The Existence and Localization of Nuclear snoRNAs in Arabidopsis thaliana Revisited
Abstract
1. Introduction
2. Results
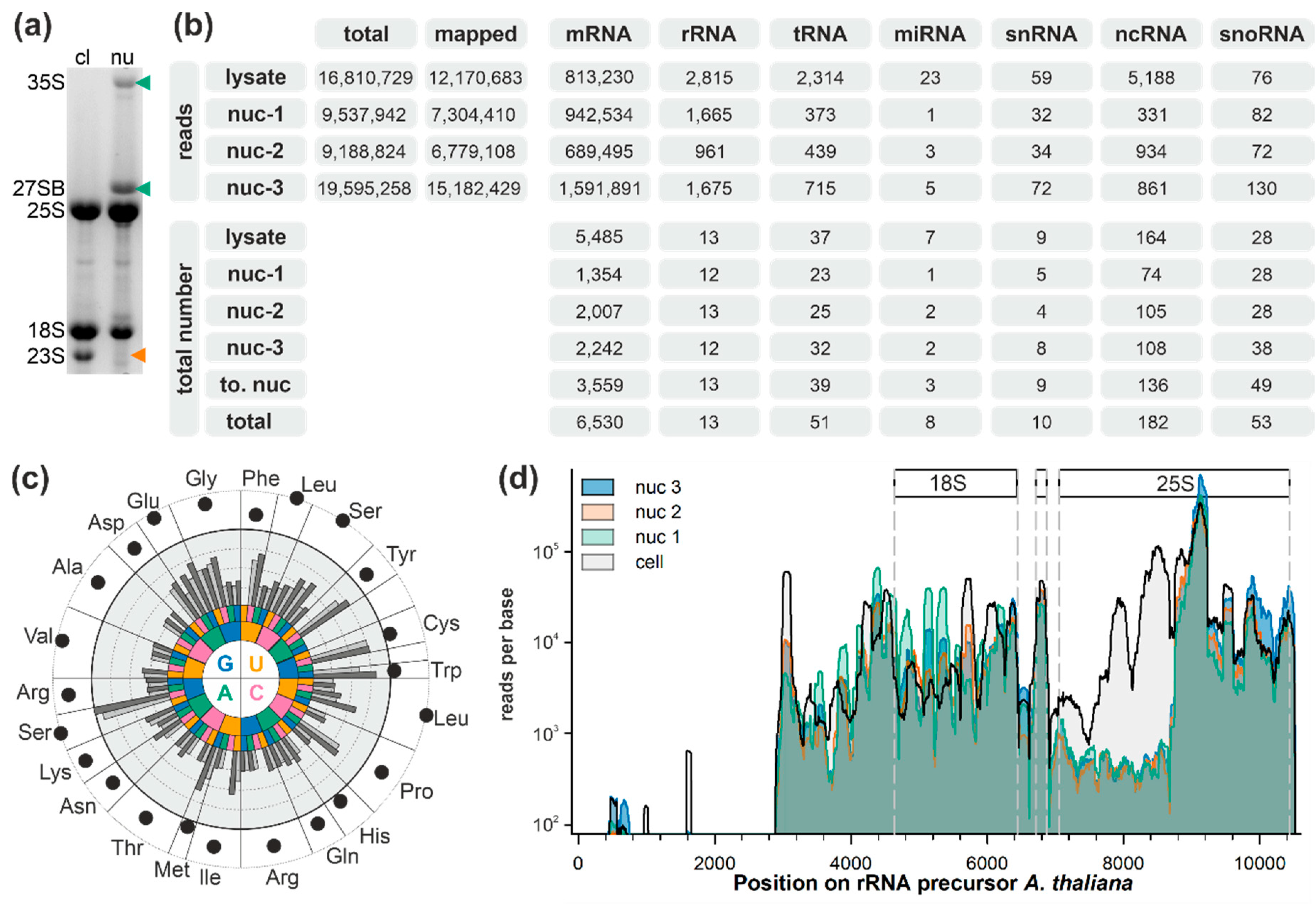
2.1. Analysis of Small RNAs in Total Cell and Nuclear Lysates
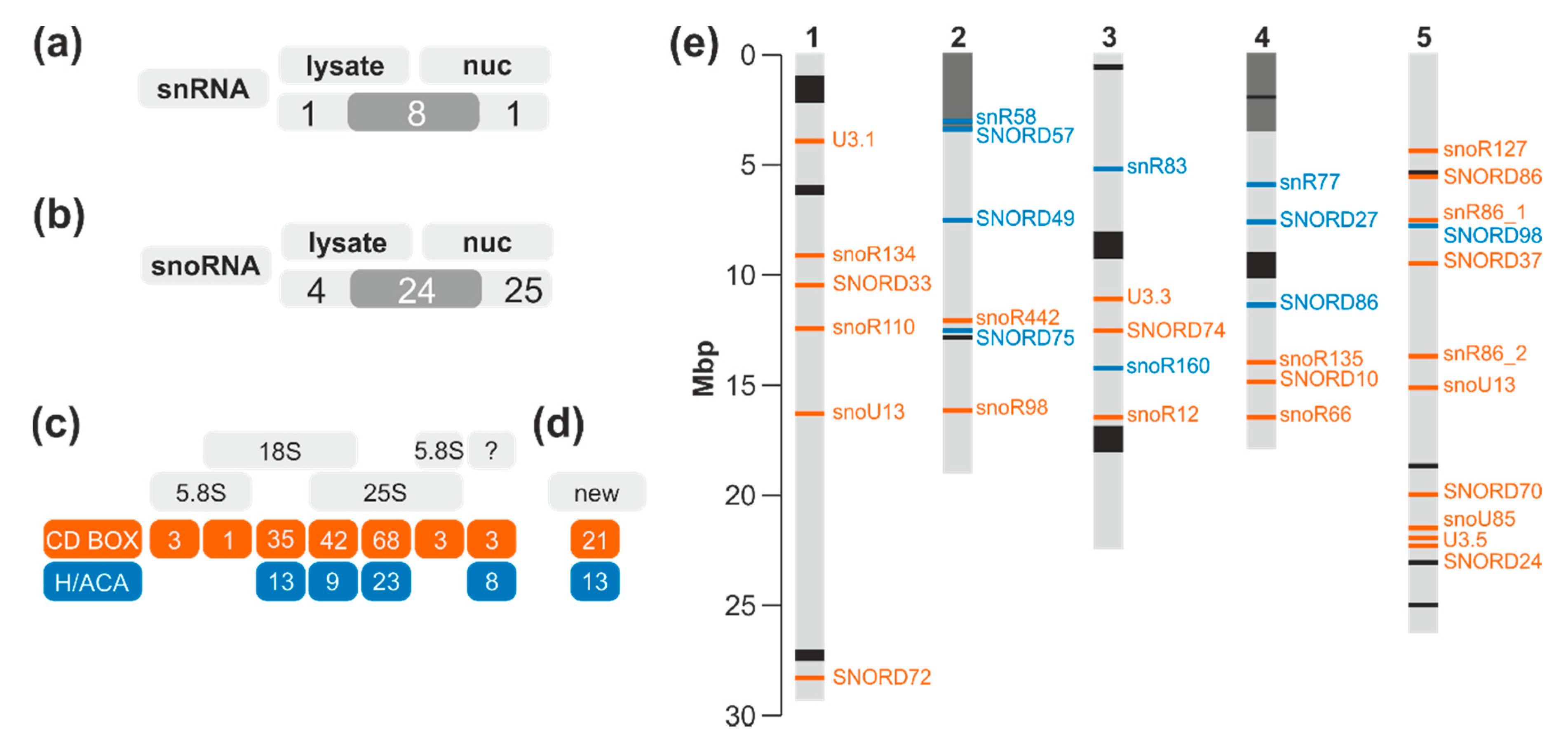
2.2. The snRNA and snoRNA Content in the Nucleus
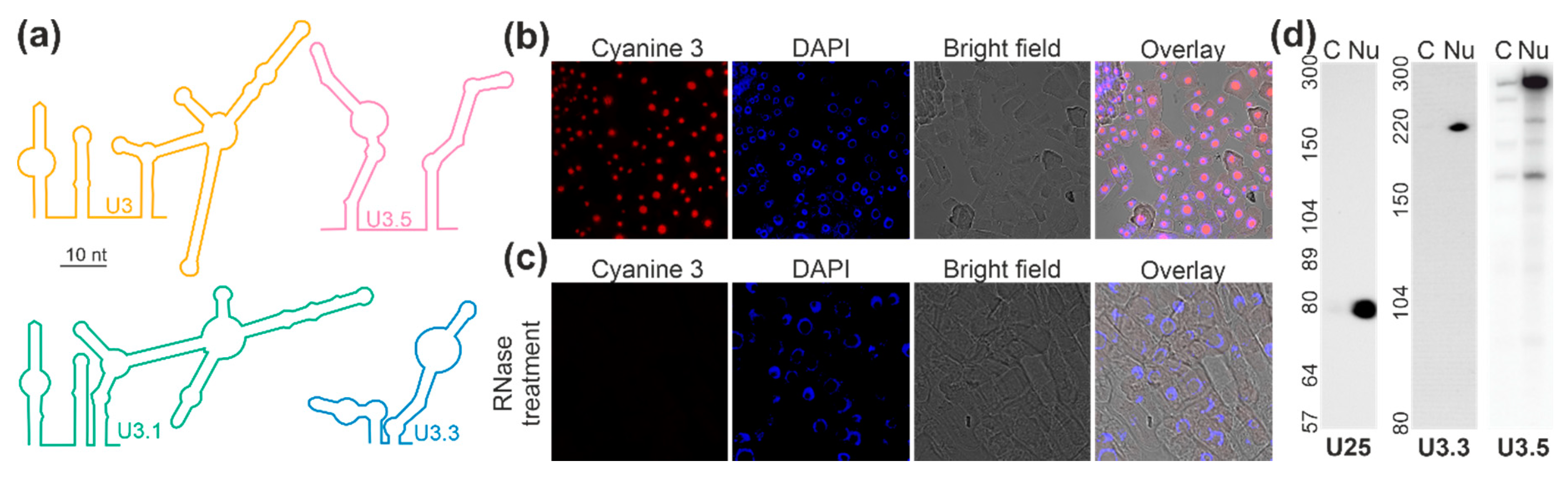
