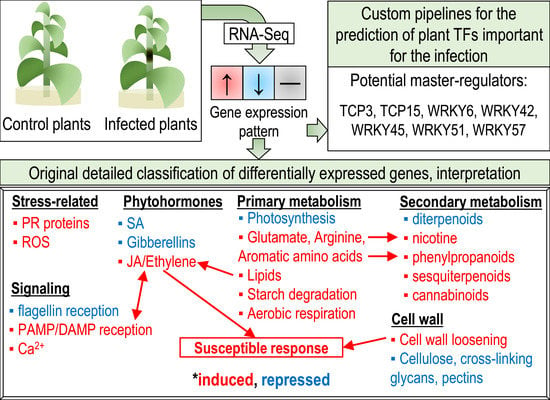
Plant Soft Rot Development and Regulation from the Viewpoint of Transcriptomic Profiling
Abstract
1. Introduction
2. Results and Discussion
2.1. Categories of DEGs
2.1.1. Cell Wall
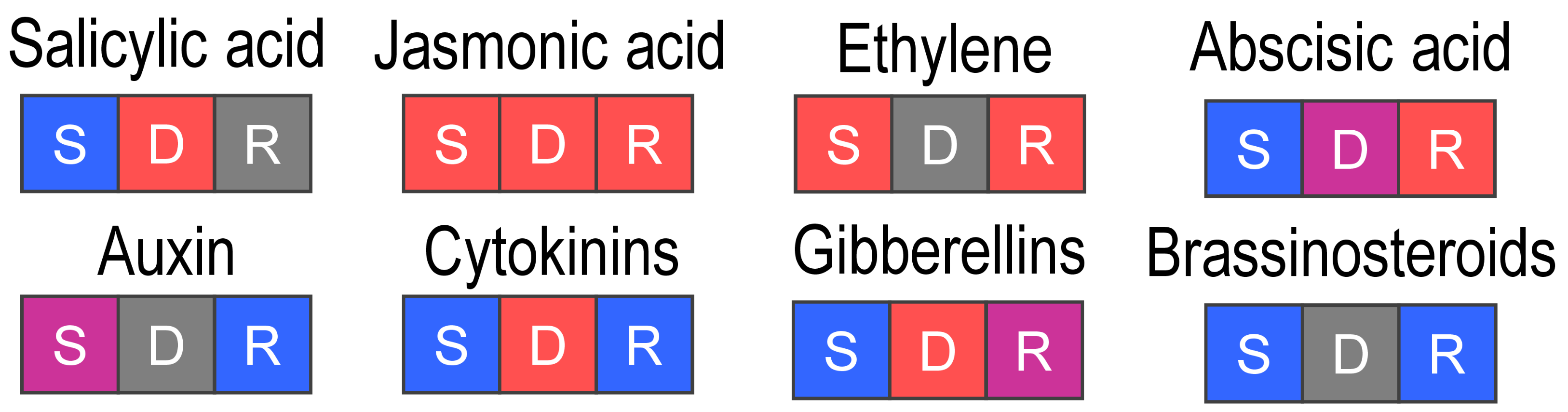
2.1.2. Phytohormones
2.1.3. Primary Metabolism
2.1.4. Secondary Metabolism
2.1.5. Stress-Related Genes
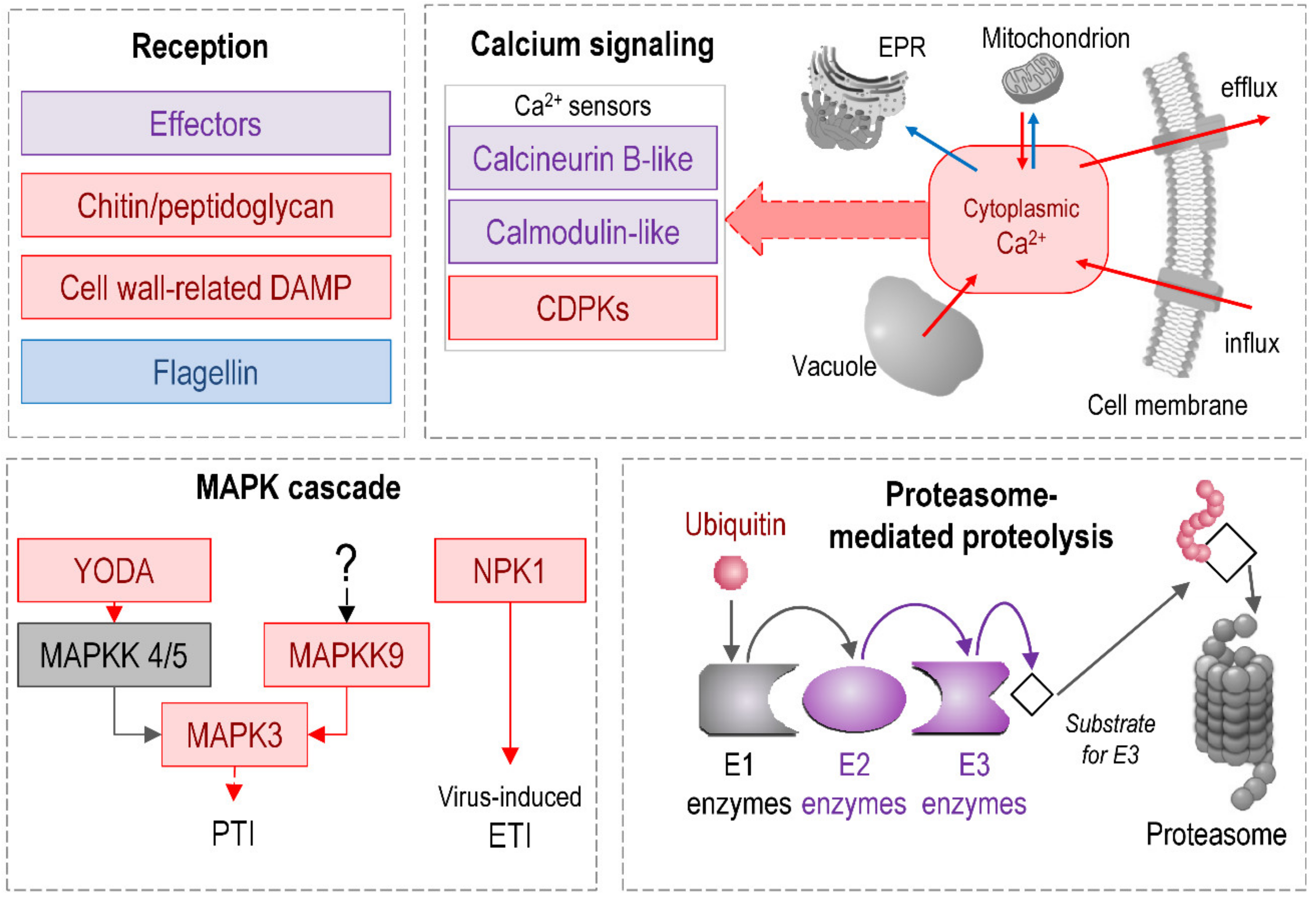
2.1.6. Signaling
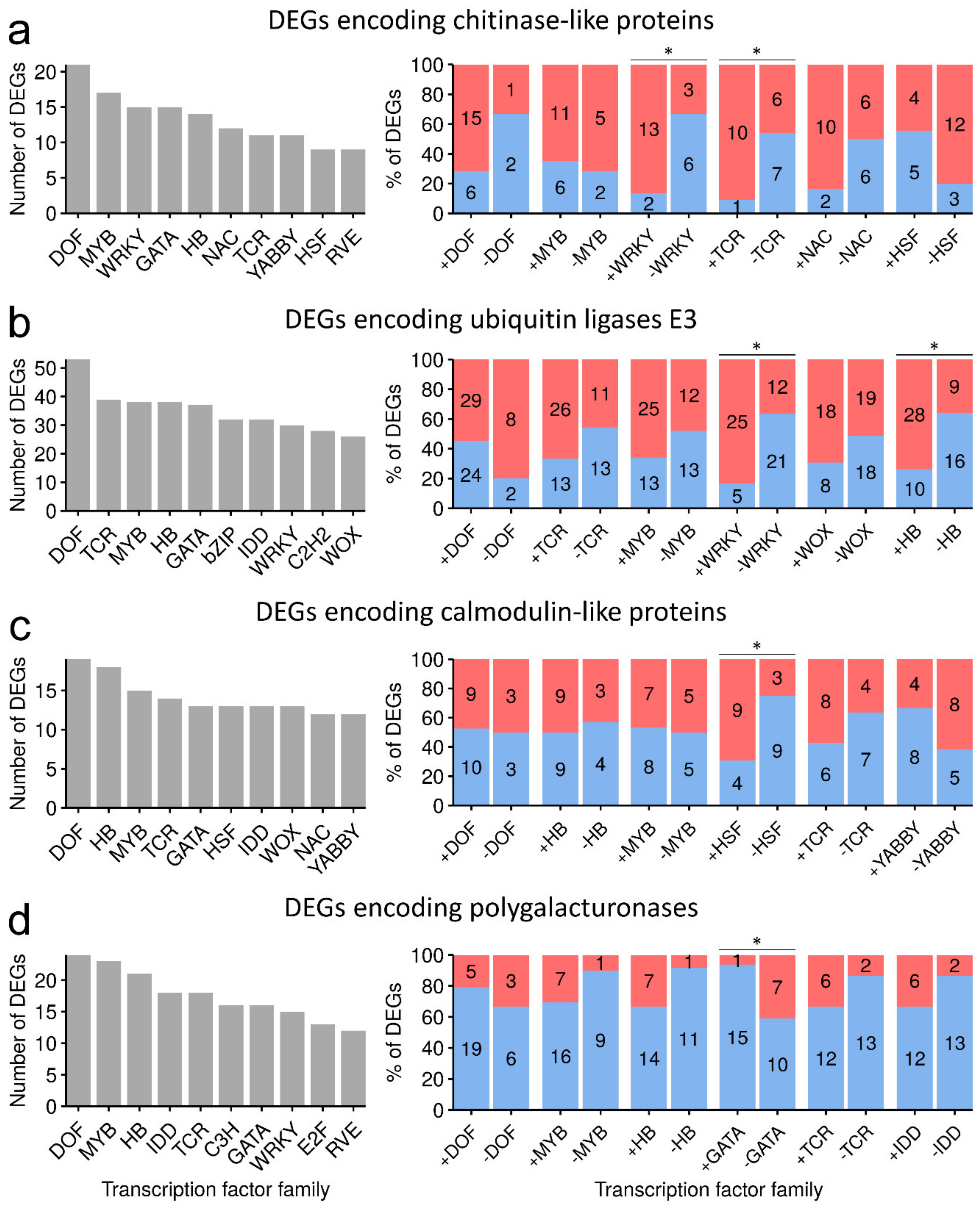
2.2. Transcription Factors
3. Materials and Methods
3.1. Bacteria and Plant Growth Conditions, Plant Inoculation, and Sample Collection
3.2. RNA Extraction and cDNA Library Preparation
3.3. Identification and Classification of DEGs
3.4. Prediction of the Transcription Factors Involved in Regulation of Pba-Induced Plant Responses
3.5. Verification of RNA-Seq by qRT-PCR
3.6. Lipoxygenase and Divinyl Ether Synthase Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Charkowski, A.; Blanco, C.; Condemine, G.; Expert, D.; Franza, T.; Hayes, C.; Hugouvieux-Cotte-Pattat, N.; López Solanilla, E.; Low, D.; Moleleki, L.; et al. The role of secretion systems and small molecules in soft-rot Enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 2012, 50, 425–449. [Google Scholar] [CrossRef] [PubMed]
- Kraepiel, Y.; Barny, M.-A. Gram-negative phytopathogenic bacteria, all hemibiotrophs after all? Mol. Plant Pathol. 2016, 17, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Holeva, M.C.; Bell, K.S.; Hyman, L.J.; Avrova, A.O.; Whisson, S.C.; Birch, P.R.J.; Toth, I.K. Use of a Pooled Transposon Mutation Grid to Demonstrate Roles in Disease Development for Erwinia carotovora subsp. Atroseptica Putative Type III Secreted Effector (DspE/A) and Helper (HrpN) Proteins. MPMI 2004, 17, 943–950. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Toth, I.K.; Birch, P.R.J. Rotting softly and stealthily. Curr. Opin. Plant Biol. 2005, 8, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Mattinen, L.; Tshuikina, M.; Mäe, A.; Pirhonen, M. Identification and Characterization of Nip, Necrosis-Inducing Virulence Protein of Erwinia carotovora subsp. carotovora. MPMI 2004, 17, 1366–1375. [Google Scholar] [CrossRef]
- Liu, H.; Coulthurst, S.J.; Pritchard, L.; Hedley, P.E.; Ravensdale, M.; Humphris, S.; Burr, T.; Takle, G.; Brurberg, M.-B.; Birch, P.R.J.; et al. Quorum Sensing Coordinates Brute Force and Stealth Modes of Infection in the Plant Pathogen Pectobacterium atrosepticum. PLoS Pathog. 2008, 4, e1000093. [Google Scholar] [CrossRef]
- Bell, K.S.; Sebaihia, M.; Pritchard, L.; Holden, M.T.G.; Hyman, L.J.; Holeva, M.C.; Thomson, N.R.; Bentley, S.D.; Churcher, L.J.C.; Mungall, K.; et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. PNAS 2004, 101, 11105–11110. [Google Scholar] [CrossRef]
- Panda, P.; Vanga, B.R.; Lu, A.; Fiers, M.; Fineran, P.C.; Butler, R.; Armstrong, K.; Ronson, C.W.; Pitman, A.R. Pectobacterium atrosepticum and Pectobacterium carotovorum Harbor Distinct, Independently Acquired Integrative and Conjugative Elements Encoding Coronafacic Acid that Enhance Virulence on Potato Stems. Front. Microbiol. 2016, 7, 397. [Google Scholar] [CrossRef]
