RUST: A Robust, User-Friendly Script Tool for Rapid Measurement of Rust Disease on Cereal Leaves
Abstract
1. Introduction
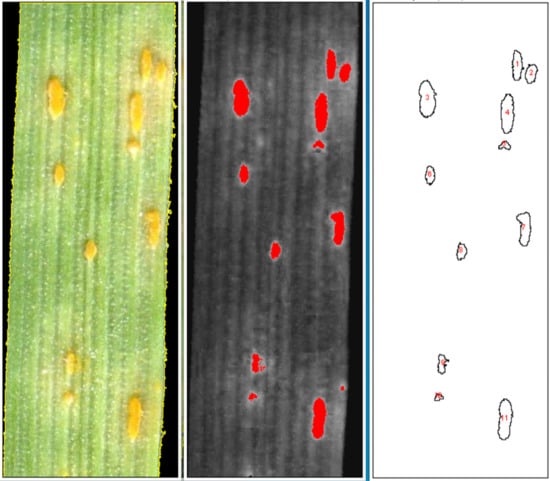
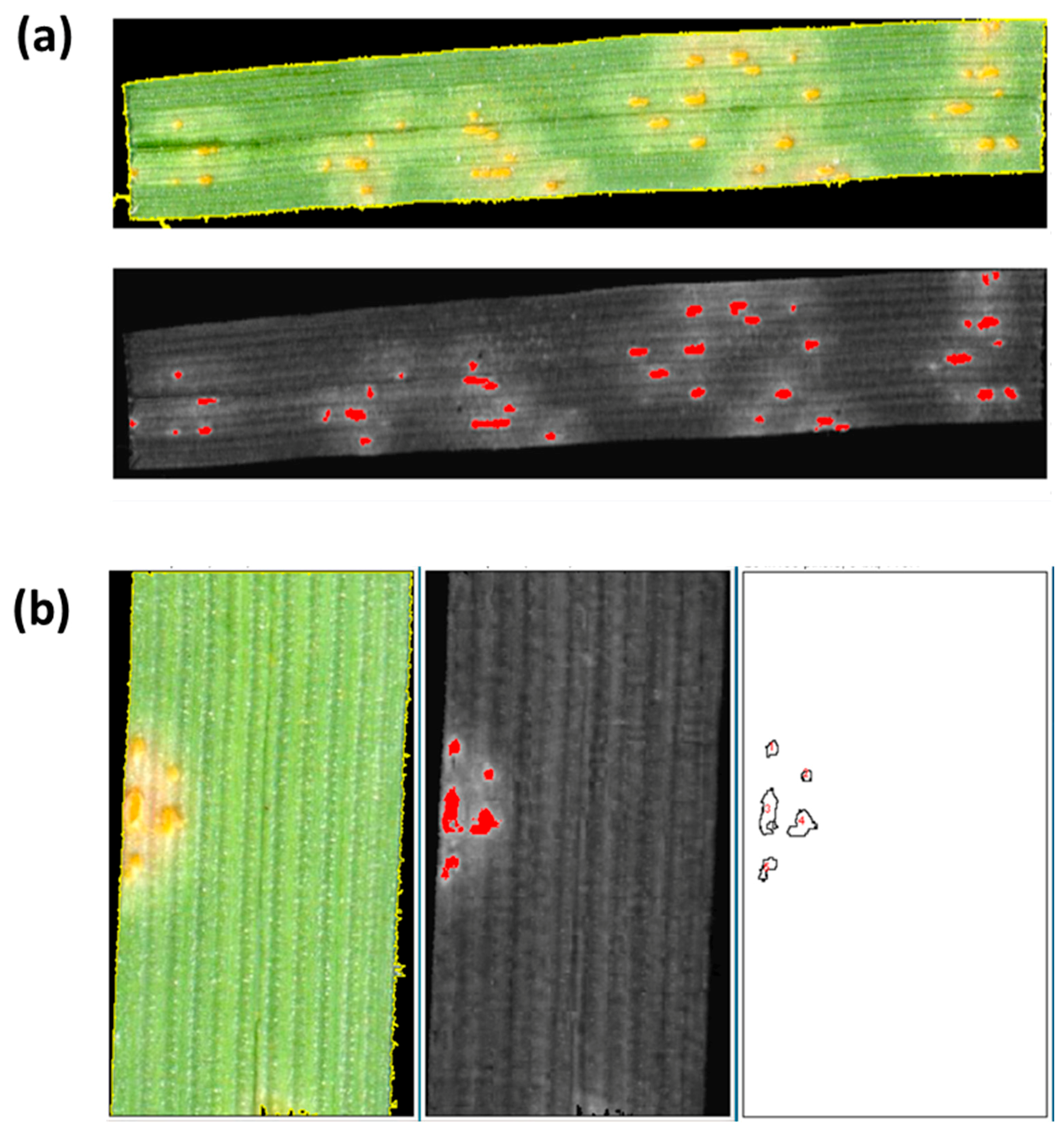
2. Results
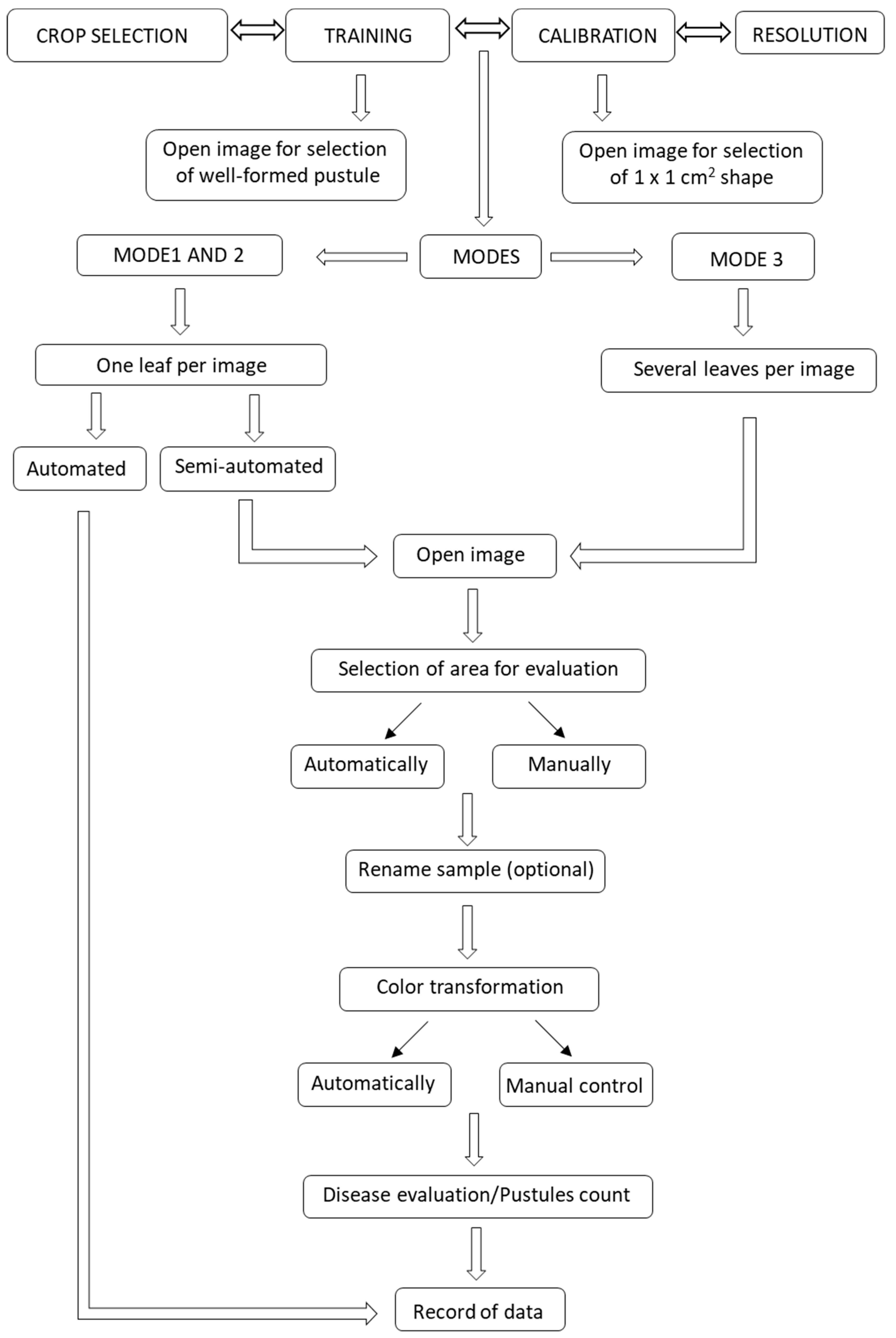
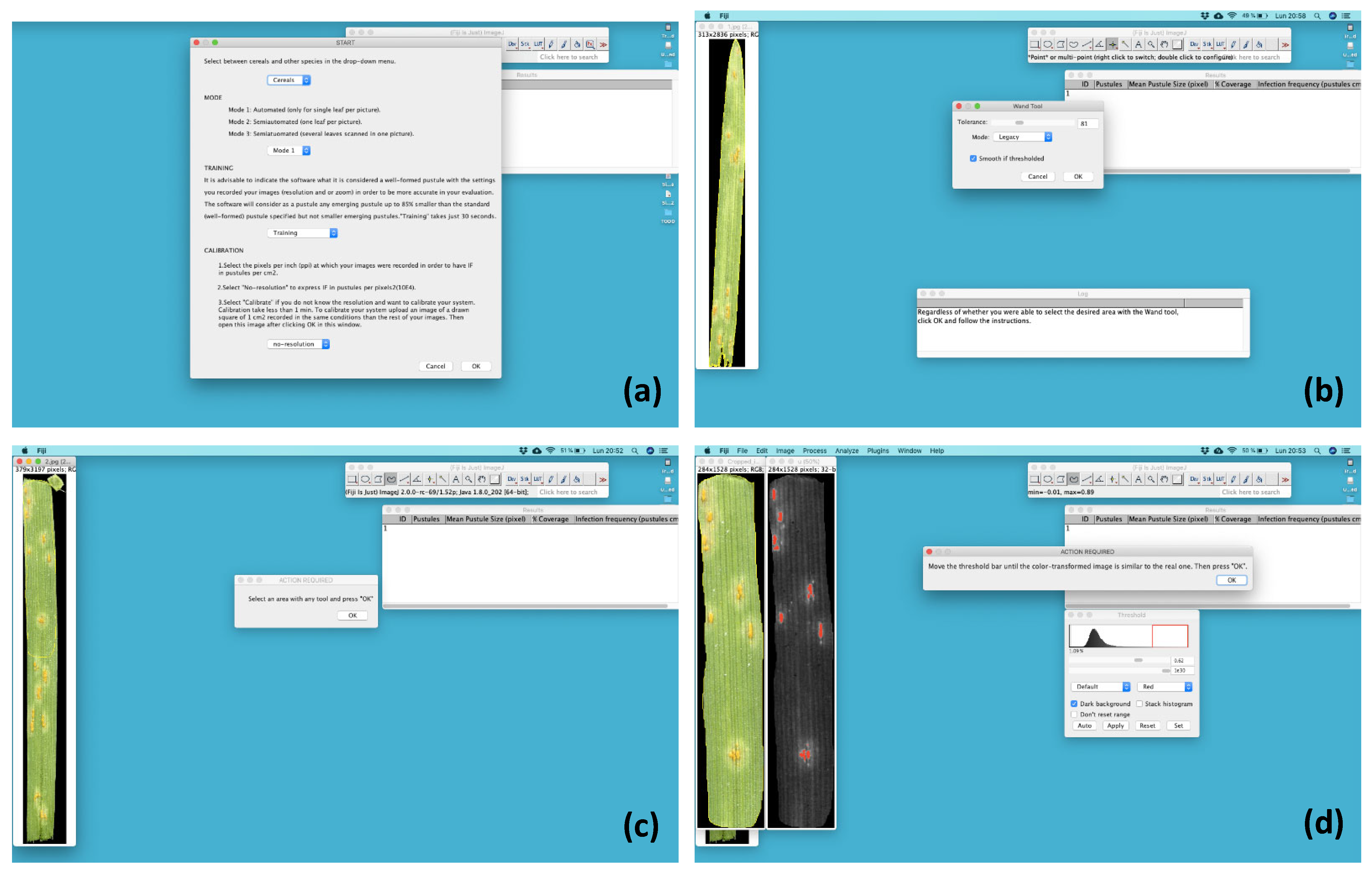
2.1. Running the Script
2.2. Validation of Script
3. Discussion
4. Materials and Methods
4.1. Plant Material and Inoculation
4.2. Image Recording and Analysis
4.3. Statistical Analysis
4.4. RUST Script
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Doehlert, D.C.; McMullen, M.S.; Jannink, J.L.; Panigrahi, S.; Gu, H.Z.; Riveland, N.R. Evaluation of oat kernel size uniformity. Crop Sci. 2004, 44, 1178–1186. [Google Scholar] [CrossRef]
- Holland, J.B.; Munkvold, G.P. Genetic relationships of crown rust resistance, grain yield, test weight, and seed weight in oat. Crop Sci. 2001, 41, 1041–1050. [Google Scholar] [CrossRef]
- Humphreys, D.G.; Mather, D.E. Heritability of beta-glucan, groat percentage, and crown rust resistance in two oat crosses. Euphytica 1996, 91, 359–364. [Google Scholar] [CrossRef]
- Endo, R.M.; Boewe, G.H. Losses caused by crown rust of oats in 1956 and 1957. Plant Dis. Rep. 1958, 42, 1126–1128. [Google Scholar]
- Herrmann, M.; Roderick, H.W. Characterization of new oat germplasm for resistance to powdery mildew. Euphytica 1996, 89, 405–410. [Google Scholar]
- Prats, E.; Sanchez-Martin, J.; Montilla-Bascon, G.; Rubiales, D.; Rispail, N. Overview and perspectives of the oat crop in Spain. Oat Newsl. 2015, 51, N9. [Google Scholar]
