F-Box Family Genes, LTSF1 and LTSF2, Regulate Low-Temperature Stress Tolerance in Pepper (Capsicum chinense)
Abstract
:1. Introduction
2. Results
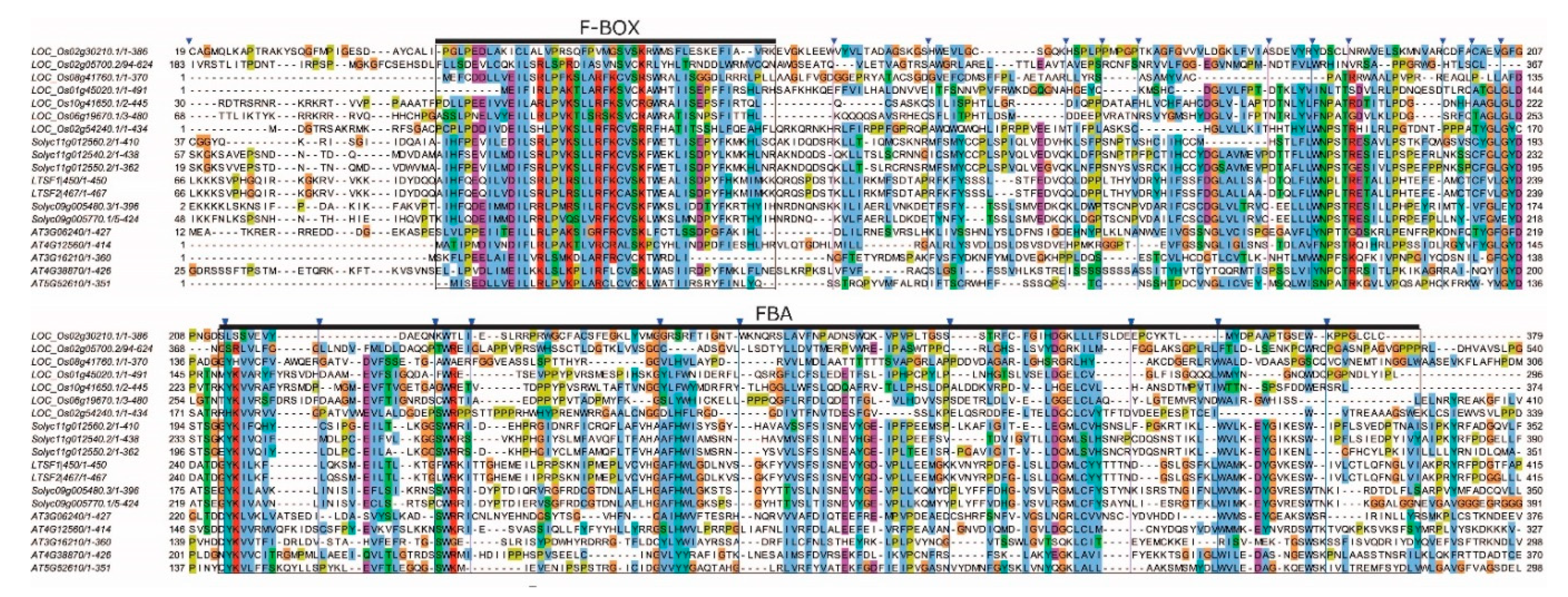
2.1. LTSF1 and LTSF2 Sequence Analysis
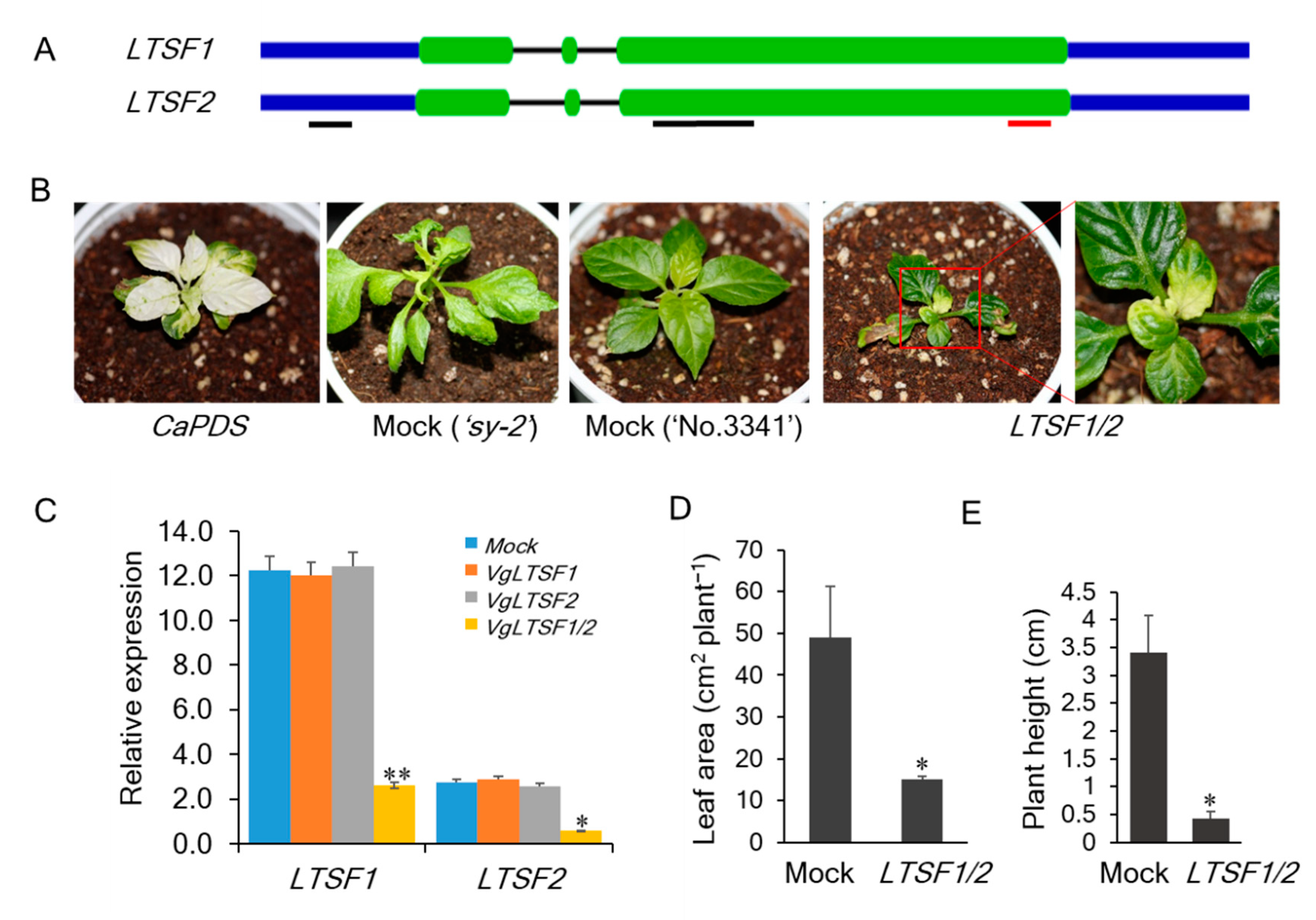
2.2. VIGS Analysis
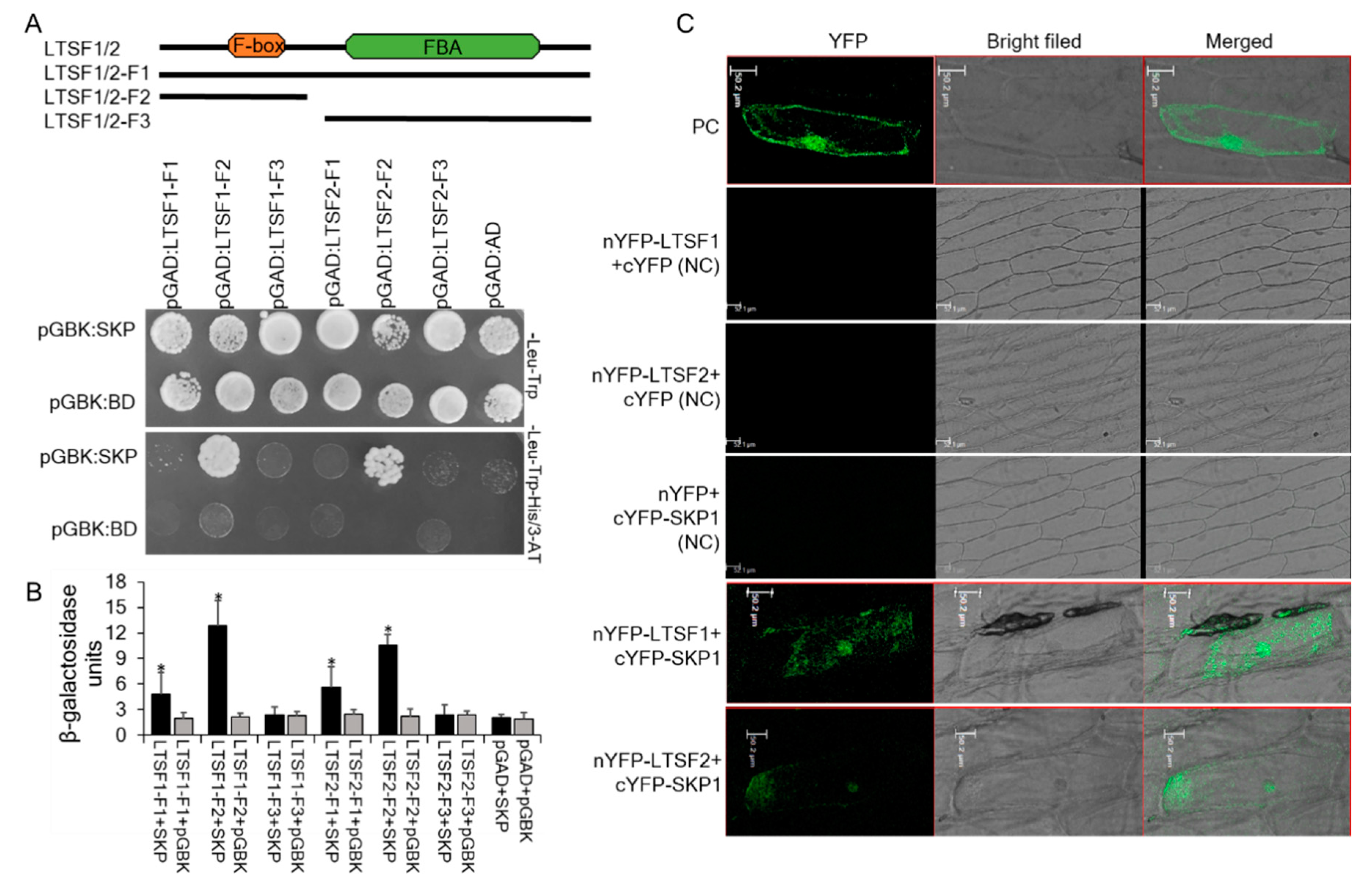
2.3. LTSF1 and LTSF2 Interaction with SKP1
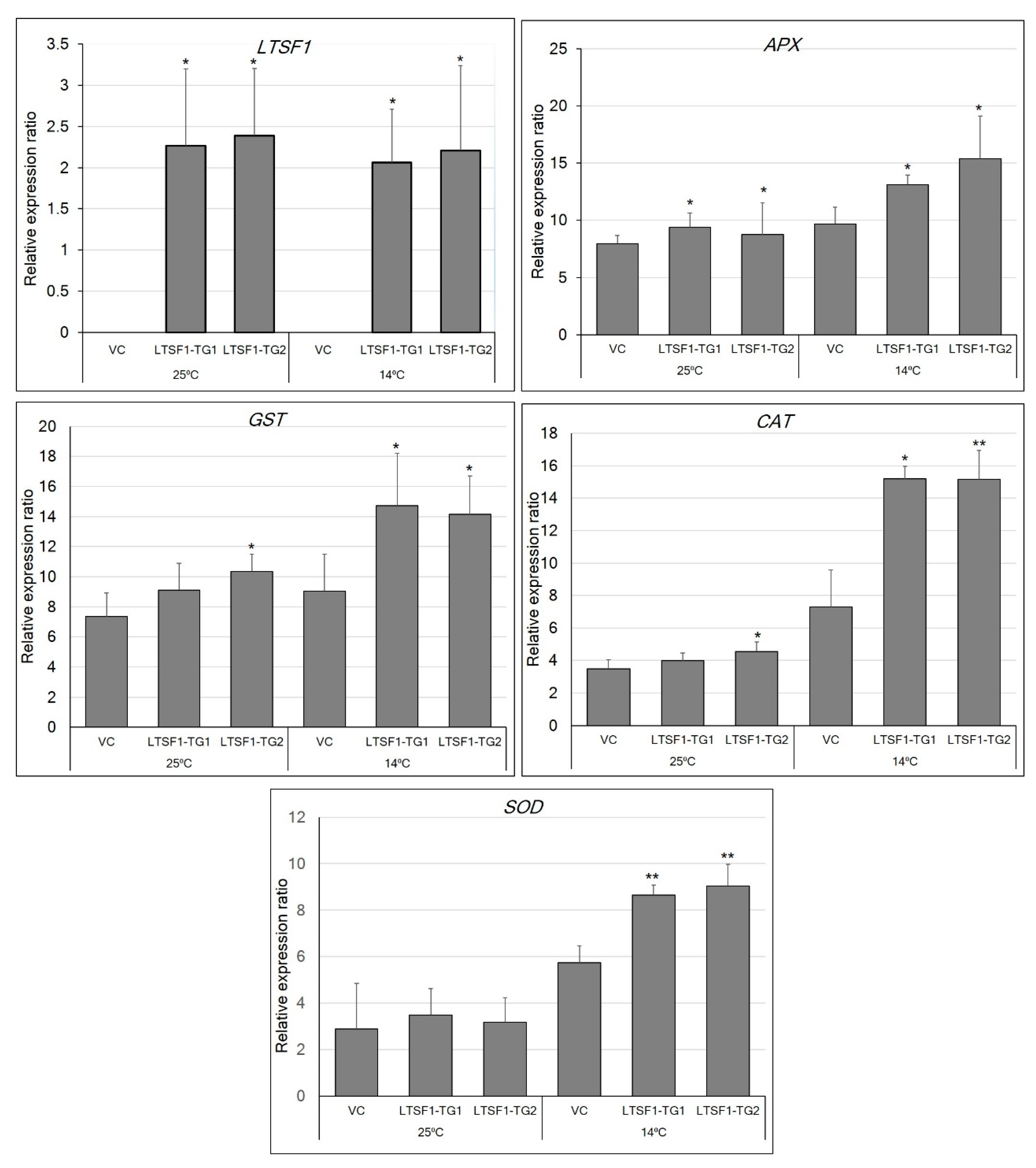
2.4. The Transgenic Expression of LTSF1 Promotes Low-Temperature Stress Tolerance in N. benthamiana Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Phylogenetic Analysis and Identification of Conserved Motifs
4.3. VIGS
4.4. Subcellular Localization of the LTSF1 and LTSF2 Proteins
4.5. Y2H Assays
4.6. Bimolecular Fluorescence Complementation (BiFC) Analysis
4.7. Genomic DNA Extraction
4.8. Agrobacterium-Mediated Transformation of N. benthamiana
4.9. Low-Temperature Stress Treatments
4.10. Total RNA Extraction and Reverse-Transcription Quantitative PCR (RT-qPCR)
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chinnusamy, V.; Zhu, J.K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 2010, 639, 39–55. [Google Scholar] [PubMed] [Green Version]
- Venkatesh, J.; Kang, B.C. Current views on temperature-modulated R gene-mediated plant defense responses and tradeoffs between plant growth and immunity. Curr. Opin. Plant Biol. 2019, 50, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Korres, N.E.; Norsworthy, J.K.; Tehranchian, P.; Gitsopoulos, T.K.; Loka, D.A.; Oosterhuis, D.M.; Gealy, D.R.; Moss, S.R.; Burgos, N.R.; Miller, M.R.; et al. Cultivars to face climate change effects on crops and weeds: A review. Agr. Sust. Devel. 2016, 36, 12. [Google Scholar] [CrossRef] [Green Version]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Heerden, P.D.; Strasser, R.J.; Kruger, G.H. Reduction of dark chilling stress in N-fixing soybean by nitrate as indicated by chlorophyll a fluorescence kinetics. Physiol. Plant. 2004, 121, 239–249. [Google Scholar] [CrossRef]
