Population Structure and Genetic Diversity in Korean Cowpea Germplasm Based on SNP Markers
Abstract
:1. Introduction
2. Results
2.1. SNP Analysis
2.2. Population Structure in the Global Cowpea Germplasm
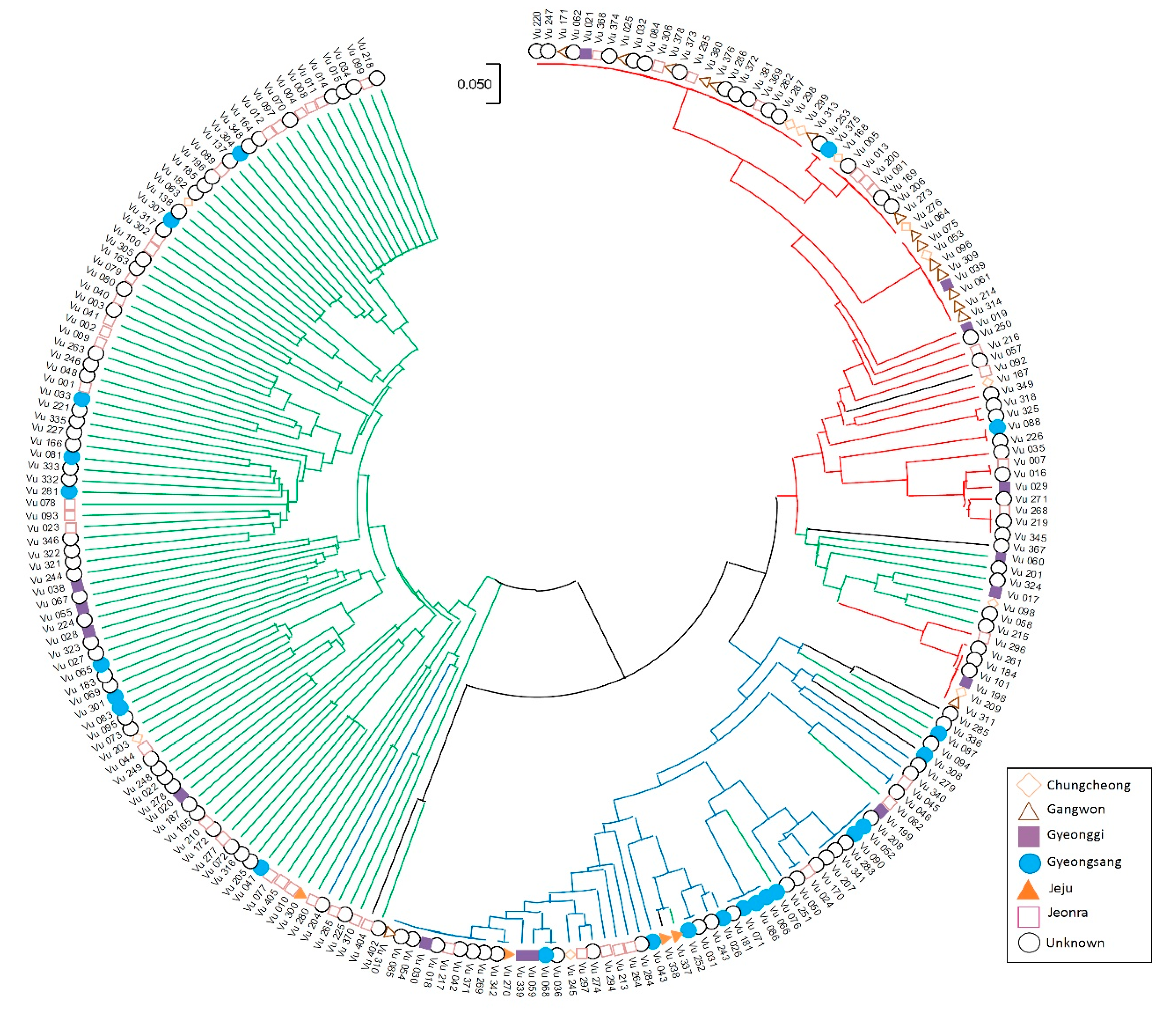
2.3. Population Structure in the Korean Cowpea Accessions
2.4. Analysis of Genetic Diversity
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Phenotypic Evaluation
4.2. DNA Isolation and SNP Genotyping
4.3. Filtering SNP Data
4.4. Population Structure Analysis
4.5. Principal Component Analysis (PCA) and Phylogeny Tree Analysis
4.6. Genetic Diversity Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Coulibaly, S.; Pasquet, R.S.; Papa, R.; Gepts, P. AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theor. Appl. Genet. 2002, 104, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Boukar, O.; Belko, N.; Chamarthi, S.; Togola, A.; Batieno, J.; Owusu, E.; Haruna, M.; Diallo, S.; Umar, M.L.; Olufajo, O.; et al. Cowpea (Vigna unguiculata): Genetics, genomics and breeding. Plant Breed. 2019, 138, 415–424. [Google Scholar] [CrossRef] [Green Version]
- Boukar, O.; Fatokun, C.A.; Huynh, B.-L.; Roberts, P.A.; Close, T.J. Genomic Tools in Cowpea Breeding Programs: Status and Perspectives. Front. Plant Sci. 2016, 7, 757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.; Ajeigbe, H.A.; Tarawali, S.A.; Fernandez-Rivera, S.; Abubakar, M. Improving the production and utilization of cowpea as food and fodder. Field Crop. Res. 2003, 84, 169–177. [Google Scholar] [CrossRef]
- Xiong, H.; Shi, A.; Mou, B.; Qin, J.; Motes, D.; Lu, W.; Ma, J.; Weng, Y.; Yang, W.; Wu, D. Genetic diversity and population structure of cowpea (Vigna unguiculata L. Walp). PLoS ONE 2016, 11, e0160941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehlers, J.; Hall, A. Cowpea (Vigna unguiculata L. Walp.). Field Crop. Res. 1997, 53, 187–204. [Google Scholar] [CrossRef]
