloux, J.; Braybrook, S.A.; Müller, K. Tuning of pectin methylesterification: Consequences for cell wall biomechanics and development. Planta 2015, 242, 791–811. [Google Scholar] [CrossRef] [PubMed]
- Hongo, S.; Sato, K.; Yokoyama, R.; Nishitani, K. Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell 2012, 24, 2624–2634. [Google Scholar] [CrossRef] [PubMed]
- Negi, J.; Moriwaki, K.; Konishi, M.; Yokoyama, R.; Nakano, T.; Kusumi, K.; Hashimoto-Sugimoto, M.; Schroeder, J.I.; Nishitani, K.; Yanagisawa, S.; et al. A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 2013, 23, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Amsbury, S.; Hunt, L.; Elhaddad, N.; Baillie, A.; Lundgren, M.; Verhertbruggen, Y.; Scheller, H.V.; Knox, J.P.; Fleming, A.J.; Gray, J.E. Stomatal function requires pectin de-methyl-esterification of the guard cell wall. Curr. Biol. 2016, 26, 2899–2906. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.J.; Smith, B.G. Plant cell walls and cell-wall polysaccharides: Structures, properties and uses in food products. Int. J. Food Sci. Technol. 2006, 41, 129–143. [Google Scholar] [CrossRef]
- Lee, C.; Teng, Q.; Huang, W.; Zhong, R.; Ye, Z.-H. The Arabidopsis family GT43 glycosyltransferases form two functionally nonredundant groups essential for the elongation of glucuronoxylan backbone. Plant Physiol. 2010, 153, 526–541. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.M.; Hörnblad, E.; Voxeur, A.; Gerber, L.; Rihouey, C.; Lerouge, P.; Marchant, A. Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveal critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 2010, 153, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Anders, N.; Wilkinson, M.D.; Lovegrove, A.; Freeman, J.; Tryfona, T.; Pellny, T.K.; Weimar, T.; Mortimer, J.C.; Stott, K.; Baker, J.M.; et al. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc. Natl. Acad. Sci. USA 2012, 109, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, J.; Lück, S.; Rajaraman, J.; Douchkov, D.; Shirley, N.J.; Schwerdt, J.G.; Schweizer, P.; Fincher, G.B.; Burton, R.A.; Little, A. Altered Expression of Genes Implicated in Xylan Biosynthesis Affects Penetration Resistance against Powdery Mildew. Front. Plant Sci. 2017, 8, 445. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.C.; Carpita, N.C. Designing the deconstruction of plant cell walls. Curr. Opin. Plant Biol. 2008, 11, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Penning, B.W.; McCann, M.C.; Carpita, N.C. Evolution of the Cell Wall Gene Families of Grasses. Front. Plant Sci. 2019, 10, 1205. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, L.V.; Nazipova, A.R.; Gorshkov, O.V.; Petrova, A.A.; Gorshkova, T.A. Elongating maize root: Zone-specific combinations of polysaccharides from type I and type II primary cell walls. Sci. Rep. 2020, 10, 10956. [Google Scholar] [CrossRef] [PubMed]
- Chiniquy, D.; Sharma, V.; Schultink, A.; Baidoo, E.E.; Rautengarten, C.; Cheng, K.; Carroll, A.; Ulvskov, P.; Harholt, J.; Keasling, J.D.; et al. XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc. Natl. Acad. Sci. USA 2012, 109, 17117–17122. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Teng, Q.; Zhong, R.; Yuan, Y.; Ye, Z.-H. Functional roles of rice glycosyltransferase family GT43 in xylan biosynthesis. Plant Signal. Behav. 2014, 9, e27809. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Mizuno, H.; Kawahara, Y.; Wakimoto, H.; Ikawa, H.; Kawahigashi, H.; Kanamori, H.; Matsumoto, T.; Itoh, T.; Gaut, B.S. Retrogenes in rice (Oryza sativa L. ssp. japonica) exhibit correlated expression with their source genes. Genome Biol. Evol. 2011, 3, 1357–1368. [Google Scholar] [CrossRef] [PubMed]