- Gorshkov, V.; Daminova, A.; Ageeva, M.; Petrova, O.; Gogoleva, N.; Tarasova, N.; Gogolev, Y. Dissociation of a population of Pectobacterium atrosepticum SCRI1043 in tobacco plants: Formation of bacterial emboli and dormant cells. Protoplasma 2014, 251, 499–510. [Google Scholar] [CrossRef]
- Gorshkov, V.Y.; Daminova, A.G.; Mikshina, P.V.; Petrova, O.E.; Ageeva, M.V.; Salnikov, V.V.; Gorshkova, T.A.; Gogolev, Y.V. Pathogen-induced conditioning of the primary xylem vessels—A prerequisite for the formation of bacterial emboli by Pectobacterium atrosepticum. Plant. Biol. (Stuttg.) 2016, 18, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Kwenda, S.; Motlolometsi, T.V.; Birch, P.R.J.; Moleleki, L.N. RNA-seq Profiling Reveals Defense Responses in a Tolerant Potato Cultivar to Stem Infection by Pectobacterium carotovorum ssp. brasiliense. Front. Plant Sci. 2016, 7, 1905. [Google Scholar] [CrossRef] [PubMed]
- Chittem, K.; Yajima, W.R.; Goswami, R.S.; Del Río Mendoza, L.E. Transcriptome analysis of the plant pathogen Sclerotinia sclerotiorum interaction with resistant and susceptible canola (Brassica napus) lines. PLoS ONE 2020, 15, e0229844. [Google Scholar] [CrossRef] [PubMed]
- Wallace, I.S.; Somerville, C.R. A Blueprint for Cellulose Biosynthesis, Deposition, and Regulation in Plants. In Plant Cell Wall Patterning and Cell Shape; Wiley: Hoboken, NJ, US, 2014; pp. 65–95. ISBN 978-1-118-64736-3. [Google Scholar]
- Thompson, J.E.; Fry, S.C. Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant J. 2001, 26, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Sharmin, S.; Azam, M.S.; Islam, M.S.; Sajib, A.A.; Mahmood, N.; Hasan, A.M.M.; Ahmed, R.; Sultana, K.; Khan, H. Xyloglucan endotransglycosylase/hydrolase genes from a susceptible and resistant jute species show opposite expression pattern following Macrophomina phaseolina infection. Commun. Integr. Biol. 2012, 5, 598–606. [Google Scholar] [CrossRef]
- Vogel, J.P.; Raab, T.K.; Schiff, C.; Somerville, S.C. PMR6, a Pectate Lyase–Like Gene Required for Powdery Mildew Susceptibility in Arabidopsis. Plant Cell 2002, 14, 2095–2106. [Google Scholar] [CrossRef]
- An, S.H.; Sohn, K.H.; Choi, H.W.; Hwang, I.S.; Lee, S.C.; Hwang, B.K. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 2008, 228, 61–78. [Google Scholar] [CrossRef]
- Raiola, A.; Lionetti, V.; Elmaghraby, I.; Immerzeel, P.; Mellerowicz, E.J.; Salvi, G.; Cervone, F.; Bellincampi, D. Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol. Plant Microbe Interact. 2011, 24, 432–440. [Google Scholar] [CrossRef]
- Simpson, C.; Thomas, C.; Findlay, K.; Bayer, E.; Maule, A.J. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 2009, 21, 581–594. [Google Scholar] [CrossRef]
- Fan, J.; Ma, L.; Zhao, C.; Yan, J.; Che, S.; Zhou, Z.; Wang, H.; Yang, L.; Hu, B. Transcriptome of Pectobacterium carotovorum subsp. carotovorum PccS1 infected in calla plants in vivo highlights a spatiotemporal expression pattern of genes related to virulence, adaptation, and host response. Mol. Plant Pathol. 2020, 21, 871–891. [Google Scholar] [CrossRef]