- Stevens, E.J.; Armstrong, K.W.; Bezaar, H.J.; Griffin, W.B.; Hampton, J.G. Fodder oats: An overview. In Fodder Oats: A World Overview; Suttie, J.M., Reynolds, S.G., Eds.; Plant Production and Protection Series; FAO: Rome, Italy, 2004; p. 33. [Google Scholar]
- Sanchez-Martin, J.; Rubiales, D.; Sillero, J.C.; Prats, E. Identification and characterization of sources of resistance in Avena sativa, A. byzantina and A. strigosa germplasm against a pathotype of Puccinia coronata f. sp avenae with virulence against the Pc94 resistance gene. Plant Pathol. 2012, 61, 315–322. [Google Scholar] [CrossRef]
- Leonard, K.J.; Szabo, L.J. Pathogen profile. Stem rust of small grains and grasses caused by Puccinia graminis. Mol. Plant Pathol. 2005, 6, 99–111. [Google Scholar] [CrossRef]
- Nazareno, E.S.; Li, F.; Smith, M.; Park, R.F.; Kianian, S.F.; Figueroa, M. Puccinia coronata f. sp avenae: A threat to global oat production. Mol. Plant Pathol. 2018, 19, 1047–1060. [Google Scholar] [CrossRef]
- Hoch, H.C.; Staples, R.C. Structural and chemical changes among the rust fungi during appressorium development. Annu. Rev. Phytopathol. 1987, 25, 231–247. [Google Scholar] [CrossRef]
- Prats, E.; Rubiales, D.; Jorrin, J. Acibenzolar-S-methyl-induced resistance to sunflower rust (Puccinia helianthi) is associated with an enhancement of coumarins on foliar surface. Physiol. Mol. Plant Pathol. 2002, 60, 155–162. [Google Scholar] [CrossRef]
- Parlevliet, J.E.; Kievit, C. Development of barley leaf rust, Puccinia hordei, infections in barley. 1. Effect of partial resistance and plant stage. Euphytica 1986, 35, 953–959. [Google Scholar] [CrossRef]
- Harder, D.E.; Harber, S. Oat diseases and pathological techniques. In Oat Science and Technology, Agronomy Monograph; Marshall, H.G., Sorrells, M.E., Eds.; American Society of Agronomy: Madison, WI, USA, 1992; p. 33. [Google Scholar]
