Genome-Wide Association Mapping for Stripe Rust Resistance in Pakistani Spring Wheat Genotypes
Abstract
:1. Introduction
2. Results
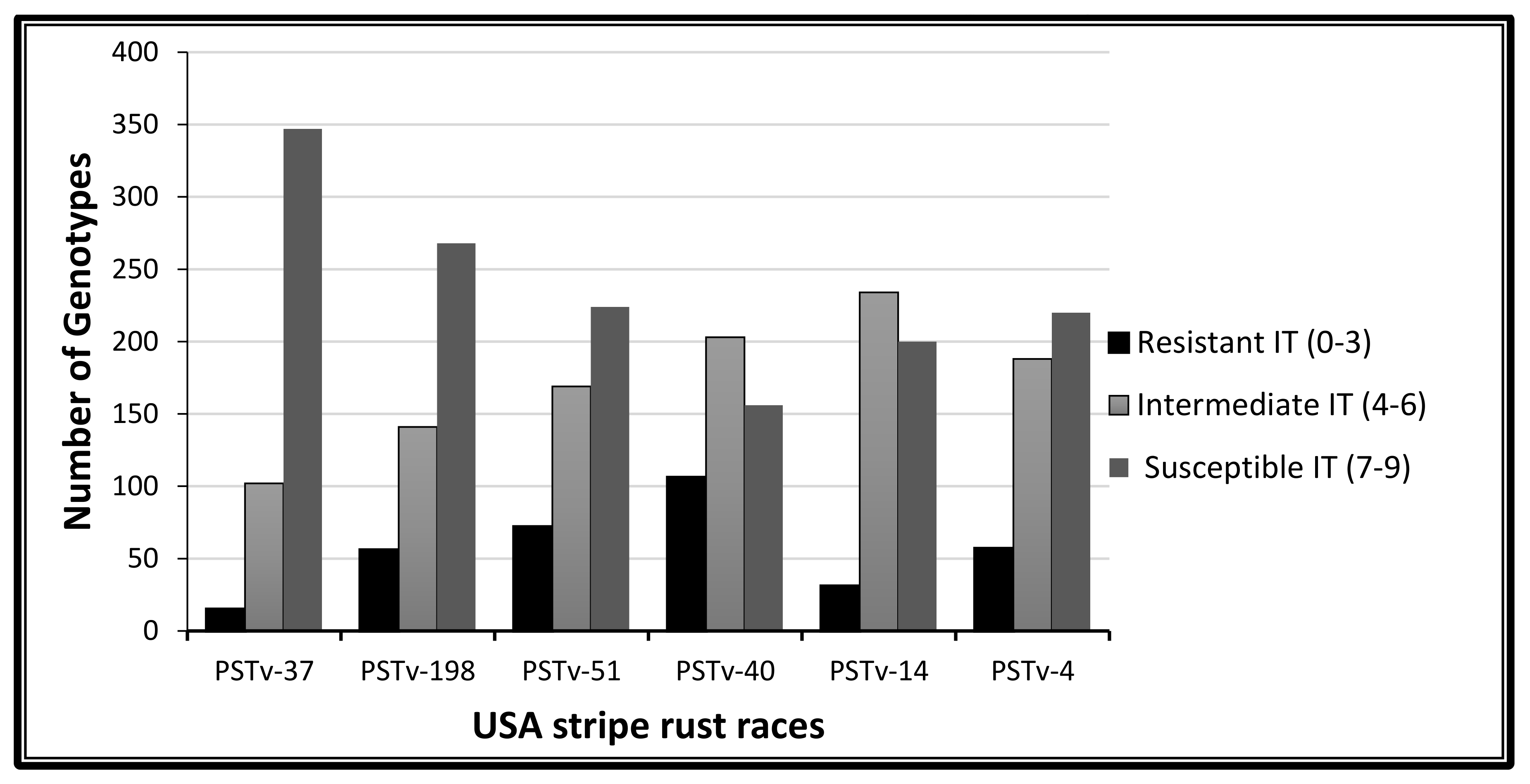
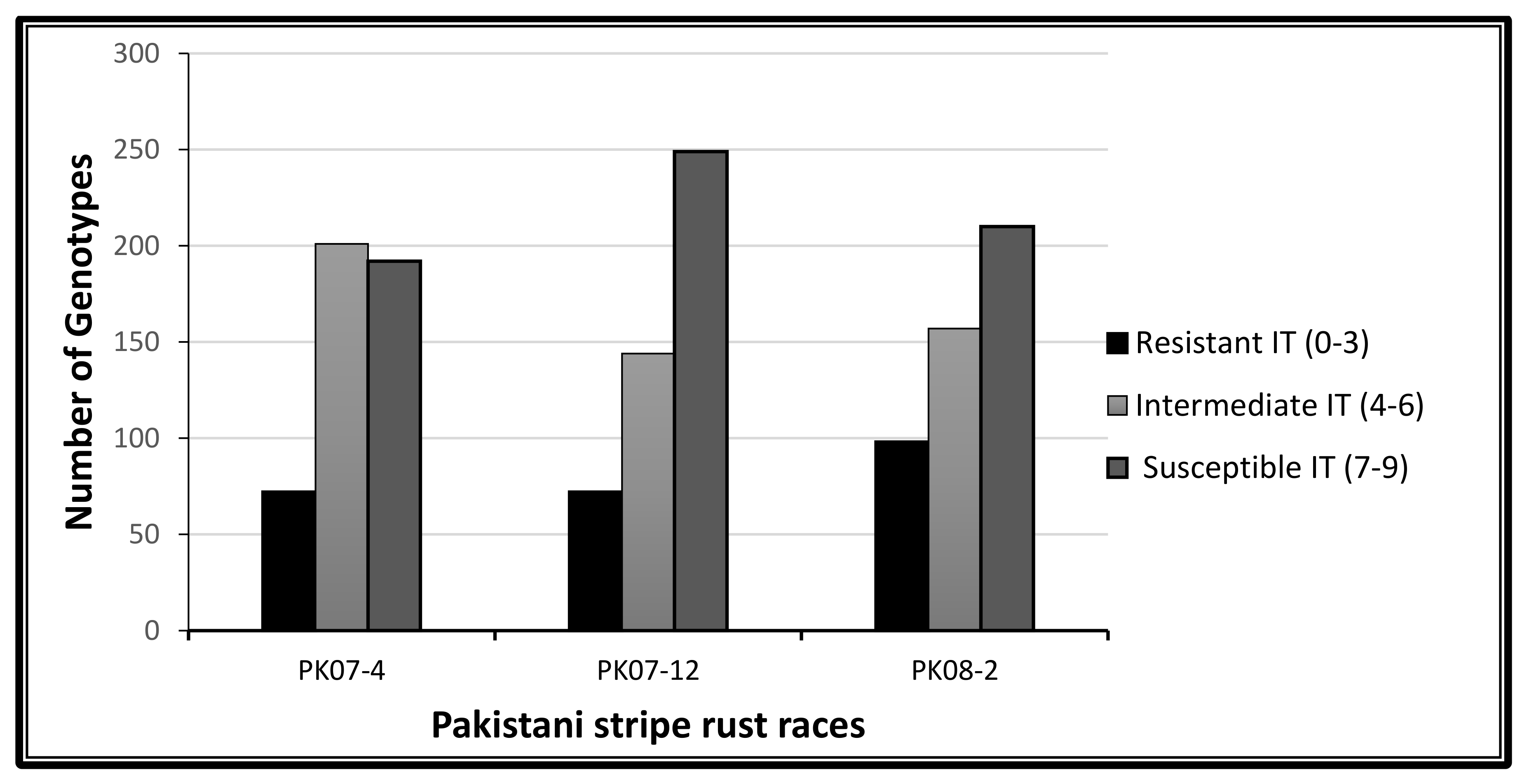
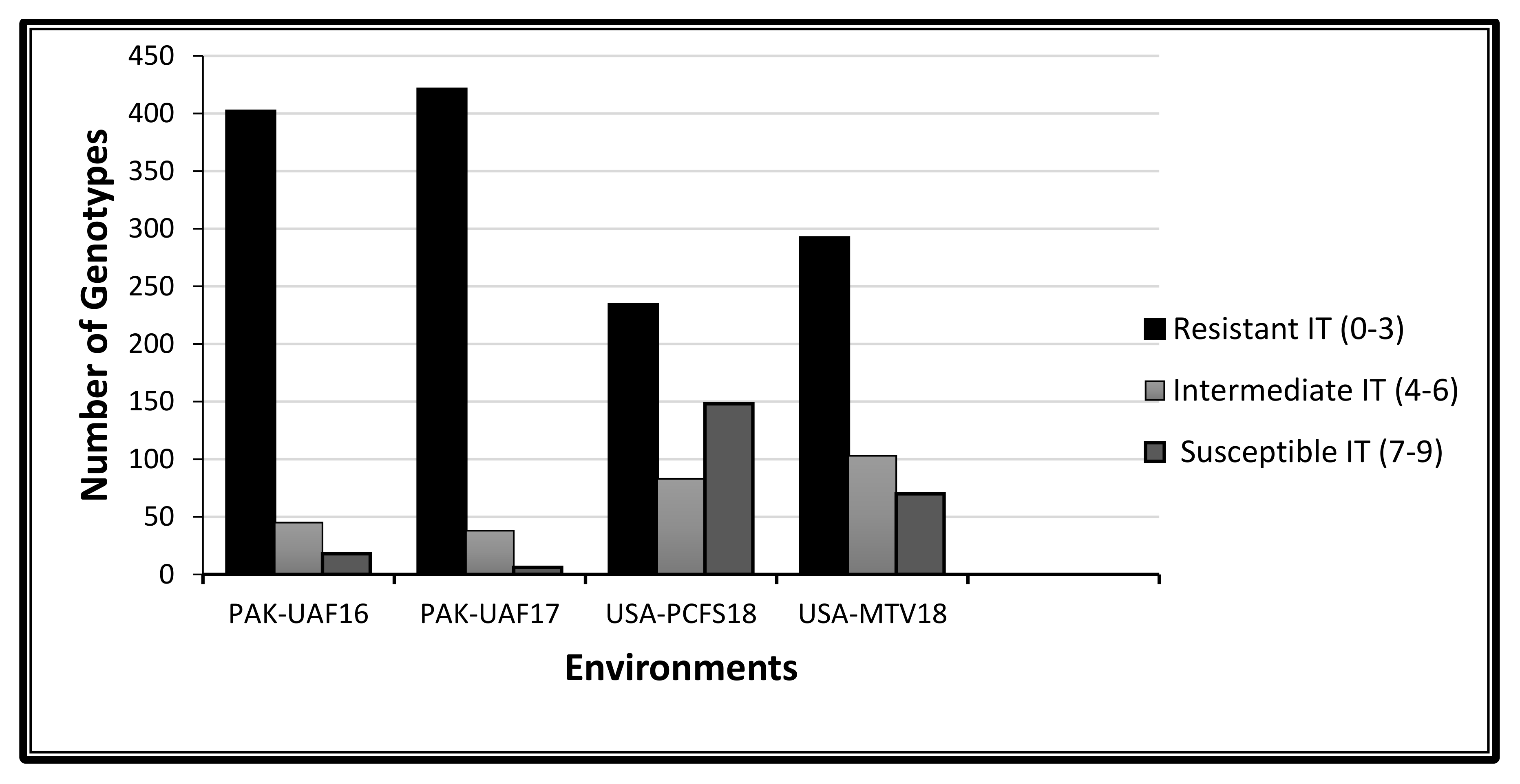
2.1. Phenotypic Variation to Stripe Rust Response
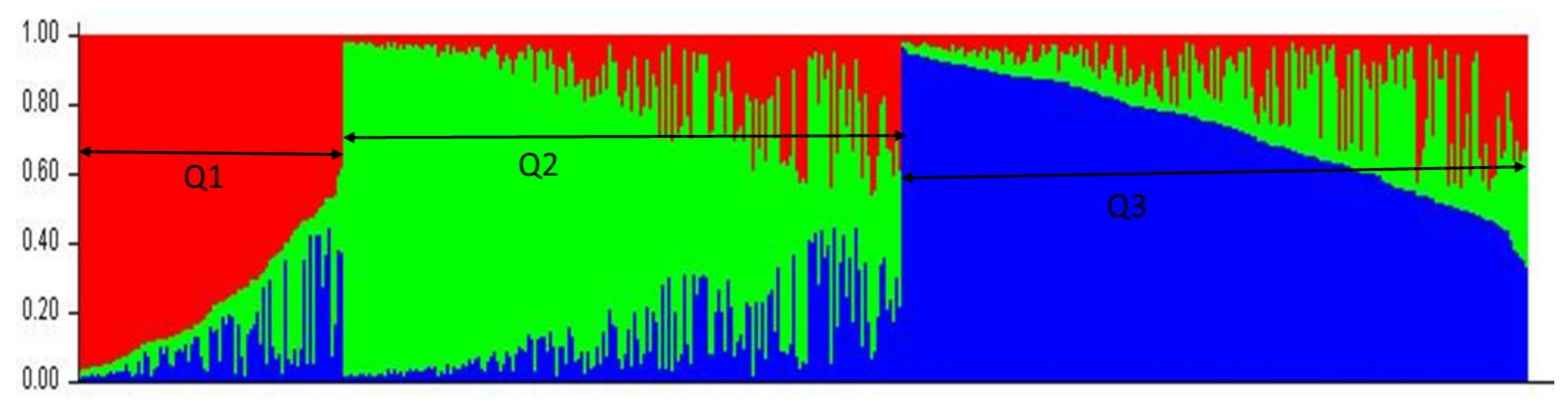
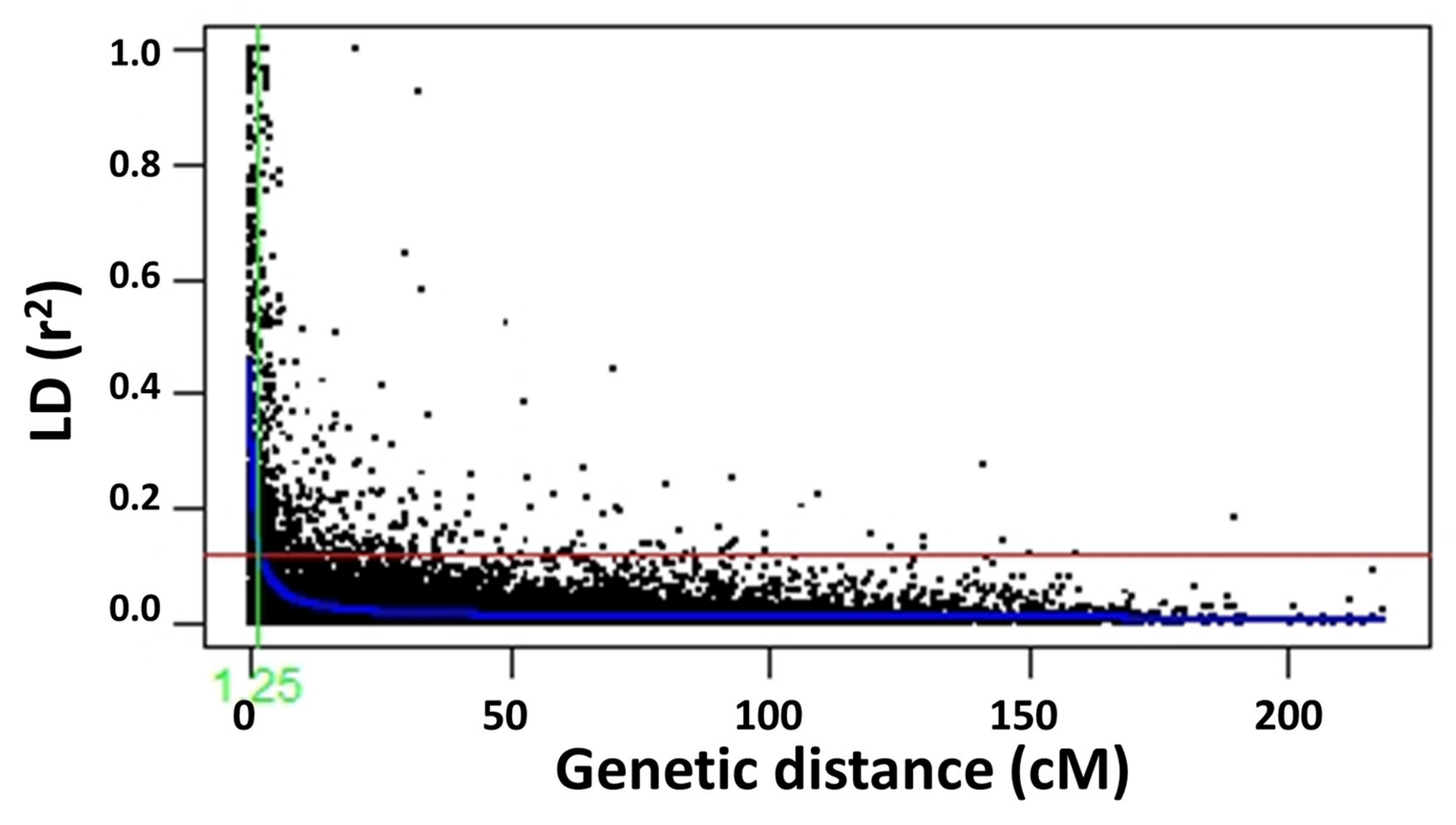
2.2. Population Structure and Linkage Disequilibrium
2.3. Marker–Trait Association at Seedling Stage
2.4. Marker–Trait Association at Adult Plant Stage
3. Discussion
3.1. Phenotypic Variation to Stripe Rust Response
3.2. Genome-Wide Association Analysis with Stripe Rust Response
3.3. Genome-Wide Association Analysis for Adult Plant Response
4. Materials and Methods
4.1. Collection of Genetic Material
4.2. Field Based Resistance Screening to Puccinia striiformis (Pst)
4.3. Greenhouse-Based Resistance Screening to Puccinia striiformis (Pst)
4.4. Statistical Analysis
4.5. DNA Extraction, SNP Genotyping
4.6. Population Structure and Linkage Disequilibrium (LD)
4.7. Genome-Wide Association (GWA) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Pst | Puccinia striiformis f. sp. tritici |
| MH | Million hectare |
| Yr | Yellow rust |
| USA | United State of America |
| APR | Adult plant resistance |
| MAS | Marker assisted selection |
| LD | Linkage disequilibrium |
| AM | Association mapping |
| GWAS | Genome-wide association mapping |
| QTL | Quantitative trait loci |
| SNP | Single nucleotide polymorphism |
| SEV | Severity |
| DArT | Diversity Arrays Technology |
| SSR | Simple sequence repeats |
| PCFS | Palouse conservation field station |
| MTV | Mount Vernon |
| UAF | University of Agriculture, Faisalabad |
| AvS | Avocet susceptible |
| MAF | Minor allelic frequency |
| TAS | Targeted amplicon sequencing |
| MLM | Mixed linear model |
| MTA | Marker–trait association |
| TASSEL | Trait analysis by association, evolution and linkage |
References
- Ogbonnaya, F.; Abdalla, O.; Mujeeb-Kazi, A.; Kazi, A.G.; Xu, S.S.; Gosman, N.; Lagudah, E.; Bonnett, D.; Sorrells, M.E.; Tsujimoto, H. Synthetic Hexaploids: Harnessing Species of the Primary Gene Pool for Wheat Improvement. Plant Breed. 2013, 37, 35–122. [Google Scholar] [CrossRef]
- Farooq, J.; Khaliq, I.; Akbar, M.; Petrescu-mag, I.V.; Hussain, M. Genetic analysis of some grain yield and its attributes at high temperature stress in wheat (Triticum aestivum L.). Ann. RSCB 2015, 19, 71–81. [Google Scholar] [CrossRef]
- Bhanupriya, B.; Satyanarayana, N.H.; Mukherjee, S.; Sarkar, K.K. Genetic diversity of wheat genotypes based on principal component analysis in Gangetic alluvial soil of West Bengal. J. Crop. Weed. 2014, 10, 104–107. [Google Scholar]
- Wellings, C. Global status of stripe rust: A review of historical and current threats. Euphytica 2011, 179, 129–141. [Google Scholar] [CrossRef]