- An, S.J.; Pandeya, D.; Park, S.W.; Li, J.J.; Kwon, J.K.; Koeda, S.; Hosokawa, M.; Paek, N.C.; Choi, D.; Kang, B.C. Characterization and genetic analysis of a low-temperature-sensitive mutant, sy-2, in Capsicum chinense. Theor. Appl. Genet. 2011, 122, 459–470. [Google Scholar] [CrossRef]
- Koeda, S.; Hosokawa, M.; Kang, B.C.; Tanaka, C.; Choi, D.; Sano, S.; Shiina, T.; Doi, M.; Yazawa, S. Defense response of a pepper cultivar cv. Sy-2 is induced at temperatures below 24 °C. J. Plant Res. 2012, 125, 137–145. [Google Scholar]
- Koeda, S.; Hosokawa, M.; Saito, H.; Doi, M. Temperature-sensitive phenotype caused by natural mutation in Capsicum latescent in two tropical regions. J. Plant Res. 2013, 126, 675–684. [Google Scholar] [CrossRef]
- Almadanim, M.C.; Alexandre, B.M.; Rosa, M.T.; Sapeta, H.; Leitão, A.E.; Ramalho, J.C.; Lam, T.T.; Negrão, S.; Abreu, I.A.; Oliveira, M.M. Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 2017, 40, 1197–1213. [Google Scholar] [CrossRef]
- Domon, J.M.; Baldwin, L.; Acket, S.; Caudeville, E.; Arnoult, S.; Zub, H.; Gillet, F.; Lejeune-Hénaut, I.; Brancourt-Hulmel, M.; Pelloux, J.; et al. Cell wall compositional modifications of Miscanthus ecotypes in response to cold acclimation. Phytochemistry 2013, 85, 51–61. [Google Scholar] [CrossRef]
- Kielbowicz-Matuk, A.; Rey, P.; Rorat, T. The organ-dependent abundance of a Solanum lipid transfer protein is up-regulated upon osmotic constraints and associated with cold acclimation ability. J. Exp. Bot. 2008, 59, 2191–2203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orvar, B.L.; Sangwan, V.; Omann, F.; Dhindsa, R.S. Early steps in cold sensing by plant cells: The role of actin cytoskeleton and membrane fluidity. Plant J. 2000, 23, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Miquel, M.F.; Browse, J.A. High-oleate oilseeds fail to develop at low temperature. Plant Physiol. 