- Elowad, H.O.; Hall, A.E. Influences of early and late nitrogen fertilization on yield and nitrogen fixation of cowpea under well-watered and dry field conditions. Field Crop. Res. 1987, 15, 229–244. [Google Scholar] [CrossRef]
- Kouam, E.B.; Pasquet, R.S.; Campagne, P.; Tignegre, J.-B.; Thoen, K.; Gaudin, R.; Ouedraogo, J.T.; Salifu, A.B.; Muluvi, G.M.; Gepts, P. Genetic structure and mating system of wild cowpea populations in West Africa. BMC Plant Biol. 2012, 12, 113. [Google Scholar] [CrossRef] [Green Version]
- Pasquet, R.S. Cultivated cowpea (Vigna unguiculata): Genetic organization and domestication. In Advances in Legume systematics 8: Legumes of Economic Importance; Pickersgill, B., Lock, J.M., Eds.; Royal Botanic Gardens, Kew.: London, UK, 1996; pp. 101–108. [Google Scholar]
- Pasquet, R.S. Morphological study of cultivated cowpea Vigna unguiculata (L.) Walp. Importance of ovule number and definition of cv gr Melanophthalmus. Agronomie 1998, 18, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Xu, P. The Genetic Driving Force for the Grain-Vegetable Cowpea Diversification: A Focus on the Pod Length. J. Cell Signal. 2017, 2, 1–2. [Google Scholar] [CrossRef]
- Xu, P.; Wu, X.; Muñoz-Amatriaín, M.; Wang, B.; Wu, X.; Hu, Y.; Huynh, B.L.; Close, T.J.; Roberts, P.A.; Zhou, W. Genomic regions, cellular components and gene regulatory basis underlying pod length variations in cowpea (V. unguiculata L. Walp). Plant Biotechnol. J. 2017, 15, 547–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muruli, B.; Pathak, R.; Mukunya, D.; Karel, A.; Keya, S.; Ssali, H. Cowpea research in Kenya. Trop. Grain Legume Bull. 1980, 19, 13–16. [Google Scholar]