- Mboup, M.; Leconte, M.; Gautier, A.; Wan, A.; Chen, W.; De Vallavieille-Pope, C.; Enjalbert, J. Evidence of genetic recombination in wheat yellow rust populations of a Chinese oversummering area. Fungal Genet. Biol. 2009, 46, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Hovmøller, M.; Walter, S.; Justesen, A.F. Escalating Threat of Wheat Rusts. Science 2010, 329, 369. [Google Scholar] [CrossRef] [Green Version]
- Prescott, J.M.; Burnett, P.A.; Saari, E.E.; Ransom, J.K.; Bowman, J.; De Milliano, W.; Singh, R.P.; Abeyo Bekele, G. Wheat Diseases and Pests: A Guide for Field Identification; CIMMYT: Ciudad de México, Mexico, 1986; p. 135. [Google Scholar]
- Ma, H.; Singh, R.P. Expression of adult resistance to yellow rust at different growth stages of wheat. Plant Dis. 1996, 80, 375–379. [Google Scholar] [CrossRef]
- Statista. 2019. Available online: https://www.statista.com/statistics/237912/global-top-wheat-producing-countries/ (accessed on 5 November 2019).
- Singh, R.P.; William, H.M.; Huerta-Espino, J.; Rosewarne, G. Wheat rust in Asia: Meeting the challenges with old and new technologies. In Proceedings of the 4th International Crop Science Congress, Brisbane, QLD, Australia, 26 September–1 October 2004; Volume 26. [Google Scholar]
- Solh, M.; Nazari, K.; Tadesse, W.; Wellings, C.R. The growing threat of stripe rust worldwide. In Proceedings of the Borlaug Global Rust Initiative (BGRI) Conference, Beijing, China, 1–4 September 2012; pp. 1–4. [Google Scholar]
- Duveiller, E.; Singh, R.P.; Nicol, J.M. The challenges of maintaining wheat productivity: Pests, diseases, and potential epidemics. Euphytica 2007, 157, 417–430. [Google Scholar] [CrossRef]
- Murray, G.M.; Brennan, J.P. The Current and Potential Costs from Diseases of Wheat in Australia; Grains Research and Development Corporation: Canberra, Australia, 2009; p. 70. [Google Scholar]
- Rahmatov, M. Sources of resistance to yellow rust and stem rust in wheat-alien introgressions. In Introductory paper at the Faculty of Landscape Planning. Hortic. Agric. Sci. 2013, 3, 1–64. [Google Scholar]
- Chen, X. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Plant Pathol. 2005, 27, 314–337. [Google Scholar] [CrossRef]
- Godoy, J.G.; Rynearson, S.; Chen, X.M.; Pumphrey, M. Genome-Wide Association Mapping of Loci for Resistance to Stripe Rust in North American Elite Spring Wheat Germplasm. Phytopathology 2018, 108, 234–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roelfs, A.P.; Bushnell, W.R. The Cereal Rusts: Diseases, Distribution, Epidemiology and Control; Academic Press: Orlando, FL, USA; London, UK, 1985; Volume 2, p. 606. [Google Scholar]
- Kolmer, J. Tracking wheat rust on a continental scale. Curr. Opin. Plant Biol. 2005, 8, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Maccaferri, M.; Rynearson, S.; Letta, T.; Zegeye, H.; Tuberosa, R.; Chen, X.; Pumphrey, M. Novel Sources of Stripe Rust Resistance Identified by Genome-Wide Association Mapping in Ethiopian Durum Wheat (Triticum turgidum ssp. durum). Front. Plant Sci. 2017, 8, 774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Line, R.F. Stripe rust of wheat and barley in North America: A retrospective historical review. Annu. Rev. Phytopathol. 2002, 40, 75–118. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Huerta-Espino, J.; William, H.M. Genetics and breeding for durable resistance to leaf and stripe rusts in wheat. Turk. J. Agric. 2005, 29, 121–127. [Google Scholar]