2.3. Localization of the U3 snoRNAs of the C/D Box Family
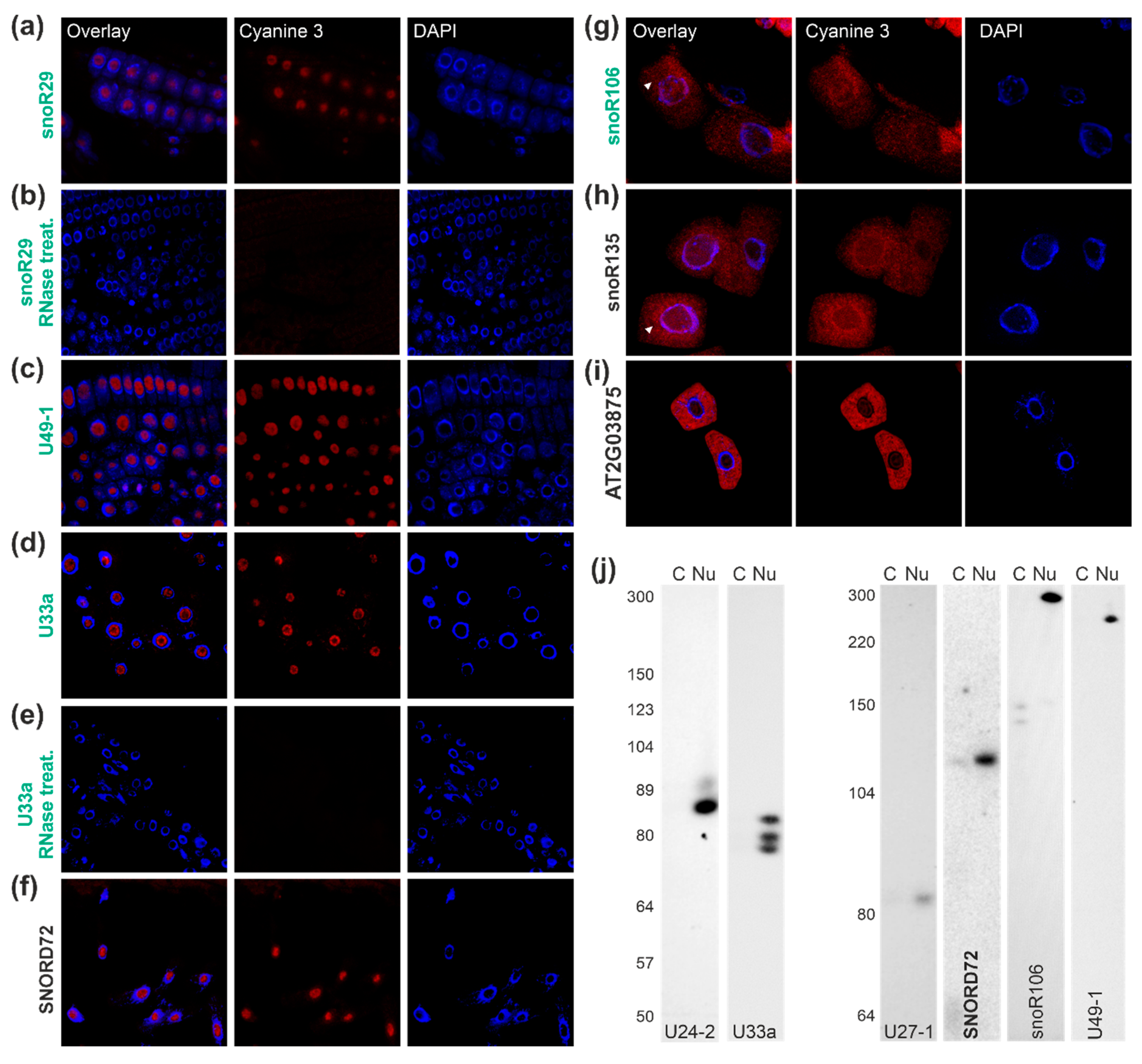
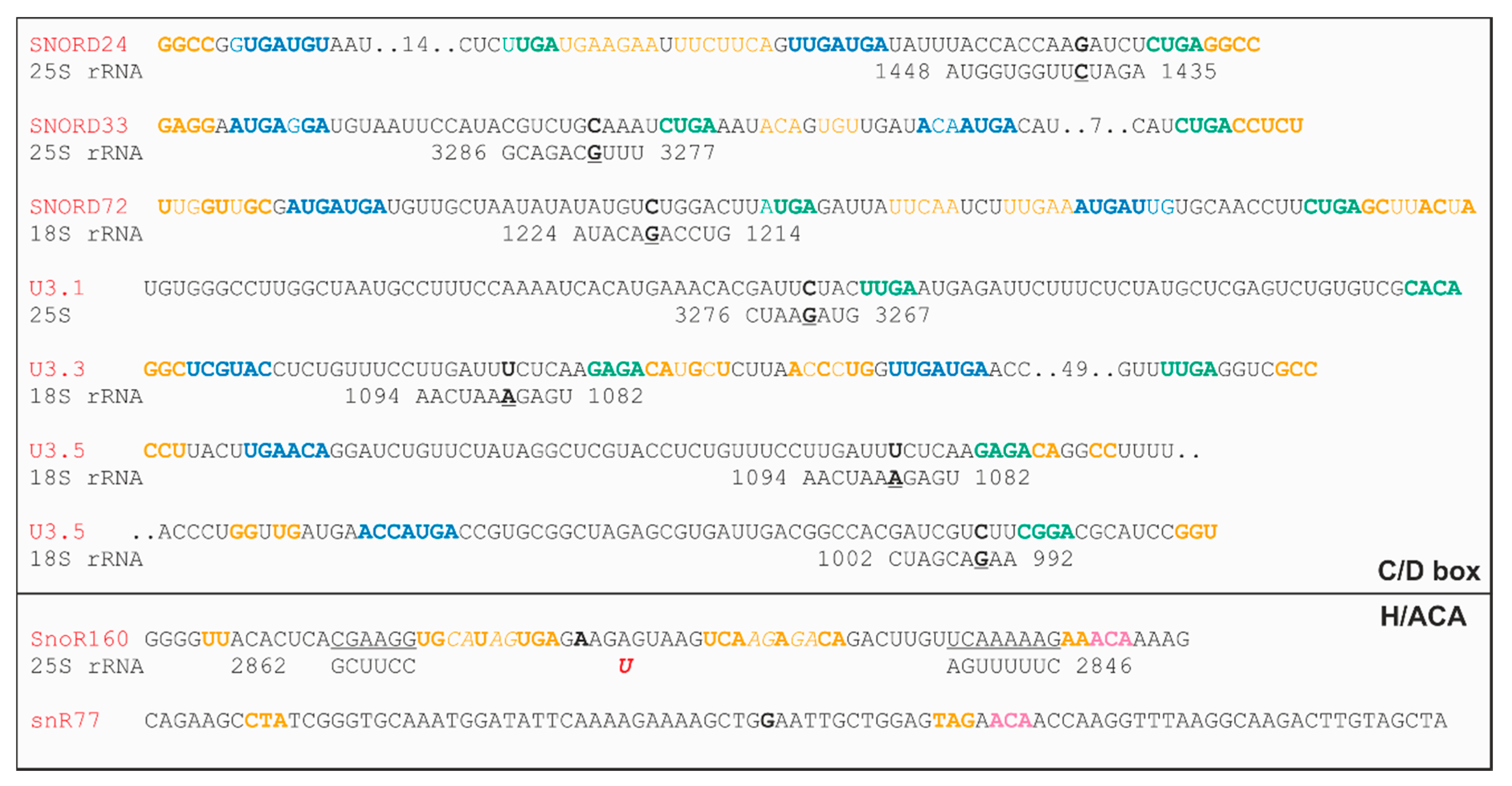
2.4. Localization of snoRNAs of the C/D Box Family
2.5. Localization of Two snoRNAs of the H/ACA Box Family
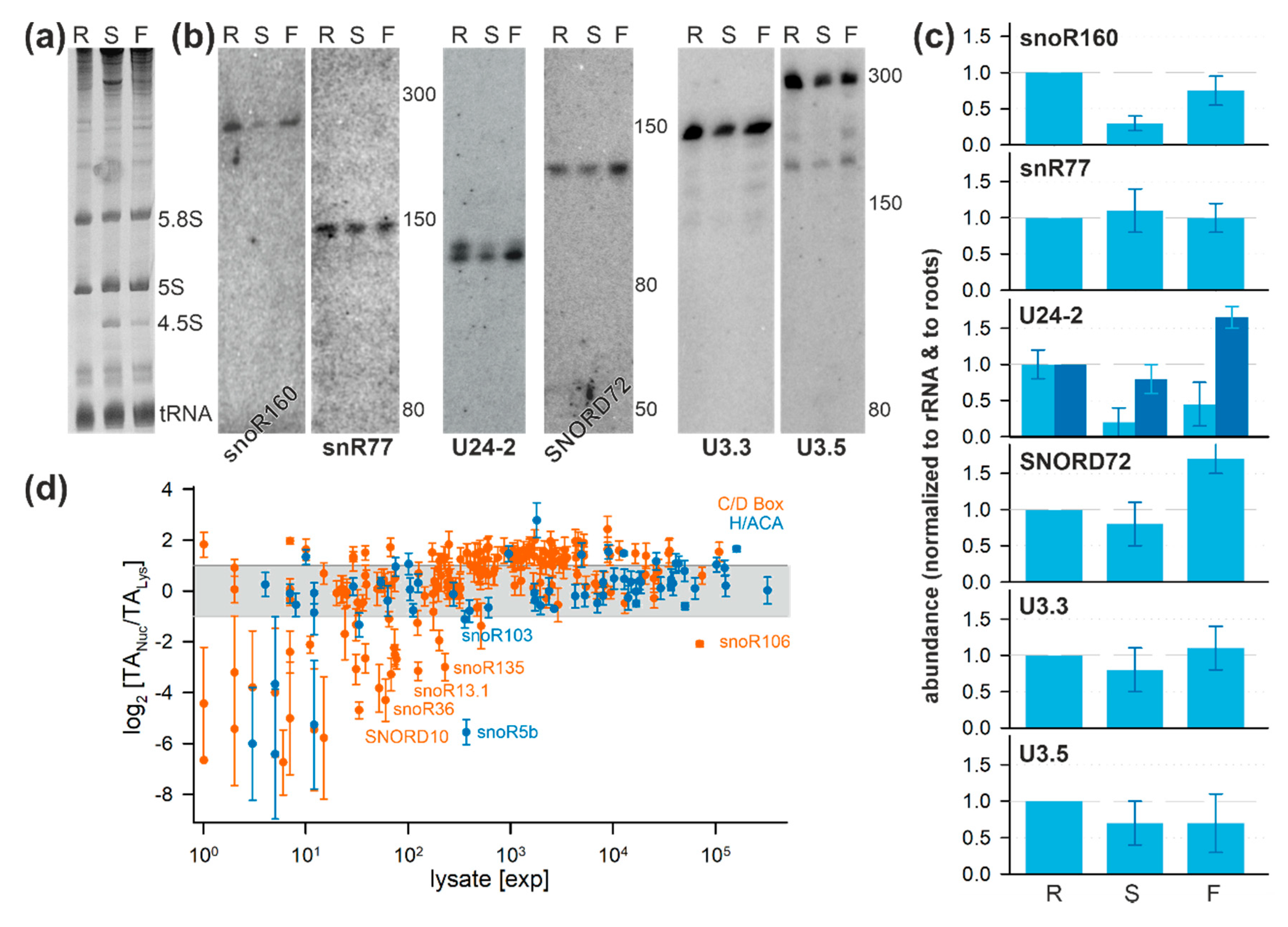
2.6. Tissue-Specific Localization of Selected snoRNAs