- Jackson, E.W.; Obert, D.E.; Chong, J.; Avant, J.B.; Bonman, J.M. Detached-leaf method for propagating Puccinia coronata and assessing crown rust resistance in oat. Plant Dis. 2008, 92, 1400–1406. [Google Scholar] [CrossRef][Green Version]
- Simons, M.D. Crown Rust of Oats and Grasses; Simons, M.D., Ed.; Heffernan Press: Worcester, MA, USA, 1970. [Google Scholar]
- Staples, R.C.; Macko, V. Germination of urediospores and differentiation of infection structures. In The Cereals Rusts; Bushnell, W.R., Roelfs, A.P., Eds.; Academic Press: Orlando, FL, USA, 1984; Volume 1, pp. 255–289. [Google Scholar]
- Diaz-Lago, J.E.; Stuthman, D.D.; Leonard, K.J. Evaluation of components of partial resistance to oat crown rust using digital image analysis. Plant Dis. 2003, 87, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Pujari, J.D.; Siddarammayya Yakkundimath, R.; Jahagirdar, S.; Byadgi, A. Quantitative detection of soybean rust using image processing techniques. Crop Prot. 2015, 5, 75–87. [Google Scholar] [CrossRef]
- Bock, C.H.; Barbedo, J.G.A.; Del Ponte, E.M.; Bohnenkamp, D.; Mahlein, A.K. From visual estimates to fully automated sensor-based measurements of plant disease severity: Status and challenges for improving accuracy. Phytopathol. Res. 2020, 2, 1–30. [Google Scholar] [CrossRef]
- Mutka, A.M.; Bart, R.S. Image-based phenotyping of plant disease symptoms. Front. Plant Sci. 2015, 5, 734. [Google Scholar] [CrossRef]
- Courjon, D. Scanning tunneling optical microscopy. In Scanning Tunneling Microscopy; Behm, R.J., Garcia, N., Rohrer, H., Eds.; Springer: Heidelberg, Germany, 1990; pp. 497–505. [Google Scholar]