- Flint-Garcia, S.; Thornsberry, J.M.; Buckler, E.S.; Iv, B. Structure of Linkage Disequilibrium in Plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef] [Green Version]
- Zondervan, K.T.; Cardon, L.R. The complex interplay among factors that influence allelic association. Nat. Rev. Genet. 2004, 5, 89–100. [Google Scholar] [CrossRef]
- Yu, J.; Buckler, E.S. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 2006, 17, 155–160. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant. Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Tadesse, W.; Ogbonnaya, F.; Jighly, A.; Nazari, K.; Rajaram, S.; Baum, M. Association Mapping of Resistance to Yellow Rust in Winter Wheat Cultivars and Elite Genotypes. Crop. Sci. 2014, 54, 607–616. [Google Scholar] [CrossRef]
- Zegeye, H.; Rasheed, A.; Makdis, F.; Badebo, A.; Ogbonnaya, F.C. Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLoS ONE 2014, 9, e0105593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muleta, K.T.; Bulli, P.; Rynearson, S.; Chen, X.; Pumphrey, M. Loci associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a core collection of spring wheat (Triticum aestivum). PLoS ONE 2017, 12, e0179087. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Maccaferri, M.; Chen, X.; Laghetti, G.; Pignone, D.; Pumphrey, M.; Tuberosa, R. Genome-wide association mapping reveals a rich genetic architecture of stripe rust resistance loci in emmer wheat (Triticum turgidum ssp. dicoccum). Theor. Appl. Genet. 2017, 130, 2249–2270. [Google Scholar] [CrossRef] [Green Version]
- Tengberg, M. Crop husbandry at Miri Qalat, Makran, SW Pakistan (4000–2000 BC). Veg. Hist. Archeobot. 1999, 8, 3–12. [Google Scholar] [CrossRef]
- Maccaferri, M.; Zhang, J.; Bulli, P.; Abate, Z.; Chao, S.; Cantu, D.; Bossolini, E.; Chen, X.; Pumphrey, M.; Dubcovsky, J. A Genome-Wide Association Study of Resistance to Stripe Rust (Puccinia striiformisf. sp.tritici) in a Worldwide Collection of Hexaploid Spring Wheat (Triticum aestivum L.). G3 Genes Genomes Genet. 2015, 5, 449–465. [Google Scholar] [CrossRef] [Green Version]
- Joukhadar, R.; El-Bouhssini, M.; Jighly, A.; Ogbonnaya, F. Genome-wide association mapping for five major pest resistances in wheat. Mol. Breed. 2013, 32, 943–960. [Google Scholar] [CrossRef]
- Jighly, A.; Oyiga, B.C.; Makdis, F.; Nazari, K.; Youssef, O.; Tadesse, W.; Abdalla, O.; Ogbonnaya, F. Genome-wide DArT and SNP scan for QTL associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in elite ICARDA wheat (Triticum aestivum L.) germplasm. Theor. Appl. Genet. 2015, 128, 1277–1295. [Google Scholar] [CrossRef]