1994, 106, 421–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.; Zhang, H. Arabidopsis Fatty Acid Desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef]
- Havaux, M.; Kloppstech, K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta 2001, 213, 953–966. [Google Scholar] [CrossRef]
- Hua, J.; Grisafi, P.; Cheng, S.H.; Fink, G.R. Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 2001, 15, 2263–2272. [Google Scholar] [CrossRef] [Green Version]
- Millerd, A.; McWilliam, J.R. Studies on a maize mutant sensitive to low temperature I. Influence of temperature and light on the production of chloroplast pigments. Plant Physiol. 1968, 43, 1967–1972. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Zhang, Y.; Lv, J.; Zhang, J.; Li, P.; Shi, X.; Wang, Y.; Zhang, H.; He, Z.; Teng, S. Characterization and fine mapping of a novel rice albino mutant low temperature albino 1. J. Genet. Genom. 2012, 39, 385–396. [Google Scholar] [CrossRef]
- Liu, L.; Venkatesh, J.; Jo, Y.D.; Koeda, S.; Hosokawa, M.; Kang, J.H.; Goritschnig, S.; Kang, B.C. Fine mapping and identification of candidate genes for the sy-2 locus in a temperature-sensitive chili pepper (Capsicum chinense). Theor. Appl. Genet. 2016, 4, 1–6. [Google Scholar] [CrossRef]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Lechner, E.; Achard, P.; Vansiri, A.; Potuschak, T.; Genschik, P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006, 9, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Ma, H.; Nei, M.; Kong, H. Evolution of F-box genes in plants: Different modes of sequence divergence and their