- Kokko, E.G.; Conner, R.L.; Kozub, G.C.; Lee, B. Quantification by image-analysis of subcrown internode discoloration in wheat caused by common root-rot. Phytopathology 1993, 83, 976–981. [Google Scholar] [CrossRef]
- Price, T.V.; Gross, R.; Wey, J.H.; Osborne, C.F. A comparison of visual and digital image-processing methods in quantifying the severity of coffee leaf rust (Hemileia-vastatrix). Aust. J. Exp. Agric. 1993, 33, 97–101. [Google Scholar] [CrossRef]
- Kampmann, H.H.; Hansen, O.B. Using color image-analysis for quantitative assessment of powdery mildew on cucumber. Euphytica 1994, 79, 19–27. [Google Scholar] [CrossRef]
- Karisto, P.; Hund, A.; Yu, K.; Anderegg, J.; Walter, A.; Mascher, F.; McDonald, B.A.; Mikaberidze, A. Ranking Quantitative Resistance to Septoria tritici Blotch in Elite Wheat Cultivars Using Automated Image Analysis. Phytopathology 2018, 108, 568–594. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.L.; McDonald, B.A. Measuring quantitative virulence in the wheat pathogen Zymoseptoria tritici using high-throughput automated image analysis. Phytopathology 2014, 104, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.C.; Chakraborty, S. Quantitative assessment of lesion characteristics and disease severity using digital image processing. J. Phytopathol. Phytopathol. Z. 1997, 145, 273–278. [Google Scholar] [CrossRef]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant Disease Severity Estimated Visually, by Digital Photography and Image Analysis, and by Hyperspectral Imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Lamari, L. Asses 2.0: Image Analysis Software for Plant Disease Quantification; American Phytopathological Society: St. Paul, MN, USA, 2002. [Google Scholar]