- Messner, B.; Thulke, O.; Schäffner, A.R. Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 2003, 217, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Kwon, T.; Nam, J. Enzymatic and metabolic engineering for efficient production of syringin, sinapyl alcohol 4-O-glucoside, in Arabidopsis thaliana. Phytochemistry 2014, 102, 55–63. [Google Scholar] [CrossRef]
- Mareri, L.; Romi, M.; Cai, G. Arabinogalactan proteins: Actors or spectators during abiotic and biotic stress in plants? Plant Biosyst. 2019, 153, 173–185. [Google Scholar] [CrossRef]
- Bischoff, V.; Nita, S.; Neumetzler, L.; Schindelasch, D.; Urbain, A.; Eshed, R.; Persson, S.; Delmer, D.; Scheible, W.-R. TRICHOME BIREFRINGENCE and Its Homolog AT5G01360 Encode Plant-Specific DUF231 Proteins Required for Cellulose Biosynthesis in Arabidopsis. Plant Physiol. 2010, 153, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Gille, S.; de Souza, A.; Xiong, G.; Benz, M.; Cheng, K.; Schultink, A.; Reca, I.-B.; Pauly, M. O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 2011, 23, 4041–4053. [Google Scholar] [CrossRef] [PubMed]
- Roach, M.J.; Deyholos, M.K. Microarray analysis of flax (Linum usitatissimum L.) stems identifies transcripts enriched in fibre-bearing phloem tissues. Mol. Genet. Genomics 2007, 278, 149–165. [Google Scholar] [CrossRef]
- Mokshina, N.; Gorshkova, T.; Deyholos, M.K. Chitinase-like (CTL) and cellulose synthase (CESA) gene expression in gelatinous-type cellulosic walls of flax (Linum usitatissimum L.) bast fibers. PLoS ONE 2014, 9, e97949. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Kay, S.; Hahn, S.; Marois, E.; Hause, G.; Bonas, U. A Bacterial Effector Acts as a Plant Transcription Factor and Induces a Cell Size Regulator. Science 2007, 318, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Cantu, D.; Vicente, A.R.; Greve, L.C.; Dewey, F.M.; Bennett, A.B.; Labavitch, J.M.; Powell, A.L.T. The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc. Natl. Acad. Sci. USA 2008, 105, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Abuqamar, S.; Ajeb, S.; Sham, A.; Enan, M.R.; Iratni, R. A mutation in the expansin-like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana. Mol. Plant Pathol. 2013, 14, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Stegmann, M.; Villamil, J.C.M. The apoplast as battleground for plant–microbe interactions. New Phytol. 2015, 209, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Bellincampi, D.; Cervone, F.; Lionetti, V. Plant cell wall dynamics and wall-related susceptibility in plant–pathogen interactions. Front. Plant Sci. 2014, 5, 228. [Google Scholar] [CrossRef]
- Gorshkov, V.; Gubaev, R.; Petrova, O.; Daminova, A.; Gogoleva, N.; Ageeva, M.; Parfirova, O.; Prokchorchik, M.; Nikolaichik, Y.; Gogolev, Y. Transcriptome profiling helps to identify potential and true molecular switches of stealth to brute force behavior in Pectobacterium atrosepticum during systemic colonization of tobacco plants. Eur. J. Plant Pathol. 2018, 152, 957–976. [Google Scholar] [CrossRef]