- Singh, A.K.; Knox, R.E.; Depauw, R.M.; Cuthbert, R.D.; Campbell, H.L.; Shorter, S.; Bhavani, S.; Cuthbert, R. Stripe rust and leaf rust resistance QTL mapping, epistatic interactions, and co-localization with stem rust resistance loci in spring wheat evaluated over three continents. Theor. Appl. Genet. 2014, 127, 2465–2477. [Google Scholar] [CrossRef]
- Tabassum, S.; Ashraf, M.; Chen, X.M. Evaluation of Pakistani wheat germplasm for stripe rust resistance using molecular marker. Sci. China Life Sci. 2010, 53, 1–12. [Google Scholar] [CrossRef]
- Akfirat, F.S.; Ertugrul, F.; Hasancebi, S.; Aydin, Y.; Akan, K.; Mert, Z.; Cakir, M.; Uncuoglu, A.A. Chromosomal location of genomic SSR markers associated with yellow rust resistance in Turkish bread wheat (Triticum aestivum L.). J. Genet. 2013, 92, 233–240. [Google Scholar] [CrossRef]
- Naruoka, Y.; Garland-Campbell, K.A.; Carter, A.H.; Garland-Campbell, K.A. Genome-wide association mapping for stripe rust (Puccinia striiformis F. sp. tritici) in US Pacific Northwest winter wheat (Triticum aestivum L.). Theor. Appl. Genet. 2015, 128, 1083–1101. [Google Scholar] [CrossRef]
- Shewaye, Y.; Taddesse, W. Association Mapping of Seedling and Adult Plant Resistance for Stripe Rust Resistance in Spring Bread Wheat (Triticum aestivum L.). Adv. Crop. Sci. Technol. 2018, 6, 1–10. [Google Scholar] [CrossRef]
- Chen, J.; Chu, C.; Souza, E.J.; Guttieri, M.J.; Chen, X.; Xu, S.; Hole, D.; Zemetra, R. Genome-wide identification of QTL conferring high-temperature adult-plant (HTAP) resistance to stripe rust (Puccinia striiformis f. sp. tritici) in wheat. Mol. Breed. 2011, 29, 791–800. [Google Scholar] [CrossRef]
- Quan, W.; Hou, G.; Chen, J.; Du, Z.; Lin, F.; Guo, Y.; Liu, S.; Zhang, Z. Mapping of QTL lengthening the latent period of Puccinia striiformis in winter wheat at the tillering growth stage. Eur. J. Plant. Pathol. 2013, 136, 715–727. [Google Scholar] [CrossRef]
- Saari, E.E. The current stripe rust status in south Asia. In Proceedings of Wheat Research Report of National Wheat Research Program; National Wheat Research Program: Bhairahawa, Nepal, 1995; p. 12. [Google Scholar]
- Yahyaoui, A.; Abdalla, O.; Morgonouv, H.K.; Torabi, M.; Cetin, L.; Saidov, L.M. Occurrence of stripe rust and effectiveness of resistance genes in Western and Central Asia and the Caucasus (WCAC). In Proceedings of the ASA-CSSA-SSSA International Annual Meetings, Seattle, WA, USA, 31 October–4 November 2004. [Google Scholar]
- Lowe, I.; Cantu, D.; Dubcovsky, J. Durable resistance to the wheat rusts: Integrating systems biology and traditional phenotype-based research methods to guide the deployment of resistance genes. Euphytica 2010, 179, 69–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallard, S.; Gaudet, D.; Aldeia, A.; Abelard, C.; Besnard, A.L.; Sourdille, P.; Dedryver, F. Genetic analysis of durable resistance to yellow rust in bread wheat. Theor. Appl. Genet. 2005, 110, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Lan, C.; Rosewarne, G.M.; Singh, R.P.; Herrera-Foessel, S.A.; Huerta-Espino, J.; Basnet, B.; Zhang, Y.; Yang, E. QTL characterization of resistance to leaf rust and stripe rust in the spring wheat line Francolin# 1. Mol. Breed. 2014, 34, 789–803. [Google Scholar] [CrossRef]