- Padulosi, S.; Ng, N. Origin, taxonomy, and morphology of Vigna unguiculata (L.) Walp. In Advances in Cowpea Research; Singh, B.B., Mohan Raj, D.R., Dashiell, K.E., Jackai, L.E.N., Eds.; Copublication Intl Inst Tropical Agric (IITA): Ibadan, Nigeria; Japan Intl Res Center Agric Sci (JIRCAS): Ibaraki, Japan; Sayce: Devon, UK, 1997; pp. 1–12. [Google Scholar]
- Singh, B. Recent genetic studies in cowpea. In Challenges and Opportunities for Enhancing Sustainable Cowpea Production; Fatokun, C.A., Tarawali, S.A., Singh, B.B., Singh, B., Kormawa, P.M., Eds.; Intl Inst Tropical Agric (IITA): Ibadan, Nigeria, 2002; pp. 3–13. [Google Scholar]
- Egbadzor, K.F.; Ofori, K.; Yeboah, M.; Aboagye, L.M.; Opoku-Agyeman, M.O.; Danquah, E.Y.; Offei, S.K. Diversity in 113 cowpea [Vigna unguiculata (L.) Walp] accessions assessed with 458 SNP markers. SpringerPlus 2014, 3, 541. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, M.; Lino-Neto, T.; Rosa, E.; Carnide, V. Cowpea: A legume crop for a challenging environment. J. Sci. Food Agric. 2017, 97, 4273–4284. [Google Scholar] [CrossRef]
- Carvalho, M.; Muñoz-Amatriaín, M.; Castro, I.; Lino-Neto, T.; Matos, M.; Egea-Cortines, M.; Rosa, E.; Close, T.; Carnide, V. Genetic diversity and structure of Iberian Peninsula cowpeas compared to world-wide cowpea accessions using high density SNP markers. BMC Genom. 2017, 18, 891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynh, B.L.; Close, T.J.; Roberts, P.A.; Hu, Z.; Wanamaker, S.; Lucas, M.R.; Chiulele, R.; Cissé, N.; David, A.; Hearne, S. Gene pools and the genetic architecture of domesticated cowpea. Plant Genome 2013, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ganal, M.W.; Altmann, T.; Röder, M.S. SNP identification in crop plants. Curr. Opin. Plant Biol. 2009, 12, 211–217. [Google Scholar] [CrossRef]