- Grechkin, A. Recent developments in biochemistry of the plant lipoxygenase pathway. Prog. Lipid Res. 1998, 37, 317–352. [Google Scholar] [CrossRef]
- Prost, I.; Dhondt, S.; Rothe, G.; Vicente, J.; Rodriguez, M.J.; Kift, N.; Carbonne, F.; Griffiths, G.; Esquerré-Tugayé, M.-T.; Rosahl, S.; et al. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 2005, 139, 1902–1913. [Google Scholar] [CrossRef]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A.M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef]
- Zeilmaker, T.; Ludwig, N.R.; Elberse, J.; Seidl, M.F.; Berke, L.; Doorn, A.V.; Schuurink, R.C.; Snel, B.; Ackerveken, G.V. den DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015, 81, 210–222. [Google Scholar] [CrossRef]
- Strompen, G.; Grüner, R.; Pfitzner, U.M. An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol. Biol. 1998, 37, 871–883. [Google Scholar] [CrossRef]
- Sanchez Carranza, A.P.; Singh, A.; Steinberger, K.; Panigrahi, K.; Palme, K.; Dovzhenko, A.; Dal Bosco, C. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci. Rep. 2016, 6, 24212. [Google Scholar] [CrossRef]
- Wang, X.; Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 2006, 313, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Aalto, M.K.; Helenius, E.; Kariola, T.; Pennanen, V.; Heino, P.; Hõrak, H.; Puzõrjova, I.; Kollist, H.; Palva, E.T. ERD15—An attenuator of plant ABA responses and stomatal aperture. Plant Science 2012, 182, 19–28. [Google Scholar] [CrossRef]
- Van Gijsegem, F.; Pédron, J.; Patrit, O.; Simond-Côte, E.; Maia-Grondard, A.; Pétriacq, P.; Gonzalez, R.; Blottière, L.; Kraepiel, Y. Manipulation of ABA Content in Arabidopsis thaliana Modifies Sensitivity and Oxidative Stress Response to Dickeya dadantii and Influences Peroxidase Activity. Front. Plant Sci. 2017, 8, 456. [Google Scholar] [CrossRef]
- Palva, T.K. Salicylic Acid Induced Resistance to Erwinia carotovora subsp. carotovora in Tobacco. MPMI 1994, 7, 356–363. [Google Scholar] [CrossRef]
- Vidal, S.; de León, I.P.; Denecke, J.; Palva, E.T. Salicylic acid and the plant pathogen Erwinia carotovora induce defense genes via antagonistic pathways. Plant J. 1997, 11, 115–123. [Google Scholar] [CrossRef]
- Czajkowski, R.; van der Wolf, J.M.; Krolicka, A.; Ozymko, Z.; Narajczyk, M.; Kaczynska, N.; Lojkowska, E. Salicylic acid can reduce infection symptoms caused by Dickeya solani in tissue culture grown potato (Solanum tuberosum L.) plants. Eur. J. Plant Pathol. 2015, 141, 545–558. [Google Scholar] [CrossRef][Green Version]
- Thaler, J.S.; Fidantsef, A.L.; Bostock, R.M. Antagonism Between Jasmonate- and Salicylate-Mediated Induced Plant Resistance: Effects of Concentration and Timing of Elicitation on Defense-Related Proteins, Herbivore, and Pathogen Performance in Tomato. J. Chem. Ecol. 2002, 28, 1131–1159. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Körbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef]