relationships. Proc. Natl. Acad. Sci. USA 2009, 106, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Kalluri, U.C.; Jawdy, S.; Gunter, L.E.; Yin, T.; Tschaplinski, T.J.; Weston, D.J.; Ranjan, P.; Tuskan, G.A. The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol. 2008, 148, 1189–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagne, J.M.; Downes, B.P.; Shiu, S.H.; Durski, A.M.; Vierstra, R.D. The F-box subunit of the SCF E3 complex is encoded by a diverse super family of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 11519–11524. [Google Scholar] [CrossRef] [Green Version]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef] [Green Version]
- Stefanowicz, K.; Lannoo, N.; Van Damme, E.J. Plant F-box proteins-judges between life and death. Crit. Rev. Plant Sci. 2015, 34, 523–552. [Google Scholar] [CrossRef]
- Chae, E.; Tan, Q.K.; Hill, T.A.; Irish, V.F. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 2008, 135, 1235–1245. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Xu, Y.Y.; Luo, W.; Li, W.X.; Chen, N.; Zhang, D.J.; Chong, K. The F-Box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 2013, 163, 1673–1685. [Google Scholar] [CrossRef] [Green Version]
- Potuschak, T.; Lechner, E.; Parmentier, Y.; Yanagisawa, S.; Grava, S.; Koncz, C.; Genschik, P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 2003, 115, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Sonnberg, S.; Fleming, S.B.; Mercer, A.A. A truncated two-alpha-helix F-box present in poxvirus ankyrin-repeat proteins is sufficient for binding the SCF1 ubiquitin ligase complex. J. Gen. Virol. 2009, 90, 1224–1228. [Google Scholar] [CrossRef]