- Asare, A.T.; Gowda, B.S.; Galyuon, I.K.; Aboagye, L.L.; Takrama, J.F.; Timko, M.P. Assessment of the genetic diversity in cowpea (Vigna unguiculata L. Walp.) germplasm from Ghana using simple sequence repeat markers. Plant Genet. Resour. 2010, 8, 142–150. [Google Scholar] [CrossRef]
- Deulvot, C.; Charrel, H.; Marty, A.; Jacquin, F.; Donnadieu, C.; Lejeune-Hénaut, I.; Burstin, J.; Aubert, G. Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genom. 2010, 11, 468. [Google Scholar] [CrossRef] [Green Version]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Int. J. Plant Genom. 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munoz-Amatriain, M.; Mirebrahim, H.; Xu, P.; Wanamaker, S.I.; Luo, M.; Alhakami, H.; Alpert, M.; Atokple, I.; Batieno, B.J.; Boukar, O.; et al. Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J. 2017, 89, 1042–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, S.; Muñoz-Amatriaín, M.; Boukar, O.; Herniter, I.; Cisse, N.; Guo, Y.-N.; Roberts, P.A.; Xu, S.; Fatokun, C.; Close, T.J. Identification of QTL controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattarai, G.; Shi, A.; Qin, J.; Weng, Y.; Morris, J.B.; Pinnow, D.L.; Buckley, B.; Ravelombola, W.; Yang, W.; Dong, L. Association analysis of cowpea mosaic virus (CPMV) resistance in the USDA cowpea germplasm collection. Euphytica 2017, 213, 230. [Google Scholar] [CrossRef]