- Bacete, L.; Hamann, T. The role of mechanoperception in plant cell wall integrity maintenance. Plants 2020, 9, 574. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2014, 5, 771. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The plant cell wall: A complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [PubMed]
- Novaković, L.; Guo, T.; Bacic, A.; Sampathkumar, A.; Johnson, K.L. Hitting the wall-sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plants 2018, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Chinnappa, C.C.; Staal, M.; Elzenga, J.T.; Yokoyama, R.; Nishitani, K.; Voesenek, L.A.C.J.; Pieriket, R. Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by XTHs. Plant Physiol. 2010, 154, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Minami, A.; Yano, K.; Gamuyao, R.; Nagai, K.; Kuroha, T.; Ayano, M.; Nakamori, M.; Koike, M.; Kondo, Y.; Niimi, Y.; et al. Time-Course Transcriptomics Analysis Reveals Key Responses of Submerged Deepwater Rice to Flooding. Plant Physiol. 2018, 176, 3081–3102. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Jung, N.U.; Giarola, V.; Bartels, D. The Dynamic Responses of Cell Walls in Resurrection Plants during Dehydration and Rehydration. Front. Plant Sci. 2020, 10, 1698. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.P.; Nguema-Ona, E.E.; Vicre-Gibouin, M.; Sorensen, I.; Willats, W.G.; Driouich, A.; Farrant, J.M. Arabinose-rich polymers as an evolutionary strategy to plasticize resurrection plant cell walls against desiccation. Planta 2013, 237, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Gorka, M.; Erban, A.; Graf, A.; Kopka, J.; Zuther, E.; Hincha, D.K. Both cold and sub-zero acclimation induce cell wall modification and changes in the extracellular proteome in Arabidopsis thaliana. Sci. Rep. 2019, 9, 2289. [Google Scholar] [CrossRef] [PubMed]
- Bellincampi, D.; Cervone, F.; Lionetti, V. Plant cell wall dynamics and wall- related susceptibility in plant-pathogen interactions. Front. Plant Sci. 2014, 5, 228. [Google Scholar] [CrossRef] [PubMed]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed]
- Chialva, M.; Fangel, J.U.; Novero, M.; Zouari, I.; Salvioli di Fossalunga, A.; Willats, W.G.T.; Bonfante, P.; Balestrini, R. Understanding changes in tomato cell walls in roots and fruits: The contribution of arbuscular mucorrhizal colonization. Int. J. Mol. Sci. 2019, 20, 415. [Google Scholar] [CrossRef] [PubMed]
- Quist, C.W.; Smant, G.; Helder, J. Evolution of plant parasitism in the phylum nematoda. Annu. Rev. Phytopathol. 2015, 53, 289–310. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Suzuki, R.; Nakagami, S.; Kuroha, T.; Sakamoto, S.; Nakata, M.T.; Yokoyama, R.; Kimura, S.; Mitsuda, N.; Nishitani, K.; et al. Root-knot nematodes modulate cell walls during root-knot formation in Arabidopsis roots. J. Plant Res. 2020, 133, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, H.R.; Striberny, B.; Olsen, S.; Vidal-Melgosa, S.; Fangel, J.U.; Willats, W.G.; Rose, J.K.; Krause, K. Cell wall composition profiling of parasitic giant dodder (Cuscuta reflexa) and its hosts: A priori differences and induced changes. New Phytol. 2015, 207, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Mutuku, J.M.; Cui, S.; Hori, C.; Takeda, Y.; Tobimatsu, Y.; Nakabayashi, R.; Mori, T.; Saito, K.; Demura, T.; Umezawa, T.; et al. The structural integrity of lignin is crucial for resistance against Striga hermonthica parasitism in rice. Plant Physiol. 2019, 179, 1796–1809. [Google Scholar] [CrossRef] [PubMed]