- Song, J.B.; Wang, Y.X.; Li, H.B.; Li, B.W.; Zhou, Z.S.; Gao, S.; Yang, Z.M. The F-box family genes as key elements in response to salt, heavy mental, and drought stresses in Medicago truncatula. Funct. Integr. Genomic. 2015, 15, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Takahara, M.; Magori, S.; Soyano, T.; Okamoto, S.; Yoshida, C.; Yano, K.; Sato, S.; Tabata, S.; Yamaguchi, K.; Shigenobu, S.; et al. Too much love, a novel Kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume-Rhizobium symbiosis. Plant Cell Physiol. 2013, 54, 433–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takase, T.; Nishiyama, Y.; Tanihigashi, H.; Ogura, Y.; Miyazaki, Y.; Yamada, Y.; Kiyosue, T. LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 2011, 67, 608–621. [Google Scholar] [CrossRef]
- Zhang, X.; Gou, M.; Liu, C.J. Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonialyase. Plant Cell 2013, 25, 4994–5010. [Google Scholar] [CrossRef] [Green Version]
- Rao, V.; Petla, B.P.; Verma, P.; Salvi, P.; Kamble, N.U.; Ghosh, S.; Kaur, H.; Saxena, S.C.; Majee, M. Arabidopsis SKP1-like protein13 (ASK13) positively regulates seed germination and seedling growth under abiotic stress. J. Exp. Bot. 2018, 69, 3899–3915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.S.; Chen, X.Y.; Yang, K.; Sun, Z.X.; Fu, Y.P.; Zhang, Y.M.; Fang, R.X. Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant 2011, 4, 190–197. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.M.; Kong, X.Z.; Kang, H.H.; Sun, X.D.; Wang, W. The involvement of wheat F-box protein gene TaFBA1 in the oxidative stress tolerance of plants. PLoS ONE 2015, 10, e0122117. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.M.; Wang, S.H.; Lin, C.; Song, Y.Z.; Zheng, X.X.; Song, F.M.; Zhu, C.X. Molecular cloning and functional characterisation of the tomato E3 ubiquitin ligase SlBAH1 gene. Funct. Plant Biol. 2016, 43, 1091–1101. [Google Scholar] [CrossRef]