- Ravelombola, W.; Qin, J.; Shi, A.; Lu, W.; Weng, Y.; Xiong, H.; Yang, W.; Bhattarai, G.; Mahamane, S.; Payne, W.A. Association mapping revealed SNP markers for adaptation to low phosphorus conditions and rock phosphate response in USDA cowpea (Vigna unguiculata (L.) Walp.) germplasm. Euphytica 2017, 213, 183. [Google Scholar] [CrossRef]
- Pan, L.; Wang, N.; Wu, Z.; Guo, R.; Yu, X.; Zheng, Y.; Xia, Q.; Gui, S.; Chen, C. A high density genetic map derived from RAD sequencing and its application in QTL analysis of yield-related traits in Vigna unguiculata. Front. Plant Sci. 2017, 8, 1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Baek, H.-J.; Yoon, M.-S.; Park, S.-K.; Cho, Y.-H.; Kim, C.-Y. Analysis of genetic diversity of cowpea landraces from Korea determined by Simple Sequence Repeats and establishment of a core collection. Korean J. Breed. Sci. 2009, 41, 369–376. [Google Scholar]
- Kim, J.; Ko, M.; Chang, K. Studies on Genetic Analysis by the Diallel Crosses in F2 Generation of Cowpea (Vigna sinensis savi.). Korean J. Crop. Sci. 1983, 28, 216–226. [Google Scholar]
- Lee, J.; Baek, H.; Yoon, M.; Kim, C.; Cho, E. Taxonomic review and genetic diversity of cowpea species and related taxa. Korean J. Breed. 2005, 37, 187–191. [Google Scholar]
- Hegde, V.; Mishra, S. Landraces of cowpea, Vignaunguiculata (L.) Walp., as potential sources of genes for unique characters in breeding. Genet. Resour. Crop Evol. 2009, 56, 615–627. [Google Scholar] [CrossRef]
- Fang, J.; Chao, C.-C.T.; Roberts, P.A.; Ehlers, J.D. Genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] in four West African and USA breeding programs as determined by AFLP analysis. Genet. Resour. Crop Evol. 2007, 54, 1197–1209. [Google Scholar] [CrossRef]
