Document or Lose It—On the Importance of Information Management for Genetic Resources Conservation in Genebanks
Abstract
1. Introduction
2. Information Required
2.1. Basic Data
2.2. Stable and Unique Identifiers
2.3. Data for Collection Development
2.4. Securing the Legal Status of Acquisitions
2.5. Material Management Data
2.6. Data to Protect Against Losses
3. Information Management for Genetic Resources Conservation
3.1. Documentation Development
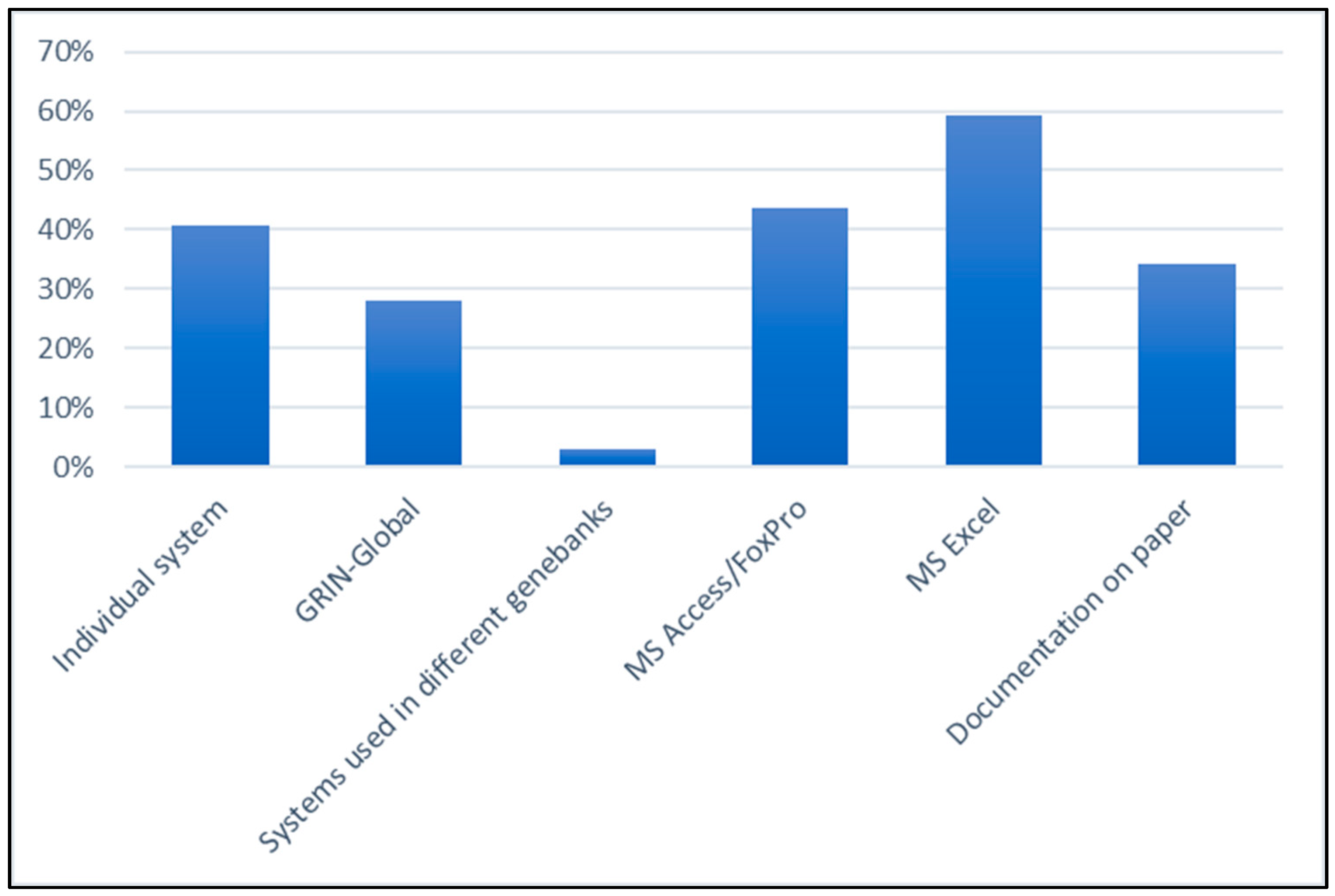
3.2. Current Situation
3.3. International Collaboration
4. Challenges
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grusak, M.A.; DellaPenna, D. Improving the Nutrient Composition of Plants to Enhance Human Nutrition and Health. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1999, 50, 133–161. [Google Scholar] [CrossRef] [PubMed]
- Metzger, J.O.; Bornscheuer, U. Lipids as renewable resources: Current state of chemical and biotechnological conversion and diversification. Appl. Microbiol. Biotechnol. 2006, 71, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Hill, J.; Lehman, C. Carbon-Negative Biofuels from Low-Input High-Diversity Grassland Biomass. Science 2006, 314, 1598–1600. [Google Scholar] [CrossRef] [PubMed]
- Tollefson, J. Humans are driving one million species to extinction. Nature 2019, 569, 171. [Google Scholar] [CrossRef] [PubMed]
- Hoisington, D.; Khairallah, M.; Reeves, T.; Ribaut, J.-M.; Skovmand, B.; Taba, S.; Warburton, M. Plant genetic resources: What can they contribute toward increased crop productivity? Proc. Natl. Acad. Sci. USA 1999, 96, 5937–5943. [Google Scholar] [CrossRef]
- Engels, J.M.M.; Maggioni, L. AEGIS: A regionally based approach to PGR conservation. In Agrobiodiversity Conservation: Securing the Diversity of Crop Wild Relatives and Landraces; Maxted, N., Dulloo, M.E., Ford-Lloyd, B.V., Frese, L., Iriondo, J.M., de Pinheiro Carvalho, M.A.A., Eds.; CABI: Wallingford, UK, 2012; pp. 321–326. [Google Scholar] [CrossRef]
- Keilwagen, J.; Kilian, B.; Özkan, H.; Babben, S.; Perovic, D.; Mayer, K.F.X.; Walther, A.; Poskar, C.H.; Ordon, F.; Eversole, K.; et al. Separating the wheat from the chaff—a strategy to utilize plant genetic resources from ex situ genebanks. Sci. Rep. 2014, 4, 5231. [Google Scholar] [CrossRef]
- Tanksley, S.D.; McCouch, S.R. Seed Banks and Molecular Maps: Unlocking Genetic Potential from the Wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef]
- Díez, M.J.; De la Rosa, L.; Martín, I.; Guasch, L.; Cartea, M.E.; Mallor, C.; Casals, J.; Simó, J.; Rivera, A.; Anastasio, G.; et al. Plant Genebanks: Present Situation and Proposals for Their Improvement. the Case of the Spanish Network. Front. Plant. Sci. 2018, 9, 1794. [Google Scholar] [CrossRef]
- Fowler, C.; Hodgkin, T. Plant Genetic Resources for Food and Agriculture: Assessing Global Availability. Annu. Rev. Environ. Resour. 2004, 29, 143–179. [Google Scholar] [CrossRef]