- Guo, Q.; Zhang, Z.J.; Xu, Y.B.; Li, G.; Feng, J.; Zhou, Y. Quantitative Trait Loci for High-Temperature Adult-Plant and Slow-Rusting Resistance to Puccinia striiformis f. sp. tritici in Wheat Cultivars. Phytopathology 2008, 98, 803–809. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.S.; Wang, M.N.; Cheng, P.; Kang, Z.S.; Hulbert, S.H.; Chen, X.M. Molecular mapping of Yr53, a new gene for stripe rust resistance in durum wheat accession PI 480148 and its transfer to common wheat. Theor. Appl. Genet. 2013, 126, 523–533. [Google Scholar] [CrossRef]
- Jagger, L.J.; Newell, C.; Berry, S.T.; MacCormack, R.; Boyd, L.A. The genetic characterisation of stripe rust resistance in the German wheat cultivar Alcedo. Theor. Appl. Genet. 2010, 122, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Melichar, J.P.E.; Berry, S.; Newell, C.; MacCormack, R.; Boyd, L.A. QTL identification and microphenotype characterisation of the developmentally regulated yellow rust resistance in the UK wheat cultivar Guardian. Theor. Appl. Genet. 2008, 117, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Christopher, M.D.; Liu, S.; Hall, M.D.; Marshall, D.S.; Fountain, M.O.; Johnson, J.W.; Milus, E.A.; Garland-Campbell, K.A.; Chen, X.; Griffey, C.A. Identification and Mapping of Adult Plant Stripe Rust Resistance in Soft Red Winter Wheat VA00W-38. Crop. Sci. 2013, 53, 871–879. [Google Scholar] [CrossRef]
- Agenbag, G.M.; Pretorius, Z.A.; Boyd, L.A.; Bender, C.M.; Prins, R. Identification of adult plant resistance to stripe rust in the wheat cultivar Cappelle-Desprez. Theor. Appl. Genet. 2012, 125, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Bariana, H.; Bansal, U.; Schmidt, A.; Lehmensiek, A.; Kaur, J.; Miah, H.; Howes, N.; McIntyre, C.L. Molecular mapping of adult plant stripe rust resistance in wheat and identification of pyramided QTL genotypes. Euphytica 2010, 176, 251–260. [Google Scholar] [CrossRef]
- Lin, F.; Chen, X.M. Genetics and molecular mapping of genes for race-specific all-stage resistance and non-race-specific high-temperature adult-plant resistance to stripe rust in spring wheat cultivar Alpowa. Theor. Appl. Genet. 2007, 114, 1277–1287. [Google Scholar] [CrossRef]
- Basnet, B.R.; Ibrahim, A.; Chen, X.; Singh, R.P.; Mason, E.R.; Bowden, R.L. Molecular mapping of stripe rust resistance in hard red winter wheat TAM 111 adapted to the US High Plains. Crop. Sci. 2014, 54, 1361–1373. [Google Scholar] [CrossRef]
- Helguera, M.; Khan, I.A.; Kolmer, J.; Lijavetzky, D.; Zhong-Qi, L.; Dubcovsky, J. PCR Assays for the Lr37-Yr17-Sr38 Cluster of Rust Resistance Genes and Their Use to Develop Isogenic Hard Red Spring Wheat Lines. Crop. Sci. 2003, 43, 1839–1847. [Google Scholar] [CrossRef]
- Friebe, B.; Badaeva, E.D.; Gill, B.S.; Tuleen, N.A. Cytogenetic identification of Triticum peregrinum chromosomes added to common wheat. Genome 1996, 39, 272–276. [Google Scholar] [CrossRef]