- Gwag, J.-G.; Dixit, A.; Park, Y.-J.; Ma, K.-H.; Kwon, S.-J.; Cho, G.-T.; Lee, G.-A.; Lee, S.-Y.; Kang, H.-K.; Lee, S.-H. Assessment of genetic diversity and population structure in mungbean. Genes Genom. 2010, 32, 299–308. [Google Scholar] [CrossRef]
- Wu, X.; Li, Y.; Li, X.; Li, C.; Shi, Y.; Song, Y.; Zheng, Z.; Li, Y.; Wang, T. Analysis of genetic differentiation and genomic variation to reveal potential regions of importance during maize improvement. BMC Plant Biol. 2015, 15, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fatokun, C.; Girma, G.; Abberton, M.; Gedil, M.; Unachukwu, N.; Oyatomi, O.; Yusuf, M.; Rabbi, I.; Boukar, O. Genetic diversity and population structure of a mini-core subset from the world cowpea (Vigna unguiculata (L.) Walp.) germplasm collection. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- AbdelGawwad, M.R.; Marić, A.; Al-Ghamdi, A.A.; Hatamleh, A.A. Interactome Analysis and Docking Sites of MutS Homologs Reveal New Physiological Roles in Arabidopsis thaliana. Molecules 2019, 24, 2493. [Google Scholar] [CrossRef] [Green Version]
- Langyintuo, A.; Lowenberg-DeBoer, J.; Faye, M.; Lambert, D.; Ibro, G.; Moussa, B.; Kergna, A.; Kushwaha, S.; Musa, S.; Ntoukam, G. Cowpea supply and demand in West and Central Africa. Field Crop. Res. 2003, 82, 215–231. [Google Scholar] [CrossRef]
- Hellens, R.P.; Moreau, C.; Lin-Wang, K.; Schwinn, K.E.; Thomson, S.J.; Fiers, M.W.E.J.; Frew, T.J.; Murray, S.R.; Hofer, J.M.I.; Jacobs, J.M.E.; et al. Identification of Mendel’s White Flower Character. PLoS ONE 2010, 5, e13230. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Wu, X.; Wang, B.; Luo, J.; Liu, Y.; Ehlers, J.; Close, T.; Roberts, P.; Lu, Z.; Wang, S. Genome wide linkage disequilibrium in Chinese asparagus bean (Vigna. unguiculata ssp. sesquipedialis) germplasm: Implications for domestication history and genome wide association studies. Heredity 2012, 109, 34–40. [Google Scholar]