- Bu, Q.; Lv, T.; Shen, H.; Luong, P.; Wang, J.; Wang, Z.; Huang, Z.; Xiao, L.; Engineer, C.; Kim, T.H.; et al. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol. 2014, 164, 424–439. [Google Scholar] [CrossRef] [Green Version]
- Gray, W.M.; del Pozo, J.C.; Walker, L.; Hobbie, L.; Risseeuw, E.; Banks, T.; Crosby, W.L.; Yang, M.; Ma, H.; Estelle, M. Identification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999, 13, 1678–1691. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.C.; Wu, Y.R.; Huang, X.H.; Sun, J.; Xie, Q. AtPUB19, a U-Box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol. Plant 2011, 4, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.G.; Guo, W.L.; Yin, Y.X.; Gong, Z.H. Novel F-box protein CaF-box is involved in responses to plant hormones and abiotic stress in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2014, 15, 2413–2430. [Google Scholar] [CrossRef]
- Zhou, S.; Sun, X.; Yin, S.; Kong, X.; Zhou, S.; Xu, Y.; Luo, Y.; Wang, W. The role of the F-box gene TaFBA1 from wheat (Triticum aestivum L.) in drought tolerance. Plant Physiol. Biochem. 2014, 84, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, G.; Zhou, S.; Ren, Y.; Wang, W. The improvement of salt tolerance in transgenic tobacco by overexpression of wheat F-box gene TaFBA1. Plant Sci. 2017, 259, 71–85. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, A.; Chang, H.Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pozo, N.; Rosli, H.G.; Martin, G.B.; Mueller, L.A. The SGN VIGS Tool: User-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol. Plant 2015, 8, 486–488. [Google Scholar] [CrossRef] [Green Version]
- Hwang, J.; Lee, S.; Lee, J.H.; Kang, W.H.; Kang, J.H.; Kang, M.Y.; Oh, C.S.; Kang, B.C. Plant translation elongation factor 1Bβ facilitates potato virus x (PVX) infection and interacts with PVX triple gene block protein 1. PLoS ONE 2015, 10, e0128014. [Google Scholar] [CrossRef]