- Rosewarne, G.M.; Singh, R.P.; Huerta-Espino, J.; Herrera-Foessel, S.A.; Forrest, K.L.; Hayden, M.; Rebetzke, G.J. Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTLs associated with resistance in an Avocet × Pastor wheat population. Theor. Appl. Genet. 2012, 124, 1283–1294. [Google Scholar] [CrossRef]
- Dedryver, F.; Paillard, S.; Mallard, S.; Robert, O.; Trottet, M.; Negre, S.; Verplancke, G.; Jahier, J. Characterization of genetic components involved in durable resistance to stripe rust in the bread wheat “Renan”. Phytopathology 2009, 99, 968–973. [Google Scholar] [CrossRef]
- Sharma-Poudyal, D.; Chen, X.M.; Wan, A.M.; Zhan, G.M.; Kang, Z.S.; Cao, S.Q.; Jin, S.L.; Morgounov, A.; Akin, B.; Mert, Z.; et al. Virulence Characterization of International Collections of the Wheat Stripe Rust Pathogen, Puccinia striiformis f. sp. tritici. Plant. Dis. 2013, 97, 379–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Line, R.F.; Qayoum, A. Virulence, Aggressiveness, Evolution and Distribution of Races of Puccinia Striiformis (The Cause of Stripe Rust of Wheat) in North America; Technical Bulletin; United State Department of Agriculture, Agricultural Research Service: Washington, DC, USA, 1992; pp. 1968–1987. [Google Scholar]
- McNeal, F.H.; Konzak, C.F.; Smith, E.P.; Tate, W.S.; Russell, T.S. A uniform system for recording and processing cereal research data US Dept. Agric. Res. Serv. ARS 1971, 34, 121–143. [Google Scholar]
- Jarana, A.P.; Carandang, S.L.; Tandayu, E.S.; Cariño, G.A.; Sanchez, G.S.B.; Llantada, K.J. High-throughput sample collection and DNA extraction using PlantTrak and oKtopure systems at IRRI’s [International Rice Research Institute] genotyping services laboratory. Philipp. J. Crop. Sci. (Philipp.) 2015, 40, 86. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association Mapping in Structured Populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Breseghello, F.; Sorrells, M.E. Association Mapping of Kernel Size and Milling Quality in Wheat (Triticum aestivum L.) Cultivars. Genetics 2005, 172, 1165–1177. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2005, 38, 203–208. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]

| Parameter | PAK-IT | PAK-SEV | USA-IT | USA-SEV | ALL-IT | ALL-SEV |
|---|---|---|---|---|---|---|
| Range | 0-8 | 0-80 | 2-8 | 1-100 | 0-8 | 0-100 |
| Mean | 2.14 | 12.32 | 4.17 | 30.85 | 3.15 | 21.53 |
| Var(G) | 1.529 ** | 175.66 ** | 3.035 ** | 440.97 ** | 0.962 ** | 119.39 ** |
| Var(E) | 0.006 ns | 0.238 ns | 0.3 ns | 0.303 ns | 1.47 ns | 114.26 ns |
| Var(G*E) | 0.446 ** | 25.54 ** | 2.094 ** | 447.7 ** | 2.64 ** | 433.91 ** |
| Var(Total) | 1.98 | 201.44 | 5.429 | 888.98 | 5.07 | 667.57 |
| R2 | 0.87 | 0.93 | 0.76 | 0.687 | 0.54 | 0.43 |
| SD | 1.418 | 14.37 | 2.38 | 30.93 | 2.21 | 25.8 |
| H2 | 87 | 93 | 71 | 66 | 48 | 46 |