- Olsen, S.; Striberny, B.; Hollmann, J.; Schwacke, R.; Popper, Z.; Krause, K. Getting ready for host invasion: Elevated expression and action of xyloglucan endotransglucosylases/hydrolases in developing haustoria of the holoparasitic angiosperm Cuscuta. J. Exp. Bot. 2015, 67, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, A.; Bera, S.; Fujiwara, D.; Obayashi, T.; Yokoyama, R.; Nishitani, K.; Aoki, K. Arabinogalactan Proteins Accumulate in the Cell Walls of Searching Hyphae of the Stem Parasitic Plants, Cuscuta campestris and Cuscuta japonica. Plant Cell Physiol. 2017, 58, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.K.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomencalture. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, J.M.; Brumer, H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.C.; Smith, R.C.; Renwick, K.F.; Martin, D.J.; Hodge, S.K.; Matthews, K.J. Xyloglucan endotransglycosylase a new wall-loosening enzyme activity from plants. Biochem. J. 1992, 282, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, K.; Tominaga, T. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J. Biol. Chem. 1992, 267, 21058–21064. [Google Scholar] [PubMed]
- Farkaš, V.; Sulová, Z.; Stratilova, E.; Hanna, R.; Maclachlan, G. Cleavage of xyloglucan by nasturtium seed xyloglucanase and transglycosylation to xyloglucan subunit oligosaccharides. Arch. Biochem. Biophys. 1992, 298, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; Farkaš, V.; Lahnstein, J.; Fincher, G.B. A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-β-d-glucans. J. Biol. Chem. 2007, 282, 12951–12962. [Google Scholar] [CrossRef] [PubMed]
- Simmons, T.J.; Mohler, K.E.; Holland, C.; Goubet, F.; Franková, L.; Houston, D.R.; Hudson, A.D.; Meulewaeter, F.; Fry, S.C. Hetero-trans-β-glucanase, an enzyme unique to Equisetum plants, functionalizes cellulose. Plant J. 2015, 83, 753–769. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, N.; Sunagawa, N.; Tamura, S.; Yokoyama, R.; Ueda, M.; Igarashi, K.; Nishitani, K. The plant cell-wall enzyme AtXTH3 catalyses covalent cross-linking between cellulose and cellooligosaccharide. Sci. Rep. 2017, 7, 46099. [Google Scholar] [CrossRef] [PubMed]
- Viborg, A.H.; Terrapon, N.; Lombard, V.; Michel, G.; Czjzek, M.; Henrissat, B.; Brumer, H. A subfamily roadmap for functional glycogenomics of the evolutionarily diverse glycoside hydrolase family 16 (GH16). J. Biol. Chem. 2019, 294, 15973–15986. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Nishitani, K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2001, 42, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Rose, J.K.C.; Nishitani, K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 2004, 134, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Uwagaki, Y.; Sasaki, H.; Harada, T.; Hiwatashi, Y.; Hasebe, M.; Nishitani, K. Biological implications of the occurrence of 32 members of the XTH (xyloglucan endotransglucosylase/hydrolase) family of proteins in the bryophyte Physcomitrella patens. Plant J. 2010, 64, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Tatematsu, K.; Hanada, K.; Duermeyer, L.; Okamoto, M.; Yonekura-Sakakibara, K.; Saito, K.; Toyoda, T.; Kawakami, N.; Kamiya, Y.; et al. Tissue-specific transcriptome analysis reveals cell wall metabolism, flavonol biosynthesis and defense responses are activated in the endosperm of germinating Arabidopsis thaliana seeds. Plant Cell Physiol. 2012, 53, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Vissenberg, K.; Oyama, M.; Osato, Y.; Yokoyama, R.; Verbelen, J.P.; Nishitani, K. Differential expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 genes in Arabidopsis roots. Physiological roles in specification in cell wall construction. Plant Cell Physiol. 