- Rose, J.K.; Lee, H.H.; Bennett, A.B. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc. Natl. Acad. Sci. USA 1997, 94, 5955–5960. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ban, Q.; Hou, Y.; Meng, K.; Suo, J.; Rao, J. Isolation and Characterization of Two Persimmon Xyloglucan Endotransglycosylase/Hydrolase (XTH) Genes That Have Divergent Functions in Cell Wall Modification and Fruit Postharvest Softening. Front. Plant Sci. 2016, 7, 624. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Donoso, A.G.; Greve, L.C.; Walton, J.H.; Shackel, K.A.; Labavitch, J.M. Xylella fastidiosa Infection and Ethylene Exposure Result in Xylem and Water Movement Disruption in Grapevine Shoots. Plant Physiol. 2007, 143, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Gullner, G.; Kömives, T. The Role of Glutathione and Glutathione-related Enzymes in Plant-pathogen Interactions. In Significance of Glutathione to Plant Adaptation to the Environment; Grill, D., Tausz, M., De Kok, L.J., Eds.; Springer: Dordrecht, Netherlands, 2001; pp. 207–239. ISBN 978-0-306-47644-0. [Google Scholar]
- Park, D.H.; Mirabella, R.; Bronstein, P.A.; Preston, G.M.; Haring, M.A.; Lim, C.K.; Collmer, A.; Schuurink, R.C. Mutations in γ-aminobutyric acid (GABA) transaminase genes in plants or Pseudomonas syringae reduce bacterial virulence. Plant, J. 2010, 64, 318–330. [Google Scholar] [CrossRef]
- Ramos-Ruiz, R.; Martinez, F.; Knauf-Beiter, G. The effects of GABA in plants. Cogent Food Agric. 2019, 5, 1670553. [Google Scholar] [CrossRef]
- Muroi, A.; Matsui, K.; Shimoda, T.; Kihara, H.; Ozawa, R.; Ishihara, A.; Nishihara, M.; Arimura, G. Acquired immunity of transgenic torenia plants overexpressing agmatine coumaroyltransferase to pathogens and herbivore pests. Sci. Rep. 2012, 2, 689. [Google Scholar] [CrossRef]
- Li, J.; Zhang, K.; Meng, Y.; Hu, J.; Ding, M.; Bian, J.; Yan, M.; Han, J.; Zhou, M. Jasmonic acid/ethylene signaling coordinates hydroxycinnamic acid amides biosynthesis through ORA59 transcription factor. Plant J. 2018, 95, 444–457. [Google Scholar] [CrossRef]
- Van Dam, N.M.; Samudrala, D.; Harren, F.J.M.; Cristescu, S.M. Real-time analysis of sulfur-containing volatiles in Brassica plants infested with root-feeding Delia radicum larvae using proton-transfer reaction mass spectrometry. AoB Plants 2012, 2012, pls021. [Google Scholar] [CrossRef]
- Wang, B.; Lewis, R.S.; Shi, J.; Song, Z.; Gao, Y.; Li, W.; Chen, H.; Qu, R. Genetic Factors for Enhancement of Nicotine Levels in Cultivated Tobacco. Sci. Rep. 2015, 5, 17360. [Google Scholar] [CrossRef]
- Jesus, J.A.; Lago, J.H.G.; Laurenti, M.D.; Yamamoto, E.S.; Passero, L.F.D. Antimicrobial Activity of Oleanolic and Ursolic Acids: An Update. Evid. Based Complement Alternat. Med. 2015, 2015. [Google Scholar] [CrossRef]
- Jadhav, S.J.; Mazza, G.; Salunkhe, D.K. Terpenoid phytoalexins in potatoes: A review. Food Chem. 1991, 41, 195–217. [Google Scholar] [CrossRef]