| QTL a | SNP Id. b | Chr. c | Position d (cm) | MAF e | −log10(P) f | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PK07-4 | PK07-12 | PK08-2 | PSTv-37 | PSTv-198 | PSTv-51 | PSTv-40 | PSTv-14 | PSTv-4 | |||||
| QYr.uaf-1A.1 | IWB3662 | 1A | 10.69 | 0.46 | 8.14 | 9.03 | 6.59 | _ | _ | _ | _ | _ | _ |
| QYr.uaf-1B.1 | IWB12258 | 1B | 28.76 | 0.24 | _ | _ | 4.77 | _ | _ | _ | _ | _ | _ |
| QYr.uaf-1D.1 | IWA7171 | 1D | 90.30 | 0.30 | _ | 4.57 | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-1D.2 | IWA4344 | 1D | 90.30 | 0.21 | _ | _ | _ | _ | _ | _ | _ | _ | 4.69 |
| QYr.uaf-2A.1 | IWB50806 | 2A | 81.49 | 0.47 | 4.42 | _ | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-2A.2 | IWA7638 | 2A | 162.89 | 0.46 | _ | _ | _ | _ | _ | _ | _ | 4.3 | _ |
| QYr.uaf-2B.1 | IWA2344 | 2B | 96.99 | 0.26 | _ | _ | _ | _ | 6.91 | 6.09 | 5.55 | _ | _ |
| QYr.uaf-2B.2 | IWA4097 | 2B | 113.86 | 0.10 | _ | _ | _ | _ | 8.36 | 7.53 | _ | _ | 5.32 |
| QYr.uaf-4A.1 | IWA3361 | 4A | 48.84 | 0.24 | _ | _ | _ | _ | _ | _ | _ | _ | 4.66 |
| QYr.uaf-4B.1 | IWB12434 | 4B | 74.62 | 0.46 | _ | 4.32 | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-4B.2 | IWA2031 | 4B | 98.65 | 0.16 | _ | _ | _ | _ | _ | _ | _ | 4.51 | _ |
| QYr.uaf-4B.3 | IWA408 | 4B | 114.87 | 0.30 | _ | _ | 4.23 | _ | _ | _ | _ | _ | _ |
| QYr.uaf-5B.1 | IWB5781 | 5B | 56.60 | 0.47 | _ | _ | _ | _ | _ | _ | _ | _ | 4.19 |
| QYr.uaf-6A.1 | IWA8028 | 6A | 77.14 | 0.27 | _ | _ | _ | _ | _ | _ | 4.06 | _ | _ |
| QYr.uaf-6A.2 | IWA3463 | 6A | 83.04 | 0.34 | _ | 4.02 | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-6A.3 | IWB25252 | 6A | 95.87 | 0.26 | _ | 4.28 | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-6B.1 | IWB7615 | 6B | 64.82 | 0.43 | _ | _ | _ | _ | _ | 4.81 | _ | _ | _ |
| QYr.uaf-6D.1 | IWB12259 | 6D | 155.10 | 0.18 | 4.51 | _ | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-7B.1 | IWA6857 | 7B | 76.31 | 0.08 | _ | 5.4 | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-7B.2 | IWB10895 | 7B | 103.21 | 0.45 | _ | _ | _ | _ | _ | _ | 4.07 | _ | _ |
| QTL a | SNP Id. b | Chr. c | Pos. d (cm) | MAF e | −log10(p) Based on IT f Score | −log10(p) Based on SEV g Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PAK-UAF16 | PAK-UAF17 | USA-MTV18 | USA-PCFS18 | BLUE-IT | PAK-UAF16 | PAK-UAF17 | USA-MTV18 | USA-PCFS18 | BLUE-SEV | |||||
| QYr.uaf-1A.2 | IWB3662 | 1A | 10.69 | 0.45 | _ | _ | _ | _ | _ | _ | _ | _ | _ | 3.24 |
| QYr.uaf-1A.3 | IWA2541 | 1A | 95.55 | 0.22 | _ | _ | _ | _ | 3.52 | _ | _ | _ | _ | _ |
| QYr.uaf-1A.4 | IWA4080 | 1A | 96.3 | 0.5 | _ | _ | 4.03 | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-1B.2 | IWA5278 | 1B | 70.08 | 0.34 | _ | _ | _ | 3.46 | _ | _ | _ | _ | 3.34 | 3.47 |
| QYr.uaf-1D.3 | IWA642 | 1D | 67.72 | 0.32 | _ | _ | _ | 3.16 | _ | _ | _ | _ | _ | _ |
| QYr.uaf-1D.2 | IWA4344 | 1D | 90.3 | 0.21 | 3.22 | _ | _ | _ | 3.27 | 3.08 | _ | _ | _ | _ |
| QYr.uaf-2A.3 | IWB11136 | 2A | 9.41 | 0.36 | _ | _ | 4.71 | 3.25 | 3 | _ | _ | 4.25 | 3.03 | 3.23 |
| QYr.uaf-2A.4 | IWB12554 | 2A | 143.65 | 0.28 | _ | _ | _ | _ | _ | _ | _ | 3.04 | _ | _ |