- Keim, P.; Olson, T.C.; Shoemaker, R.C. A rapid protocol for isolating soybean DNA. Soybean Genet. Newsl. 1988, 15, 150–154. [Google Scholar]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.-C.; et al. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Goudet, J. Hierfstat, a package for r to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef] [Green Version]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar]
- Dray, S.; Dufour, A.-B. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef] [Green Version]
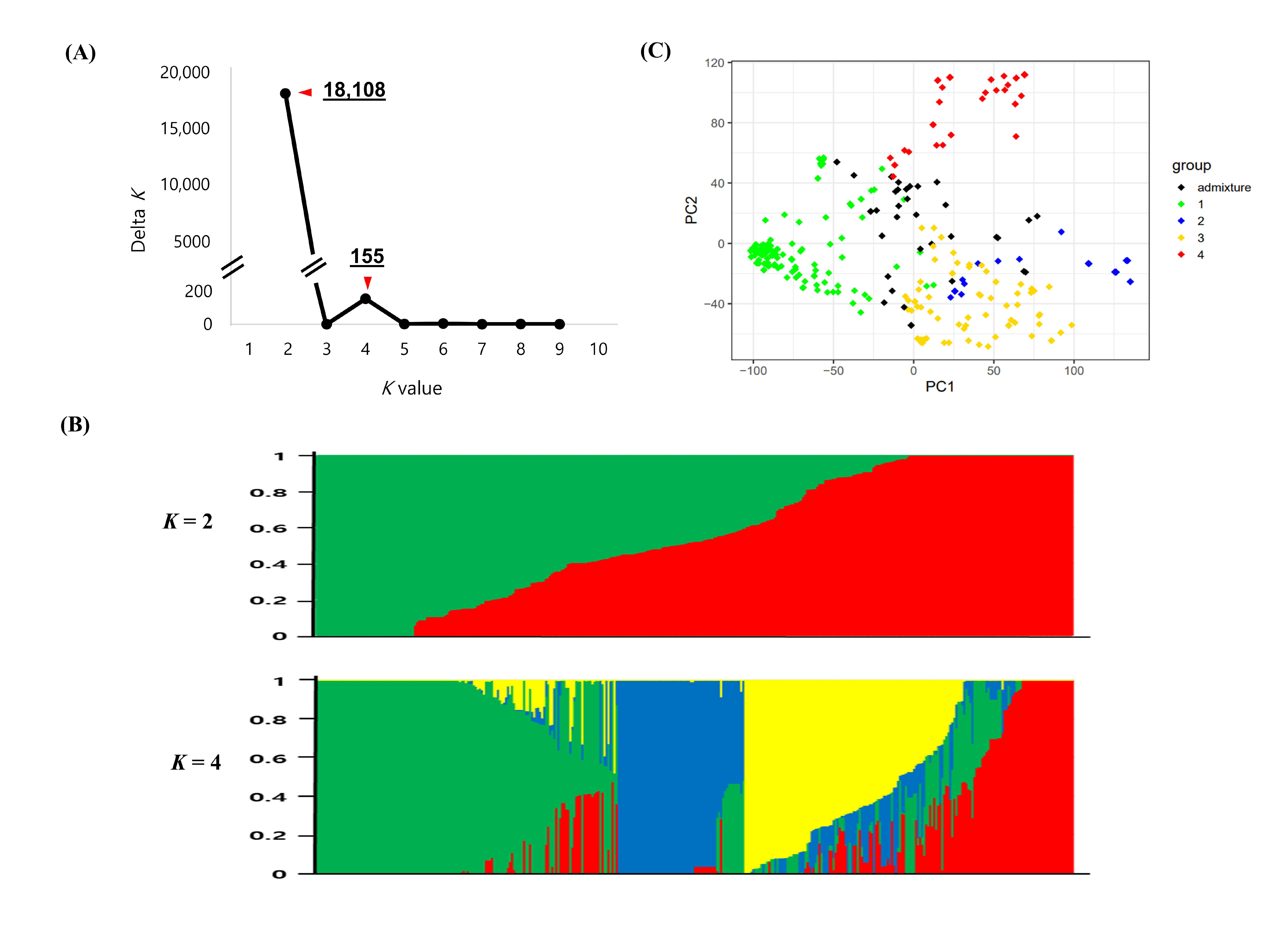
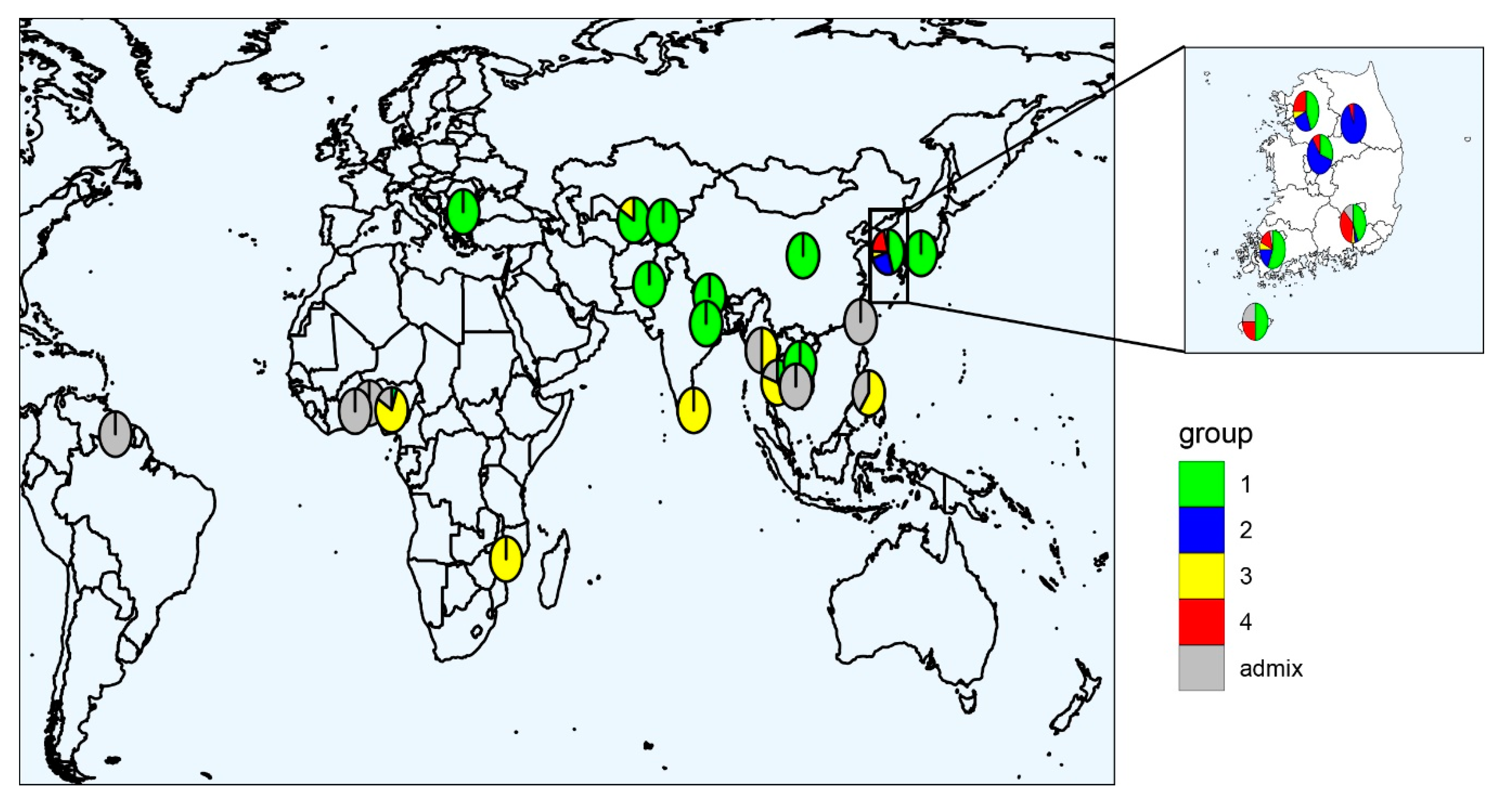
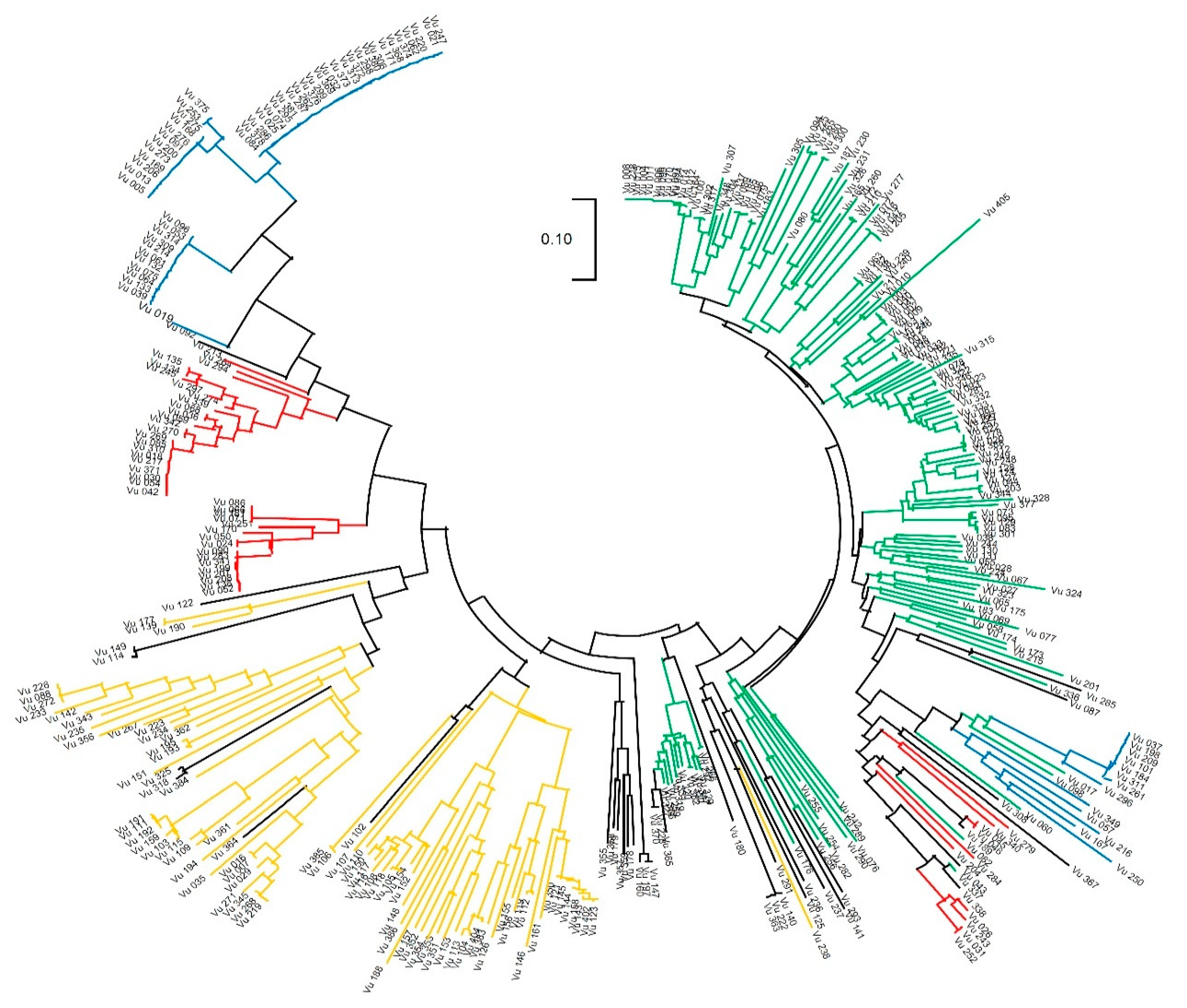
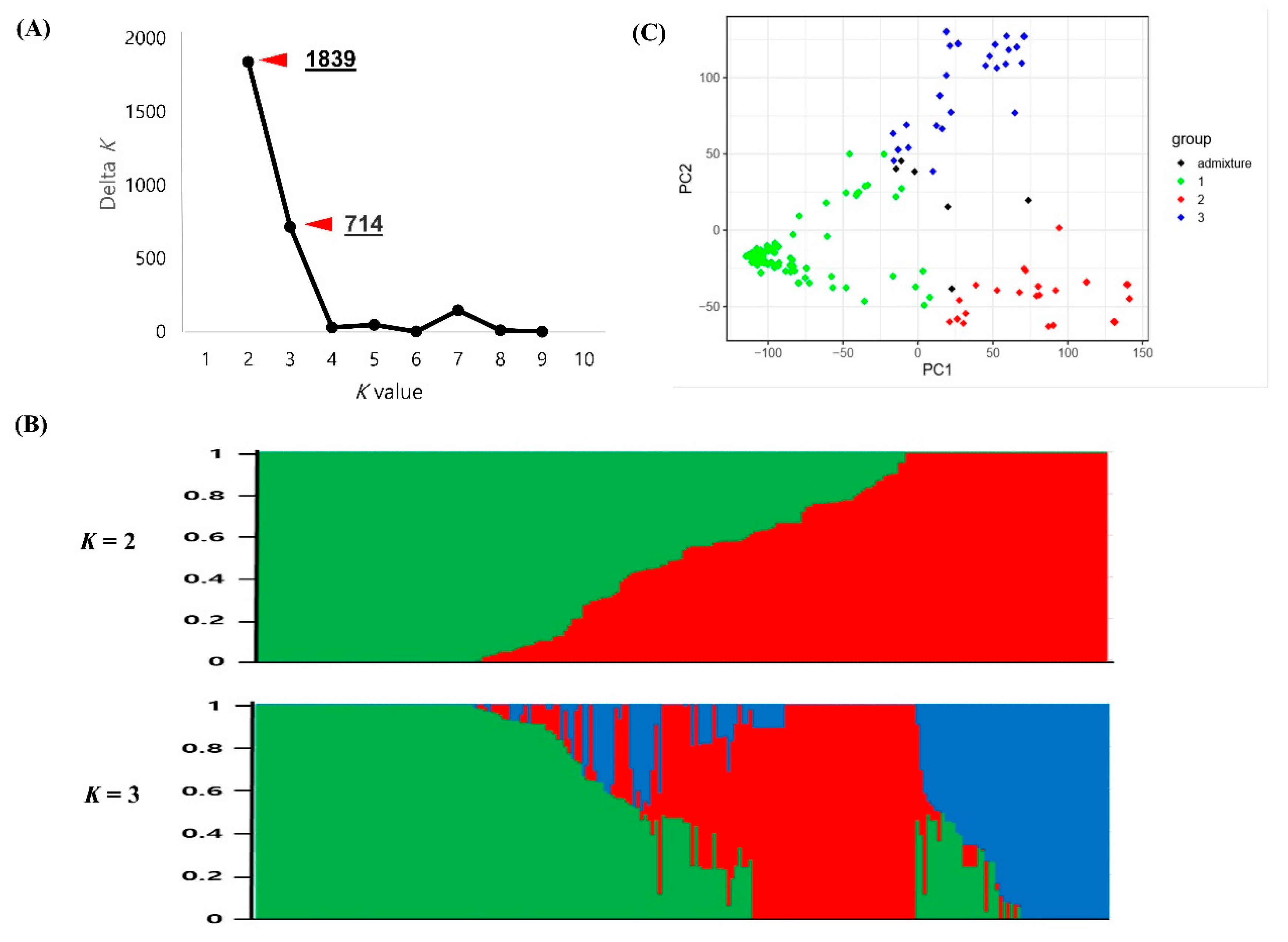
| MAF | PIC | Ho | He | Fis | |
|---|---|---|---|---|---|
| Global Accessions | |||||
| Min | 0.044 | 0.080 | 0.000 | 0.045 | –0.091 |
| Median | 0.278 | 0.321 | 0.000 | 0.257 | 1.000 |
| Max | 0.500 | 0.375 | 0.113 | 0.502 | 1.000 |
| Mean | 0.269 | 0.287 | 0.002 | 0.254 | 0.993 |
| KoreanAccessions | |||||
| Min | 0.000 | 0.000 | 0.000 | 0.000 | –0.045 |
| Median | 0.263 | 0.313 | 0.000 | 0.246 | 1.000 |
| Max | 0.500 | 0.375 | 0.138 | 0.521 | 1.000 |
| Mean | 0.254 | 0.265 | 0.003 | 0.234 | 0.988 |
| Category | Ho | He | Fis | MAF | PIC |
|---|---|---|---|---|---|
| Groups Based on Global Accessions | |||||
| 1 | 0.0016 | 0.2504 | 0.9889 | 0.1797 | 0.2027 |
| 2 | 0.0005 | 0.1421 | 0.9920 | 0.0904 | 0.1183 |
| 3 | 0.0018 | 0.3278 | 0.9928 | 0.2443 | 0.2580 |
| 4 | 0.0005 | 0.1895 | 0.9951 | 0.1328 | 0.1523 |
| Groups Based on Korean Accessions | |||||
| 1 | 0.0006 | 0.1888 | 0.9945 | 0.1355 | 0.1570 |
| 2 | 0.0017 | 0.2395 | 0.9887 | 0.1837 | 0.1939 |