- Dar, R.A.; Brahman, P.K.; Khurana, N.; Wagay, J.A.; Lone, Z.A.; Ganaie, M.A.; Pitre, K.S. Evaluation of antioxidant activity of crocin, podophyllotoxin and kaempferol by chemical, biochemical and electrochemical assays. Arab. J. Chem. 2017, 10, S1119–S1128. [Google Scholar] [CrossRef]
- Choi, J.N.; Kim, D.; Choi, H.K.; Yoo, K.M.; Kim, J.; Lee, C.H. 2′-hydroxylation of genistein enhanced antioxidant and antiproliferative activities in mcf-7 human breast cancer cells. J. Microbiol. Biotechnol. 2009, 19, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Topal, F.; Nar, M.; Gocer, H.; Kalin, P.; Kocyigit, U.M.; Gülçin, İ.; Alwasel, S.H. Antioxidant activity of taxifolin: An activity-structure relationship. J. Enzyme Inhib. Med. Chem. 2016, 31, 674–683. [Google Scholar] [CrossRef]
- Amorini, A.M.; Fazzina, G.; Lazzarino, G.; Tavazzi, B.; Di Pierro, D.; Santucci, R.; Sinibaldi, F.; Galvano, F.; Galvano, G. Activity and mechanism of the antioxidant properties of cyanidin-3-O-beta-glucopyranoside. Free Radic. Res. 2001, 35, 953–966. [Google Scholar] [CrossRef]
- Zou, S.; Kumar, U. Cannabinoid Receptors and the Endocannabinoid System: Signaling and Function in the Central Nervous System. Int. J. Mol. Sci. 2018, 19, 833. [Google Scholar] [CrossRef]
- Fernández-Ruiz, J.; Galve-Roperh, I.; Sagredo, O.; Guzmán, M. Possible therapeutic applications of cannabis in the neuropsychopharmacology field. Eur. Neuropsychopharm. 2020. [Google Scholar] [CrossRef]
- Expert, D.; Patrit, O.; Shevchik, V.E.; Perino, C.; Boucher, V.; Creze, C.; Wenes, E.; Fagard, M. Dickeya dadantii pectic enzymes necessary for virulence are also responsible for activation of the Arabidopsis thaliana innate immune system. Mol. Plant Pathol. 2017, 19, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Sherratt, P.J.; Hayes, J.D. Glutathione S-transferases. In Enzyme Systems that Metabolise Drugs and Other Xenobiotics; Wiley: Hoboken, NJ, US, 2002; pp. 319–352. ISBN 978-0-470-84630-8. [Google Scholar]
- Piffanelli, P.; Zhou, F.; Casais, C.; Orme, J.; Jarosch, B.; Schaffrath, U.; Collins, N.C.; Panstruga, R.; Schulze-Lefert, P. The Barley MLO Modulator of Defense and Cell Death Is Responsive to Biotic and Abiotic Stress Stimuli. Plant Physiol. 2002, 129, 1076–1085. [Google Scholar] [CrossRef]
- Wang, G.; Cai, G.; Kong, F.; Deng, Y.; Ma, N.; Meng, Q. Overexpression of tomato chloroplast-targeted DnaJ protein enhances tolerance to drought stress and resistance to Pseudomonas solanacearum in transgenic tobacco. Plant Physiol. Biochem. 2014, 82, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A. Peptidoglycan Perception in Plants. PLoS Pathog. 2015, 11, e1005275. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; Van Damme, E.J.M. Lectin domains at the frontiers of plant defense. Front. Plant Sci. 2014, 5, 397. [Google Scholar] [CrossRef] [PubMed]
- Sopeña-Torres, S.; Jordá, L.; Sánchez-Rodríguez, C.; Miedes, E.; Escudero, V.; Swami, S.; López, G.; Piślewska-Bednarek, M.; Lassowskat, I.; Lee, J.; et al. YODA MAP3K kinase regulates plant immune responses conferring broad-spectrum disease resistance. New Phytol. 2018, 218, 661–680. [Google Scholar] [CrossRef]