| QYr.uaf-2A.5 | IWA6798 | 2A | 150.11 | 0.4 | _ | _ | _ | _ | _ | _ | _ | 3.29 | _ | _ |
| QYr.uaf-2B.3 | IWA3478 | 2B | 130.62 | 0.31 | _ | _ | 5.19 | _ | _ | _ | _ | 5.7 | _ | 3.48 |
| QYr.uaf-2D.1 | IWA5673 | 2D | 82.82 | 0.16 | _ | _ | 3.32 | _ | _ | _ | _ | 3.32 | _ | _ |
| QYr.uaf-3A.1 | IWA288 | 3A | 49.1 | 0.32 | _ | _ | _ | 3.59 | _ | _ | _ | _ | _ | _ |
| QYr.uaf-3A.2 | IWB73771 | 3A | 66.48 | 0.37 | _ | _ | _ | 4.24 | _ | _ | _ | _ | _ | _ |
| QYr.uaf-3A.3 | IWB68593 | 3A | 195.1 | 0.36 | _ | _ | _ | _ | _ | _ | _ | 3.6 | _ | _ |
| QYr.uaf-3B.1 | IWB11085 | 3B | 25.09 | 0.15 | _ | _ | 3.41 | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-3B.2 | IWB11270 | 3B | 67.67 | 0.31 | _ | _ | 3.45 | _ | 3.57 | _ | _ | 3.8 | _ | 5.38 |
| QYr.uaf-3B.3 | IWB36652 | 3B | 71.65 | 0.39 | _ | _ | _ | 3.73 | _ | _ | _ | _ | 4.3 | _ |
| QYr.uaf-3D.1 | IWA4725 | 3D | 4.56 | 0.29 | _ | _ | _ | _ | _ | _ | 3.18 | _ | _ | _ |
| QYr.uaf-4B.4 | IWB60835 | 4B | 0 | 0.34 | 3.48 | _ | _ | _ | _ | _ | 3.9 | _ | _ | _ |
| QYr.uaf-4B.2 | IWA2031 | 4B | 98.65 | 0.16 | _ | _ | _ | 4.71 | 4.57 | _ | _ | _ | _ | 5.63 |
| QYr.uaf-5A.1 | IWB38719 | 5A | 88.7 | 0.39 | _ | _ | 3.13 | _ | 4.24 | _ | _ | _ | 4.13 | _ |
| QYr.uaf-5A.2 | IWA589 | 5A | 123.21 | 0.25 | _ | _ | _ | 3.12 | 4.87 | _ | _ | _ | 8.34 | 7.07 |
| QYr.uaf-5B.2 | IWB9459 | 5B | 110.56 | 0.25 | _ | _ | 4.2 | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-6A.4 | IWB29623 | 6A | 40.6 | 0.32 | _ | _ | 3.23 | _ | _ | _ | _ | 4.25 | _ | _ |
| QYr.uaf-6B.2 | IWB26626 | 6B | 113.67 | 0.39 | _ | _ | _ | _ | _ | _ | _ | _ | _ | 5.32 |
| QYr.uaf-6D.2 | IWA6673 | 6D | 9.47 | 0.38 | 3.05 | _ | _ | _ | _ | _ | _ | _ | _ | _ |
| QYr.uaf-7A.1 | IWA4173 | 7A | 218.7 | 0.48 | _ | _ | _ | _ | 3.56 | 3.15 | _ | _ | _ | _ |
| QYr.uaf-7B.3 | IWB22838 | 7B | 73.79 | 0.22 | _ | _ | _ | 3.58 | _ | _ | _ | _ | 6.3 | _ |
| QYr.uaf-7B.4 | IWA6401 | 7B | 77.73 | 0.34 | _ | _ | _ | _ | _ | _ | _ | _ | _ | 5.12 |
| QYr.uaf-7D.1 | IWB26628 | 7D | 144.96 | 0.44 | _ | _ | _ | _ | _ | _ | _ | _ | _ | 3.33 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habib, M.; Awan, F.S.; Sadia, B.; Zia, M.A. Genome-Wide Association Mapping for Stripe Rust Resistance in Pakistani Spring Wheat Genotypes. Plants 2020, 9, 1056. https://doi.org/10.3390/plants9091056
Habib M, Awan FS, Sadia B, Zia MA. Genome-Wide Association Mapping for Stripe Rust Resistance in Pakistani Spring Wheat Genotypes. Plants. 2020; 9(9):1056. https://doi.org/10.3390/plants9091056
Chicago/Turabian StyleHabib, Madiha, Faisal Saeed Awan, Bushra Sadia, and Muhammad Anjum Zia. 2020. "Genome-Wide Association Mapping for Stripe Rust Resistance in Pakistani Spring Wheat Genotypes" Plants 9, no. 9: 1056. https://doi.org/10.3390/plants9091056
APA StyleHabib, M., Awan, F. S., Sadia, B., & Zia, M. A. (2020). Genome-Wide Association Mapping for Stripe Rust Resistance in Pakistani Spring Wheat Genotypes. Plants, 9(9), 1056. https://doi.org/10.3390/plants9091056