- Walter, M.; Chaban, C.; Schütze, K.; Batistic, O.; Weckermann, K.; Näke, C.; Blazevic, D.; Grefen, C.; Schumacher, K.; Oecking, C.; et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004, 40, 428–438. [Google Scholar] [CrossRef]
- Benvenuto, E.; Ordàs, R.J.; Tavazza, R.; Ancora, G.; Biocca, S.; Cattaneo, A.; Galeffi, P. ‘Phytoantibodies’: A general vector for the expression of immunoglobulin domains in transgenic plants. Plant Mol. Biol. 1991, 17, 865–874. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Wang, X.; Zhang, W.; Hao, L.; Chu, X.; Guo, X. Cotton GhMPK6a negatively regulates osmotic tolerance and bacterial infection in transgenic Nicotiana benthamiana, and plays a pivotal role in development. FEBS J. 2013, 280, 5128–5144. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Sequence | Purpose |
|---|---|---|
| Y2H: LTSF1/2: NdeF1 | GAGACATATGATGCCTGTCAAAGTAGCA | Y2H vector cloning |
| Y2H: LTSF1/2: BamR1 | GAGAGGATCCTTAGATAACTAATTTTGGAGA | Y2H vector cloning |
| Y2H: LTSF1/2: NdeF2 | GAGACATATGATAGATTATGATCAGCAGGCAAT | Y2H vector cloning |
| Y2H: LTSF1/2: BamR2 | GAGAGGATCCCTATACATCCTCAAAAGTGGATA | Y2H vector cloning |
| Y2H: LTSF1/2: NdeF3 | TATACATATGGTACAACAACTCGATCCCCCT | Y2H vector cloning |
| SKP1-F-EcoRI | TCGCGAATTCATGTCTGCCCCAAAGAAAAT | Y2H vector cloning |
| SKP1-R-BamHI | TATAGGATCCTCACTCAAAGGCCCAAGCAT | Y2H vector cloning |
| SKP1:F-BamHI | CAATGGATCCATGTCTGCCCCAAAGAAAAT | BiFC vector cloning |
| SKP1:R-XhoI | ATATCTCGAGCTCAAAGGCCCAAGCA | BiFC vector cloning |
| LTSF1/2-F-BamHI | ACAGGGATCCATGCCTGTCAAAGTAGCA | BiFC vector cloning |
| LTSF1/2-R-XhoI | ACAGCTCGAGGATAACTAATTTTGGAGAA | BiFC vector cloning |
| VIGS-LTSF1UT-1F | CGACGACAAGACCCTAGGTGTTATTTTTGTCCTTTCC | TRV2 vector cloning |