2005, 46, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Osato, Y.; Yokoyama, R.; Nishitani, K. A principal role for AtXTH18 in Arabidopsis thaliana root growth: A functional analysis using RNAi plants. J. Plant Res. 2006, 119, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Yokoyama, R.; Seki, M.; Ito, T.; Shinozaki, K.; Takahashi, T.; Komeda, Y.; Nishitani, K. AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J. 2005, 42, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H.; Braam, J.; Jiang, T.; Xu, X.Y.; Mao, C.Z.; et al. XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef] [PubMed]

| Family | Subfamily | Substrate/Product 1 | Description 1 |
|---|---|---|---|
| GT2 | |||
| CesA | Cellulose | Cellulose synthase | |
| CslA, D | Mannan | Mannan synthase | |
| CslC | Xyloglucan | Xyloglucan synthase | |
| CslF, H, J | (1,3;1,4)-β-d-glucan | (1,3;1,4)-β-d-glucan synthase | |
| GT8 | |||
| GT8A | Glucuronoxylan | Glucuronoxylan glucuronosyltransferase | |
| GT8C | Xylan | Xylan primary oligopolysaccharide synthase | |
| GT8D | Xylan | Xylan primary oligopolysaccharide synthase | |
| Xylan galacturonosyltransferas | |||
| HG 2 | HG galacturonosyltransferase | ||
| GT10 | Glycoprotein α-1,3-fucosyltransferase | ||
| GT14 | AGP 3 | AGP glucuronosyltransferase | |
| GT29 | AGP | AGP galactosyltransferase | |
| GT31 | AGP | AGP galactosyltransferase | |
| GT34 | Xyloglucan | Xyloglucan α-1,6-xylosyltransferase | |
| GT37 | Xyloglucan | Xyloglucan α-1,2-fucosyltransferase | |
| AGP | AGP α-1,2-fucosyltransferase | ||
| GT43 | Xylan | Xylan xylosyltransfearse | |
| GT47 | |||
| GT47A | Xyloglucan | Xyloglucan β-1,2-galactosyltransferase | |
| GT47B | RGI 4 | RGI arabinosyltransferase | |
| GT47C | Xylogalacturonan | Xylogalacturonan β-1,3-xylosyltransferase | |
| GT47E | Xylan | Xylan xylosyltransfearse | |
| Xylan | Xylan primary oligopolysaccharide synthase | ||
| GT48 | Callose | Callose synthase | |
| GT61 | Arabinoxylan | Arabinoxylan α-1,3-arabinosyltransferase | |
| GT64 | |||
| GT68 | |||
| GT77 | RGII 5 | RGII α-1,3-d-xylosyltransferase | |
| AGP | Arabinofuranosyltransferase | ||
| GT92 | RGI | RGI galactosyltransferase | |
| GH1 | Mannan | Exo-β-1,4-mannosidase | |
| GH3 | β-glucosidase/xylosidase | ||
| GH5 | Mannan | Endo-β-mannanase | |
| GH9 | Cellulose | β-1,4-glucanase | |
| GH10 | Xylan | Endo-β-xylanase | |
| GH16 | Xyloglucan | Xyloglucan endotransglucosylase/hydrolase | |
| GH17 | Callose | β-1,3-glucanase | |
| GH18 | |||
| GH19 | Cellulose | ||
| GH28 | HG | Polygalacturonase | |
| GH31 | Xyloglucan | Xyloglucan α-1,6-xylosidase | |
| GH35 | Xyloglucan | Xyloglucan β-1,2-galactosidase | |
| GH38 | |||
| GH51 | Arabinan | Bifunctionalα-L-arabinofuranosidase/β-d-xylosidase | |
| GH95 | Xyloglucan | Xyloglucan α-1,2-fucosidase | |
| Expansin | |||
| CE8 | HG | Pectin methylesterase | |
| CE13 | RGI | Pectin acetylesterases | |
| PL1 | HG | Pectin Lyases | |
| PL4 | RGI | RGI lyses | |
| PMT | HG | HG methyltransferase |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yokoyama, R. A Genomic Perspective on the Evolutionary Diversity of the Plant Cell Wall. Plants 2020, 9, 1195. https://doi.org/10.3390/plants9091195
Yokoyama R. A Genomic Perspective on the Evolutionary Diversity of the Plant Cell Wall. Plants. 2020; 9(9):1195. https://doi.org/10.3390/plants9091195
Chicago/Turabian StyleYokoyama, Ryusuke. 2020. "A Genomic Perspective on the Evolutionary Diversity of the Plant Cell Wall" Plants 9, no. 9: 1195. https://doi.org/10.3390/plants9091195
APA StyleYokoyama, R. (2020). A Genomic Perspective on the Evolutionary Diversity of the Plant Cell Wall. Plants, 9(9), 1195. https://doi.org/10.3390/plants9091195