- FAO. The State of the World’s Plant Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 1997. [Google Scholar]
- FAO. The Second Report on the State of the World’s Plant. Genetic Resources for Food and Agriculture; Commission on Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2010. [Google Scholar]
- Alercia, A.; Diulgheroff, S.; Metz, T. FAO/IPGRI Multi-Crop. Passport Descriptors (MCPD); Food and Agriculture Organization of the United Nations (FAO): Rome, Italy; International Plant Genetic Resources Instit. (IPGRI): Rome, Italy, 2001. [Google Scholar]
- Alercia, A.; Diulgheroff, S.; Mackay, M. FAO/Bioversity Multi-Crop. Passport Descriptors, V.2.1 (MCPD V.2.1); Food and Agriculture Organization of the United Nations (FAO): Rome, Italy; Bioversity International: Rome, Italy, 2015. [Google Scholar]
- FAO. Genebank Standards for Plant. Genetic Resources for Food and Agriculture; Commission on Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014. [Google Scholar]
- Richards, K.; White, R.; Nicolson, N.; Pyle, R. A Beginner’s Guide to Persistent Identifiers; GBIF Secretariat: Copenhagen, Denmark, 2011. [Google Scholar] [CrossRef]
- Garrity, G.M.; Thompson, L.M.; Ussery, D.W.; Paskin, N.; Baker, D.; Desmeth, P.; Schindel, D.E.; Ong, P.S. Studies on Monitoring and Tracking Genetic Resources. Stand. Genom. Sci. 2009, 1, 78–86. [Google Scholar]
- Davidson, L.A.D.; Kimberly. Digital Object Identifiers: Promise and Problems for Scholarly Publishing. J. Electron. Publ. 1998, 4. [Google Scholar] [CrossRef]
- Paskin, N. Digital Object Identifier (DOI®) System. In Encyclopedia of Library and Information Sciences, 3rd ed.; Bates, M.J., Maack, M.N., Eds.; CRC Press: Boca Raton, FL, USA, 2009; pp. 1586–1592. [Google Scholar] [CrossRef]
- Alercia, A.; López, F.M.; Sackville Hamilton, N.R.; Marsella, M. Digital Object Identifiers for Food Crops—Descriptors and Guidelines of the Global Information System; Food and Agriculture Organization of the United Nations: Rome, Italy, 2018. [Google Scholar]
- Jennings, M.D. Gap analysis: Concepts, methods, and recent results. Landsc. Ecol. 2000, 15, 5–20. [Google Scholar] [CrossRef]
- Margules, C.R.; Pressey, R.L. Systematic conservation planning. Nature 2000, 405, 243–253. [Google Scholar] [CrossRef]
- Maxted, N.; Dulloo, E.V.; Ford-Lloyd, B.; Iriondo, J.M.; Jarvis, A. Gap analysis: A tool for complementary genetic conservation assessment. Divers. Distrib. 2008, 14, 1018–1030. [Google Scholar] [CrossRef]
- Aguirre-Gutiérrez, J.; van Treuren, R.; Hoekstra, R.; van Hintum, T.J.L. Crop wild relatives range shifts and conservation in Europe under climate change. Divers. Distrib. 2017, 23, 739–750. [Google Scholar] [CrossRef]