- Slice, D. Image-Pro-Plus: Image processing system. Q. Rev. Biol. 1994, 69, 143. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Wijekoon, C.P.; Goodwin, P.H.; Hsiang, T. Quantifying fungal infection of plant leaves by digital image analysis using Scion Image software. J. Microbiol. Methods 2008, 74, 94–101. [Google Scholar] [CrossRef]
- Bock, C.H.; Hotchkiss, M.W.; Wood, B.W. Assessing disease severity: Accuracy and reliability of rater estimates in relation to number of diagrams in a standard area diagram set. Plant Pathol. 2016, 65, 261–272. [Google Scholar] [CrossRef]
- Lin, L.I. A concordance correlation-coefficient to evaluate reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef]
- Madden, L.V.; Hughes, G.; Bosch, F. The Study of Plant Disease Epidemics; American Phytophatological Society: St. Paul, MN, USA, 2007. [Google Scholar]
- Jimenez Lopez, A.F.; Mauricio Salamanca, J.; Quiroz Medina, M.J.; Acevedo Perez, O.E. Crops diagnosis using digital image processing and precision agriculture technologies. Inge CUC 2015, 11, 63–71. [Google Scholar] [CrossRef]
- Omari, M.K.; Lee, J.; Faqeerzada, M.A.; Joshi, R.; Park, E.; Cho, B.-K. Digital image-based plant phenotyping: A review. Korean J. Agric. Sci. 2020, 47, 119–130. [Google Scholar]
- Pethybridge, S.J.; Nelson, S.C. The Leaf Doctor: A new application for quantifying disease severity. Phytopathology 2015, 105, 110. [Google Scholar]
- Machado, B.B.; Orue, J.P.M.; Arruda, M.S.; Santos, C.V.; Sarath, D.S.; Goncalves, W.N.; Silva, G.G.; Pistori, H.; Roel, A.R.; Rodrigues, J.F., Jr. BioLeaf: A professional mobile application to measure foliar damage caused by insect herbivory. Comput. Electron. Agric. 2016, 129, 44–55. [Google Scholar] [CrossRef]