- Jin, H.; Axtell, M.J.; Dahlbeck, D.; Ekwenna, O.; Zhang, S.; Staskawicz, B.; Baker, B. NPK1, an MEKK1-like Mitogen-Activated Protein Kinase Kinase Kinase, Regulates Innate Immunity and Development in Plants. Dev. Cell 2002, 3, 291–297. [Google Scholar] [CrossRef]
- Singh, A.; Sagar, S.; Biswas, D.K. Calcium Dependent Protein Kinase, a Versatile Player in Plant Stress Management and Development. Crit. Rev. Plant Sci. 2017, 36, 336–352. [Google Scholar] [CrossRef]
- Demidchik, V.; Bowen, H.C.; Maathuis, F.J.M.; Shabala, S.N.; Tester, M.A.; White, P.J.; Davies, J.M. Arabidopsis thaliana root non-selective cation channels mediate calcium uptake and are involved in growth. Plant, J. 2002, 32, 799–808. [Google Scholar] [CrossRef]
- Lytton, J. Na+/Ca2+ exchangers: Three mammalian gene families control Ca2+ transport. Biochem. J. 2007, 406, 365–382. [Google Scholar] [CrossRef]
- Morris, J.; Tian, H.; Park, S.; Sreevidya, C.S.; Ward, J.M.; Hirschi, K.D. AtCCX3 Is an Arabidopsis Endomembrane H+-Dependent K+ Transporter. Plant Physiol. 2008, 148, 1474–1486. [Google Scholar] [CrossRef]
- Miricescu, A.; Goslin, K.; Graciet, E. Ubiquitylation in plants: Signaling hub for the integration of environmental signals. J. Exp. Bot. 2018, 69, 4511–4527. [Google Scholar] [CrossRef]
- Trujillo, M.; Ichimura, K.; Casais, C.; Shirasu, K. Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 2008, 18, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Tsugama, D.; Liu, S.; Takano, T. A U-Box E3 ubiquitin ligase, PUB20, interacts with the Arabidopsis G-protein β subunit, AGB1. PLoS ONE 2012, 7, e49207. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, S.; Sato, T.; Asada, Y.; Yasuda, S.; Yoshida, M.; Chiba, Y.; Yamaguchi, J. The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol. Biol. 2012, 79, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Demura, T.; Ye, Z.-H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef]
- Usadel, B.; Obayashi, T.; Mutwil, M.; Giorgi, F.M.; Bassel, G.W.; Tanimoto, M.; Chow, A.; Steinhauser, D.; Persson, S.; Provart, N.J. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009, 32, 1633–1651. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.-N.; Lee, T.-Y.; Hung, Y.-C.; Li, G.-Z.; Tseng, K.-C.; Liu, Y.-H.; Kuo, P.-L.; Zheng, H.-Q.; Chang, W.-C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, D1155–D1163. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Nolan, D.; Lang, D.T. Parsing XML Content. In XML and Web Technologies for Data Sciences with R.; Nolan, D., Temple Lang, D., Eds.; Springer: New York, NY, USA, 2014; pp. 53–74. ISBN 978-1-4614-7900-0. [Google Scholar]
- Chechetkin, I.R.; Osipova, E.V.; Tarasova, N.B.; Mukhitova, F.K.; Hamberg, M.; Gogolev, Y.V.; Grechkin, A.N. Specificity of oxidation of linoleic acid homologs by plant lipoxygenases. Biochem. Mosc. 2009, 74, 855–861. [Google Scholar] [CrossRef]
- Gogolev, Y.V.; Gorina, S.S.; Gogoleva, N.E.; Toporkova, Y.Y.; Chechetkin, I.R.; Grechkin, A.N. Green leaf divinyl ether synthase: Gene detection, molecular cloning and identification of a unique CYP74B subfamily member. Biochim. Biophys. Acta 2012, 1821, 287–294. [Google Scholar] [CrossRef]