- Diederichsen, A. Duplication assessments in Nordic Avena sativa accessions at the Canadian national genebank. Genet. Resour. Crop. Evol. 2009, 56, 587–597. [Google Scholar] [CrossRef]
- Mascher, M.; Schreiber, M.; Scholz, U.; Graner, A.; Reif, J.C.; Stein, N. Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat. Genet. 2019, 51, 1076–1081. [Google Scholar] [CrossRef]
- Van Hintum, T.J.L.; Visser, D.L. Duplication within and between germplasm collections. Genet. Resour. Crop. Evol. 1995, 42, 135–145. [Google Scholar] [CrossRef]
- Van Treuren, R.; van Hintum, T.J.L.; van de Wiel, C.C.M. Marker-assisted optimization of an expert-based strategy for the acquisition of modern lettuce varieties to improve a genebank collection. Genet. Resour. Crop. Evol. 2008, 55, 319–330. [Google Scholar] [CrossRef]
- Engels, J.M.M.; Maggioni, L. Managing germplasm in a virtual European genebank (AEGIS) through networking. In Theorien der Lebendsammlung: Pflanzen, Mikroben und Tiere als Biofakte in Genbanken; Karafyllis, N.C., Ed.; Verlag Karl Alber: Freiburg, Germany, 2018; Volume 25, pp. 169–197. [Google Scholar]
- Barata, A.M.R.F.; Oliveira, J.; Lima, J.M.; Nobrega, H.; Pinheiro de Carvalho, M.A.A.; Dias, S. Implementation of a PGR global documentation system in Portugal. In Enhancing Crop Genepool Use: Capturing Wild Relative and Landrace Diversity for Crop Improvement; Maxted, N., Dulloo, M.E., Ford-Lloyd, B.V., Eds.; CABI: Wallingford, Oxfordshire, UK, 2016; pp. 441–452. [Google Scholar] [CrossRef]
- Qvenild, M. Svalbard Global Seed Vault: A ‘Noah’s Ark’ for the world’s seeds. Dev. Pract. 2008, 18, 110–116. [Google Scholar] [CrossRef]
- Fowler, C. The Svalbard Seed Vault and Crop Security. BioScience 2008, 58, 190–191. [Google Scholar] [CrossRef]
- Westengen, O.T.; Jeppson, S.; Guarino, L. Global Ex-Situ Crop Diversity Conservation and the Svalbard Global Seed Vault: Assessing the Current Status. PLoS ONE 2013, 8, e64146. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, G.; Karsch-Mizrachi, I.; Nakamura, Y.; Collaboration, o.b.o.t.I.N.S.D. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2010, 39, D15–D18. [Google Scholar] [CrossRef] [PubMed]
- Karsch-Mizrachi, I.; Nakamura, Y.; Cochrane, G.; International Nucleotide Sequence Database Collaboration. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2011, 40, D33–D37. [Google Scholar] [CrossRef]
- Seidewitz, L. Thesaurus for the International Standardisation of Gene Bank Documentation. Part, I. Cereals (incl. Maize, Rice and Sorghum); Genebank, institut fur Pflanzenbau und Saatgutgutforschung der Forschungsanstalt fur Landwirtschaft: Braunschweig, Germany, 1974. [Google Scholar]
- April, J. An information system to service the US National Germplasm System. In Technical Aspects of Information Management and Means of Communication in Plant Genetic Resources Work for a Future Utilization of Genetic Material in Plant Breeding, Braunschweig, Germany, 7–8 November 1978; Dambroth, M., Ed.; FAL: Braunschweig, Germany, 1979; pp. 73–78. [Google Scholar]