- Jeger, M.J.; Viljanen-Rollinson, S.L.H. The use of the area under the disease-progress curve (AUDPC) to assess quantitative disease resistance in crop cultivars. Theor. Appl. Genet. 2001, 102, 32–40. [Google Scholar] [CrossRef]
- Steddom, K.; McMullen, M.S.; Schatz, B.; Rush, C.M. Comparing image format and resolution for assessment of foliar diseases of wheat. Plant Health Prog. 2005, 6, 11. [Google Scholar] [CrossRef]
- Ganthaler, A.; Losso, A.; Mayr, S. Using image analysis for quantitative assessment of needle bladder rust disease of Norway spruce. Plant Pathol. 2018, 67, 1122–1130. [Google Scholar] [CrossRef]
- Bock, C.H.; Parker, P.E.; Cook, A.Z.; Gottwald, T.R. Visual rating and the use of image analysis for assessing different symptoms of citrus canker on grapefruit leaves. Plant Dis. 2008, 92, 530–541. [Google Scholar] [CrossRef]
- Moral, J.; Montilla-Bascon, G.; Canales, F.J.; Rubiales, D.; Prats, E. Cytoskeleton reorganization/disorganization is a key feature of induced inaccessibility for defence to successive pathogen attacks. Mol. Plant Pathol. 2017, 18, 662–671. [Google Scholar] [CrossRef]
- Prats, E.; Carver, T.L.W.; Lyngkjaer, M.F.; Roberts, P.C.; Zeyen, R.J. Induced inaccessibility and accessibility in the oat powdery mildew system: Insights gained from use of metabolic inhibitors and silicon nutrition. Mol. Plant Pathol. 2006, 7, 47–59. [Google Scholar] [CrossRef]
- Barilla, M.E. Color Transformer. 2007. Available online: https://imagej.nih.gov/ij/plugins/download/Color_Transformer.java (accessed on 14 January 2019).