| VIGS-LTSF1UT-1R | GAGGAGAAGAGCCCTCTGCAAACTCACTGTACAATTTAG | TRV2 vector cloning |
| VIGS-LTSF2UT-1F | CGACGACAAGACCCTGAAAAATGATTTTATGATATC | TRV2 vector cloning |
| VIGS-LTSF2UT-1R | GAGGAGAAGAGCCCTCAATGTATAATTTAGTCCACTGAAAAT | TRV2 vector cloning |
| VIGS-LTSF1/2-1F | CGACGACAAGACCCTTCCAAAACTTGGGAG | TRV2 vector cloning |
| VIGS-LTSF1/2-1R | GAGGAGAAGAGCCCTGCGGTTTCTCTTGTTAAGGGGT | TRV2 vector cloning |
| TRV2:LIC-F | TGTTACTCAAGGAAGCACGATGAGCT | LIC cloning confirmation |
| TRV2:LIC-R | CAGGCACGGATCTACTTAAAGAACGTAG | LIC cloning confirmation |
| LTSF1/2.Spe–F | GCTCACTAGTATGCCTGTCAAAGTAGCA | pMDC83 vector cloning/Transgene confirmation |
| LTSF1.Asc–R | AATATTGGCGCGCCAGATAACTAATTTTGGAGAA | |
| LTSF2.Asc–R | AATATTGGCGCGCCAAATAACTAATTTTGGAGAA | |
| Hpt-F | CCTGAACTCACCGCGACG | |
| Hpt-R | AAGACCAATGCGGAGCATAT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venkatesh, J.; Kang, M.-Y.; Liu, L.; Kwon, J.-K.; Kang, B.-C. F-Box Family Genes, LTSF1 and LTSF2, Regulate Low-Temperature Stress Tolerance in Pepper (Capsicum chinense). Plants 2020, 9, 1186. https://doi.org/10.3390/plants9091186
Venkatesh J, Kang M-Y, Liu L, Kwon J-K, Kang B-C. F-Box Family Genes, LTSF1 and LTSF2, Regulate Low-Temperature Stress Tolerance in Pepper (Capsicum chinense). Plants. 2020; 9(9):1186. https://doi.org/10.3390/plants9091186
Chicago/Turabian StyleVenkatesh, Jelli, Min-Young Kang, Li Liu, Jin-Kyung Kwon, and Byoung-Cheorl Kang. 2020. "F-Box Family Genes, LTSF1 and LTSF2, Regulate Low-Temperature Stress Tolerance in Pepper (Capsicum chinense)" Plants 9, no. 9: 1186. https://doi.org/10.3390/plants9091186
APA StyleVenkatesh, J., Kang, M.-Y., Liu, L., Kwon, J.-K., & Kang, B.-C. (2020). F-Box Family Genes, LTSF1 and LTSF2, Regulate Low-Temperature Stress Tolerance in Pepper (Capsicum chinense). Plants, 9(9), 1186. https://doi.org/10.3390/plants9091186