| 3 | 0.0005 | 0.1903 | 0.9951 | 0.1460 | 0.1527 |
| Df | SS | MS | Sigma † | Var. % | Phi ‡ | p-Value | |
|---|---|---|---|---|---|---|---|
| Global Accessions | |||||||
| 19Between pop | 3 | 1,435,162 | 478,387.3 | 5965.9055 | 41.5 | 0.4159 | <0.001 |
| Between samples | 335 | 2,806,352 | 8377.171 | 8377.1712 | 58.5 | ||
| Total | 338 | 4,241,514 | 12,548.86 | 14,343.0768 | 100.0 | ||
| Korean Accessions | |||||||
| Between pop | 2 | 955,123.1 | 477,561.535 | 6761.786 | 47.5 | 0.4747 | <0.001 |
| Between samples | 218 | 1,630,585.8 | 7479.751 | 7479.751 | 52.5 | ||
| Total | 220 | 2,585,708.9 | 11,753.222 | 14,241.537 | 100.0 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, E.; Kim, K.; Jun, T.-H.; Choi, J.; Kim, S.-H.; Muñoz-Amatriaín, M.; Sun, H.; Ha, B.-K. Population Structure and Genetic Diversity in Korean Cowpea Germplasm Based on SNP Markers. Plants 2020, 9, 1190. https://doi.org/10.3390/plants9091190
Seo E, Kim K, Jun T-H, Choi J, Kim S-H, Muñoz-Amatriaín M, Sun H, Ha B-K. Population Structure and Genetic Diversity in Korean Cowpea Germplasm Based on SNP Markers. Plants. 2020; 9(9):1190. https://doi.org/10.3390/plants9091190
Chicago/Turabian StyleSeo, Eunju, Kipoong Kim, Tae-Hwan Jun, Jinsil Choi, Seong-Hoon Kim, María Muñoz-Amatriaín, Hokeun Sun, and Bo-Keun Ha. 2020. "Population Structure and Genetic Diversity in Korean Cowpea Germplasm Based on SNP Markers" Plants 9, no. 9: 1190. https://doi.org/10.3390/plants9091190
APA StyleSeo, E., Kim, K., Jun, T.-H., Choi, J., Kim, S.-H., Muñoz-Amatriaín, M., Sun, H., & Ha, B.-K. (2020). Population Structure and Genetic Diversity in Korean Cowpea Germplasm Based on SNP Markers. Plants, 9(9), 1190. https://doi.org/10.3390/plants9091190