- Blixt, S. Germplasm Documentation: The Future. In Proceedings of the FAO/UNEP/IBPGR Technical Conference on Crop Genetic Resources, Rome, Italy, 6–10 April 1981. [Google Scholar]
- Knüpffer, H. Computer in Genbanken—Eine Übersicht. Die Kult. 1983, 31, 77–143. [Google Scholar] [CrossRef]
- Konopka, J.; Hanson, J. Information Handling Systems for Genebank Management; International Board for Plant Genetic Resources: Rome, Italy, 1985. [Google Scholar]
- Menting, F.B.J.; van Veller, M.G.P.; van Hintum, T.J.L. Genis Data Dictionary; Centre For Genetic Resources (GCN): Wageningen, DC, USA, 2007. [Google Scholar]
- Oppermann, M.; Weise, S.; Dittmann, C.; Knüpffer, H. GBIS: The information system of the German Genebank. Database 2015, 2015, bav021. [Google Scholar] [CrossRef]
- Agrawal, R.C.; Behera, D.; Saxena, S. Genebank Information Management System (GBIMS). Comput. Electron. Agric. 2007, 59, 90–96. [Google Scholar] [CrossRef]
- Khoury, C.; Laliberté, B.; Guarino, L. Trends in ex situ conservation of plant genetic resources: A review of global crop and regional conservation strategies. Genet. Resour. Crop. Evol. 2010, 57, 625–639. [Google Scholar] [CrossRef]
- Postman, J.; Hummer, K.; Ayala-Silva, T.; Bretting, P.; Franko, T.; Kinard, G.; Bohning, M.; Emberland, G.; Sinnott, Q.; Mackay, M.; et al. GRIN-Global: An international project to develop a global plant genebank information management system. Acta Hortic. 2010, 859, 49–55. [Google Scholar] [CrossRef]
- Gass, T.; Lipman, E.; Maggioni, L. The role of Central Crop Databases in the European Cooperative Programme for Crop Genetic Resources Networks (ECP/GR). In Central Crop Databases: Tools for Plant Genetic Resources Management; Lipman, E., Jongen, M.W.M., van Hintum, Th.J.L., Gass, T., Maggioni, L., Eds.; European Cooperative Programme for Crop Genetic Resources Networks (ECP/GR): Rome, Italy, 1997; pp. 22–29. [Google Scholar]
- Knüpffer, H. The European barley database of the ECP/GR: An introduction. Die Kult. 1988, 36, 135–162. [Google Scholar] [CrossRef]
- Perret, P.M. The ECP/GR Prunus Working Group: A collaborative action programme for Prunus genetic resources in Europe. Acta Hortic. 1988, 224, 19–32. [Google Scholar] [CrossRef]
- Van Hintum, T.J.L. Central Crop Databases—An overview. In Central Crop Databases: Tools for Plant Genetic Resources Management; Lipman, E., Jongen, M.W.M., van Hintum, Th.J.L., Gass, T., Maggioni, L., Eds.; European Cooperative Programme for Crop Genetic Resources Networks (ECP/GR): Rome, Italy, 1997; pp. 18–21. [Google Scholar]