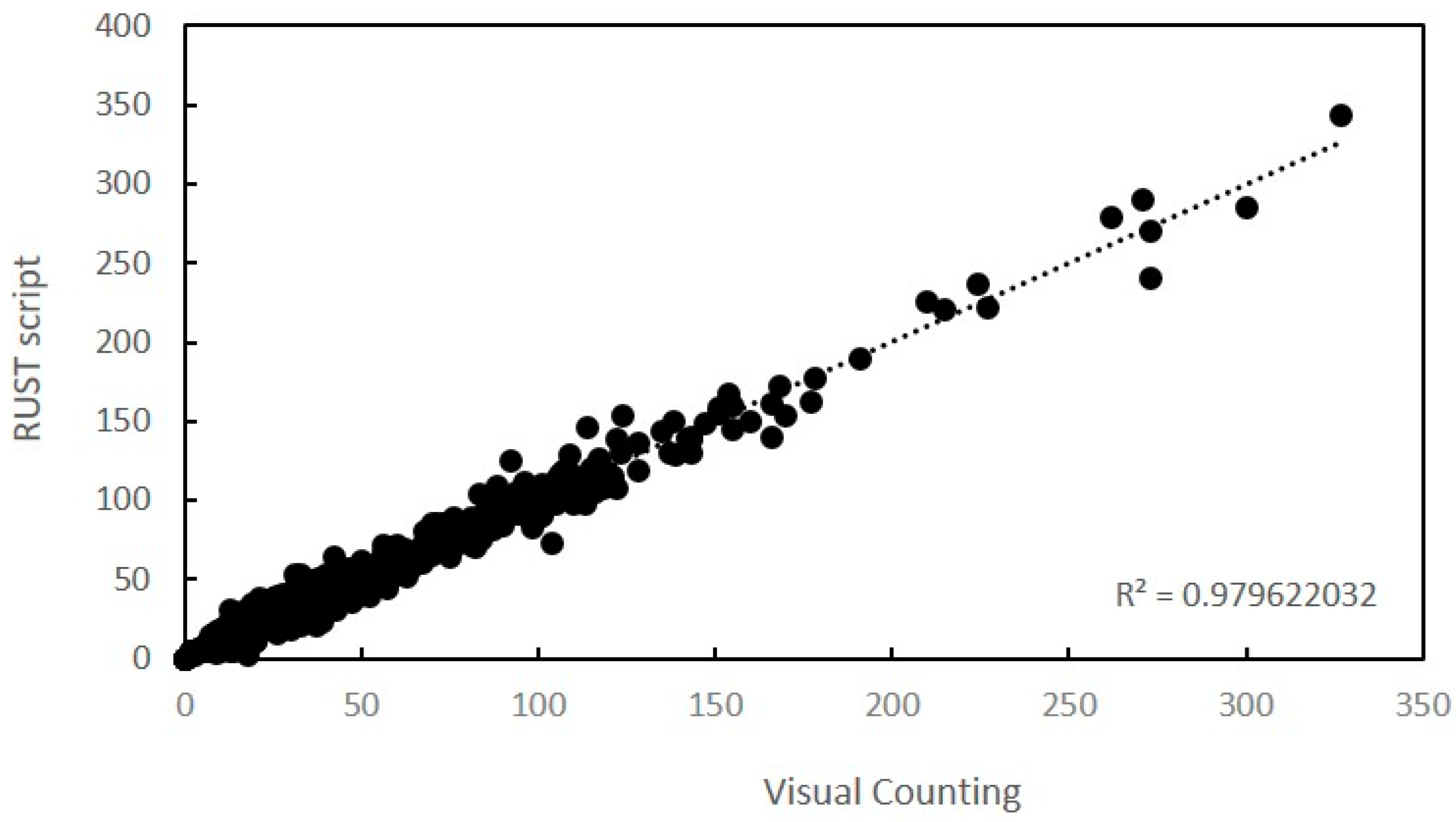
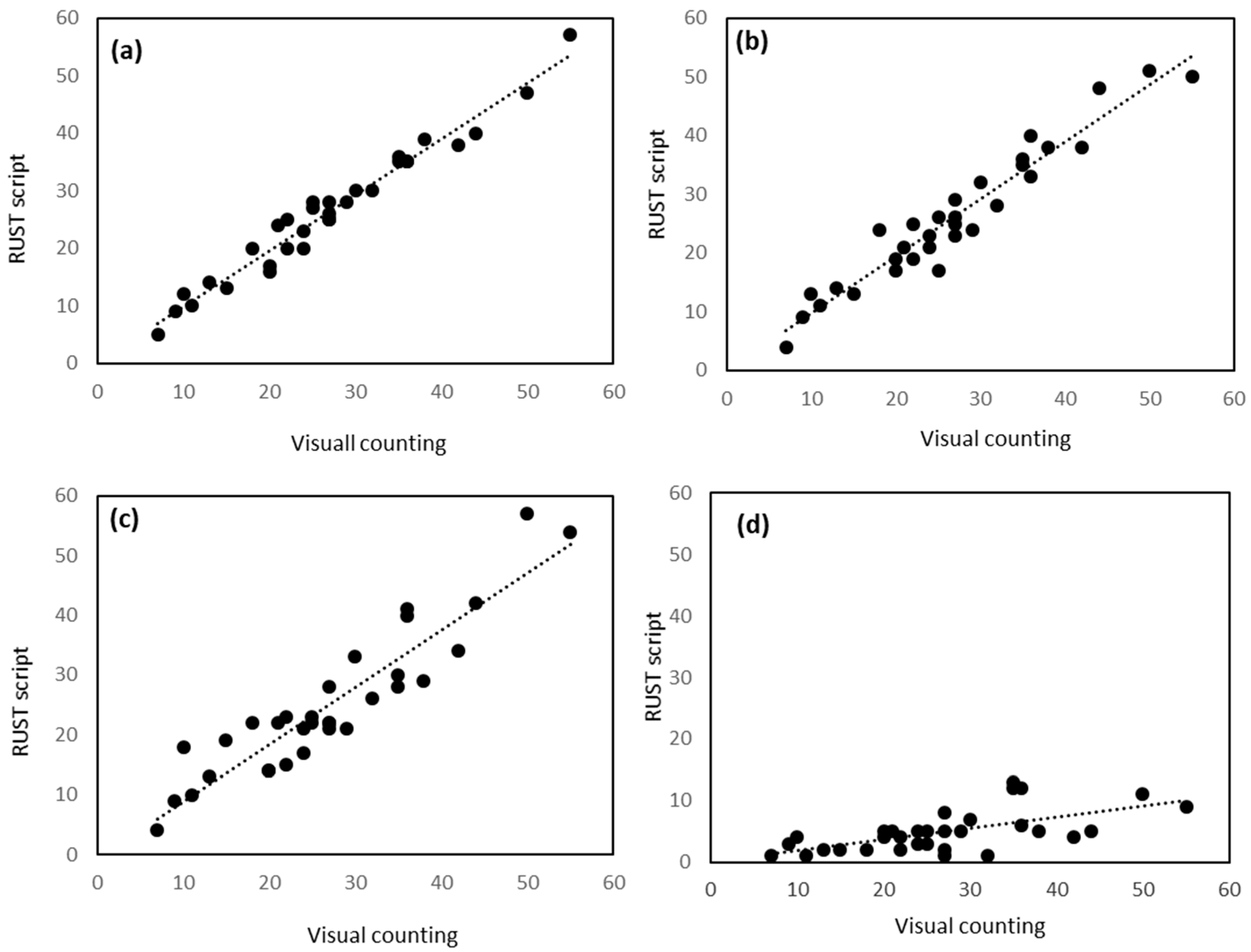
| Samples | Cb a | r b | LCCC c | R2d | Slope | Intercept | p |
|---|---|---|---|---|---|---|---|
| Complete set | 0.999 | 0.989 | 0.989 | 0.98 | 0.99 | 1.46 | <0.001 |
| Training HR | 0.998 | 0.982 | 0.980 | 0.96 | 0.97 | 0.12 | <0.001 |
| No Training HR | 0.998 | 0.964 | 0.962 | 0.93 | 0.97 | 0.08 | <0.001 |
| Training LR | 0.986 | 0.921 | 0.908 | 0.84 | 0.95 | 0.7 | <0.001 |
| No Training LR | 0.126 | 0.619 | 0.078 | 0.38 | 0.18 | 0.09 | <0.001 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallego-Sánchez, L.M.; Canales, F.J.; Montilla-Bascón, G.; Prats, E. RUST: A Robust, User-Friendly Script Tool for Rapid Measurement of Rust Disease on Cereal Leaves. Plants 2020, 9, 1182. https://doi.org/10.3390/plants9091182
Gallego-Sánchez LM, Canales FJ, Montilla-Bascón G, Prats E. RUST: A Robust, User-Friendly Script Tool for Rapid Measurement of Rust Disease on Cereal Leaves. Plants. 2020; 9(9):1182. https://doi.org/10.3390/plants9091182
Chicago/Turabian StyleGallego-Sánchez, Luis M., Francisco J. Canales, Gracia Montilla-Bascón, and Elena Prats. 2020. "RUST: A Robust, User-Friendly Script Tool for Rapid Measurement of Rust Disease on Cereal Leaves" Plants 9, no. 9: 1182. https://doi.org/10.3390/plants9091182
APA StyleGallego-Sánchez, L. M., Canales, F. J., Montilla-Bascón, G., & Prats, E. (2020). RUST: A Robust, User-Friendly Script Tool for Rapid Measurement of Rust Disease on Cereal Leaves. Plants, 9(9), 1182. https://doi.org/10.3390/plants9091182