- Geisler, M.; Kleczkowski, L.A.; Karpinski, S. A universal algorithm for genome-wide in silicio identification of biologically significant gene promoter putative cis-regulatory-elements; identification of new elements for reactive oxygen species and sucrose signaling in Arabidopsis. Plant, J. 2006, 45, 384–398. [Google Scholar] [CrossRef]
- Du, H.; Li, X.; Ning, L.; Qin, R.; Du, Q.; Wang, Q.; Song, H.; Huang, F.; Wang, H.; Yu, D. RNA-Seq analysis reveals transcript diversity and active genes after common cutworm (Spodoptera litura Fabricius) attack in resistant and susceptible wild soybean lines. BMC Genom. 2019, 20, 237. [Google Scholar] [CrossRef]
- Vercruysse, J.; Bel, M.V.; Osuna-Cruz, C.M.; Kulkarni, S.R.; Storme, V.; Nelissen, H.; Gonzalez, N.; Inzé, D.; Vandepoele, K. Comparative transcriptomics enables the identification of functional orthologous genes involved in early leaf growth. Plant Biotechnol. J. 2020, 18, 553–567. [Google Scholar] [CrossRef]
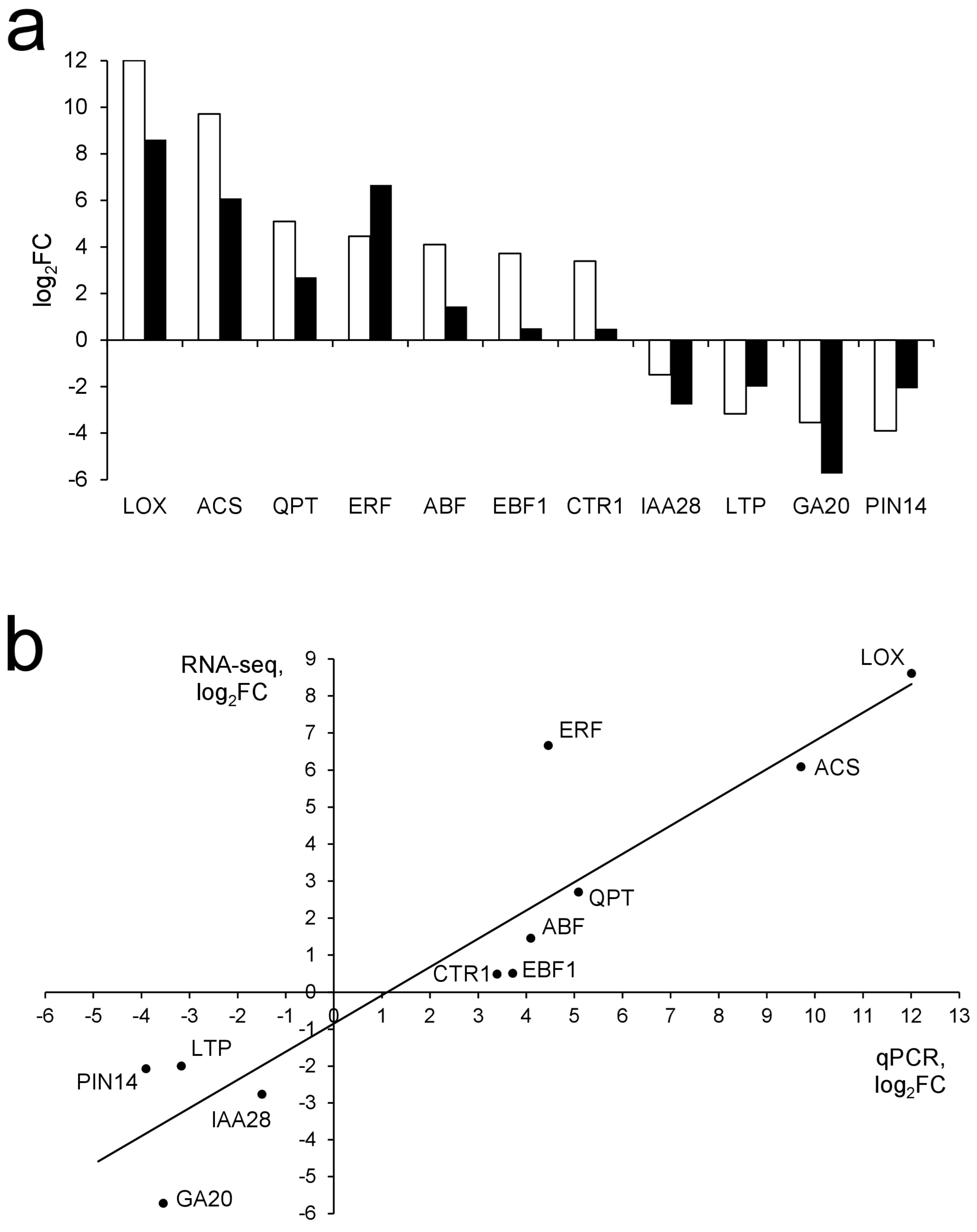




| Transcription Factor | Amount of Predicted Target Genes | FDR | GeneID (RefSeq) | Expression Pattern, log2FC | ||||
|---|---|---|---|---|---|---|---|---|
| Up Regulated DEGs | Down Regulated DEGs | Total DEGs | Total Non-DEGs | |||||
| TCP3 | 62 | 101 | 163 | 281 | 0.0405 | LOC107768765 | nonEXP | |
| TCP15 | 93 | 110 | 203 | 361 | 0.0405 | LOC107795877 | −4.424 | |
| LOC107800433 | −2.127 | |||||||
| WRKY6 | 362 | 281 | 643 | 1335 | 0.0405 | LOC107807966 | 5.332 | |
| LOC107782559 | 5.427 | |||||||
| LOC107824974 | 2.742 | |||||||
| WRKY42 | 408 | 323 | 731 | 1505 | 0.0289 | LOC107770788 | 1.352 | |
| LOC107824355 | 2.441 | |||||||
| WRKY45 | 463 | 370 | 833 | 1727 | 0.0289 | LOC107763775 | 5.993 | |
| LOC107769441 | 7.22 | |||||||
| WRKY51 | 78 | 51 | 129 | 208 | 0.0405 | LOC107787246 | 2.64 | |
| WRKY57 | 42 | 31 | 73 | 103 | 0.0405 | LOC107815490 | nonDEG | |
| LOC107788682 | nonDEG | |||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsers, I.; Gorshkov, V.; Gogoleva, N.; Parfirova, O.; Petrova, O.; Gogolev, Y. Plant Soft Rot Development and Regulation from the Viewpoint of Transcriptomic Profiling. Plants 2020, 9, 1176. https://doi.org/10.3390/plants9091176
Tsers I, Gorshkov V, Gogoleva N, Parfirova O, Petrova O, Gogolev Y. Plant Soft Rot Development and Regulation from the Viewpoint of Transcriptomic Profiling. Plants. 2020; 9(9):1176. https://doi.org/10.3390/plants9091176
Chicago/Turabian StyleTsers, Ivan, Vladimir Gorshkov, Natalia Gogoleva, Olga Parfirova, Olga Petrova, and Yuri Gogolev. 2020. "Plant Soft Rot Development and Regulation from the Viewpoint of Transcriptomic Profiling" Plants 9, no. 9: 1176. https://doi.org/10.3390/plants9091176
APA StyleTsers, I., Gorshkov, V., Gogoleva, N., Parfirova, O., Petrova, O., & Gogolev, Y. (2020). Plant Soft Rot Development and Regulation from the Viewpoint of Transcriptomic Profiling. Plants, 9(9), 1176. https://doi.org/10.3390/plants9091176