- Hazekamp, T.; Serwinski, J.; Alercia, A. Multicrop passport descriptors. In Central Crop Databases: Tools for Plant Genetic Resources Management; Lipman, E., Jongen, M.W.M., van Hintum, Th.J.L., Gass, T., Maggioni, L., Eds.; European Cooperative Programme for Crop Genetic Resources Networks (ECP/GR): Rome, Italy, 1997; pp. 40–44. [Google Scholar]
- Wieczorek, J.; Bloom, D.; Guralnick, R.; Blum, S.; Döring, M.; Giovanni, R.; Robertson, T.; Vieglais, D. Darwin Core: An Evolving Community-Developed Biodiversity Data Standard. PLoS ONE 2012, 7, e29715. [Google Scholar] [CrossRef] [PubMed]
- Endresen, D.; Knüpffer, H. The Darwin Core extension for genebanks opens up new opportunities for sharing genebank datasets. Biodivers. Inform. 2012, 8, 12–29. [Google Scholar] [CrossRef]
- Weise, S.; Oppermann, M.; Maggioni, L.; van Hintum, T.; Knüpffer, H. EURISCO: The European search catalogue for plant genetic resources. Nucleic Acids Res. 2017, 45, D1003–D1008. [Google Scholar] [CrossRef]
- Mackay, M.; Taylor, M. Crop Trait Mining Informatics Platform (Crop TMIP); The University of Queensland: Gatton, Australia, 2014. [Google Scholar] [CrossRef]
- Krajewski, P.; Chen, D.; Ćwiek, H.; van Dijk, A.D.J.; Fiorani, F.; Kersey, P.; Klukas, C.; Lange, M.; Markiewicz, A.; Nap, J.P.; et al. Towards recommendations for metadata and data handling in plant phenotyping. J. Exp. Bot. 2015, 66, 5417–5427. [Google Scholar] [CrossRef]
- International Board for Plant Genetic Resources. Plum Descriptors; Commission of the European Communities (CEC): Rome, Italy, 1984. [Google Scholar]
- Williams, P.H. Descriptors for Brassica and Raphanus; International Board for Plant Genetic Resources: Rome, Italy, 1990. [Google Scholar]
- International Plant Genetic Resources Institution. Descriptors for Allium (Allium spp.); European Cooperative Programme for Crop Genetic Resources Networks (ECP/GR): Wellesbourne, UK, 2001. [Google Scholar]
- Maggioni, L.; van Hintum, T.J.L.; Lipman, E. Tailoring the Documentation of Plant Genetic Resources in Europe to the Needs of the User. In Proceedings of the Workshop of the ECPGR Documentation and Information Working Group, Prague, Czech Republic, 20–22 May 2014. [Google Scholar]
- Ćwiek-Kupczyńska, H.; Altmann, T.; Arend, D.; Arnaud, E.; Chen, D.; Cornut, G.; Fiorani, F.; Frohmberg, W.; Junker, A.; Klukas, C.; et al. Measures for interoperability of phenotypic data: Minimum information requirements and formatting. Plant. Methods 2016, 12, 44. [Google Scholar] [CrossRef]
- Papoutsoglou, E.A.; Faria, D.; Arend, D.; Arnaud, E.; Athanasiadis, I.N.; Chaves, I.; Coppens, F.; Cornut, G.; Costa, B.V.; Ćwiek-Kupczyńska, H.; et al. Enabling reusability of plant phenomic datasets with MIAPPE 1.1. New Phytol. 2020, 227, 260–273. [Google Scholar] [CrossRef]
- Shrestha, R.; Arnaud, E.; Mauleon, R.; Senger, M.; Davenport, G.F.; Hancock, D.; Morrison, N.; Bruskiewich, R.; McLaren, G. Multifunctional crop trait ontology for breeders’ data: Field book, annotation, data discovery and semantic enrichment of the literature. Aob Plants 2010, 2010, plq008. [Google Scholar] [CrossRef]
- Shrestha, R.; Matteis, L.; Skofic, M.; Portugal, A.; McLaren, G.; Hyman, G.; Arnaud, E. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice. Front. Physiol. 2012, 3, 326. [Google Scholar] [CrossRef]
- Raubach, S.; Kilian, B.; Dreher, K.; Amri, A.; Bassi, F.M.; Boukar, O.; Cook, D.; Cruickshank, A.; Fatokun, C.; El Haddad, N.; et al. From bits to bites: Advancement of the germinate platform to support genetic resources collections and pre-breeding informatics for crop wild relatives. Crop. Sci. 2020. [Google Scholar] [CrossRef]
- Van Treuren, R.; van Hintum, T.J.L. Next-generation genebanking: Plant genetic resources management and utilization in the sequencing era. Plant. Genet. Resour. 2014, 12, 298–307. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Ndjiondjop, M.-N.; Henry, R.J. Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief. Funct. Genom. 2018, 17, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Milner, S.G.; Jost, M.; Taketa, S.; Mazón, E.R.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea, M.; König, P.; et al. Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 2019, 51, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Wu, S.; Raupp, W.J.; Sehgal, S.; Arora, S.; Tiwari, V.; Vikram, P.; Singh, S.; Chhuneja, P.; Gill, B.S.; et al. Efficient curation of genebanks using next generation sequencing reveals substantial duplication of germplasm accessions. Sci. Rep. 2019, 9, 650. [Google Scholar] [CrossRef]
- González, M.Y.; Philipp, N.; Schulthess, A.W.; Weise, S.; Zhao, Y.; Börner, A.; Oppermann, M.; Graner, A.; Reif, J.C. Unlocking historical phenotypic data from an ex situ collection to enhance the informed utilization of genetic resources of barley (Hordeum sp.). Theor. Appl. Genet. 2018, 131, 2009–2019. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weise, S.; Lohwasser, U.; Oppermann, M. Document or Lose It—On the Importance of Information Management for Genetic Resources Conservation in Genebanks. Plants 2020, 9, 1050. https://doi.org/10.3390/plants9081050
Weise S, Lohwasser U, Oppermann M. Document or Lose It—On the Importance of Information Management for Genetic Resources Conservation in Genebanks. Plants. 2020; 9(8):1050. https://doi.org/10.3390/plants9081050
Chicago/Turabian StyleWeise, Stephan, Ulrike Lohwasser, and Markus Oppermann. 2020. "Document or Lose It—On the Importance of Information Management for Genetic Resources Conservation in Genebanks" Plants 9, no. 8: 1050. https://doi.org/10.3390/plants9081050
APA StyleWeise, S., Lohwasser, U., & Oppermann, M. (2020). Document or Lose It—On the Importance of Information Management for Genetic Resources Conservation in Genebanks. Plants, 9(8), 1050. https://doi.org/10.3390/plants9081050