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weis, B.L.; Kovacevic, J.; Missbach, S.; Schleiff, E. Plant-Specific Features of Ribosome Biogenesis. Trends Plant Sci. 2015, 20, 729–740. [Google Scholar] [CrossRef]
- Tomecki, R.; Sikorski, P.J.; Zakrzewska-Placzek, M. Comparison of preribosomal RNA processing pathways in yeast, plant and human cells-focus on coordinated action of endo- and exoribonucleases. FEBS Lett. 2017, 591, 1801–1850. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Vásquez, J.; Delseny, M. Ribosome Biogenesis in Plants: From Functional 45S Ribosomal DNA Organization to Ribosome Assembly Factors. Plant Cell. 2019, 31, 1945–1967. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Klinge, S.; Woolford, J.L., Jr. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef] [PubMed]
- Baßler, J.; Hurt, E. Eukaryotic Ribosome Assembly. Annu. Rev. Biochem. 2019, 88, 281–306. [Google Scholar] [CrossRef]
- van Sluis, M.; McStay, B. Nucleolar reorganization in response to rDNA damage. Curr. Opin. Cell Biol. 2017, 46, 81–86. [Google Scholar] [CrossRef]
- Tiku, V.; Antebi, A. Nucleolar Function in Lifespan Regulation. Trends Cell Biol. 2018, 28, 662–672. [Google Scholar] [CrossRef]
- Kalinina, N.O.; Makarova, S.; Makhotenko, A.; Love, A.J.; Taliansky, M. The Multiple Functions of the Nucleolus in Plant Development, Disease and Stress Responses. Front. Plant Sci. 2018, 9, 132. [Google Scholar] [CrossRef]
- Ohbayashi, I.; Sugiyama, M. Plant Nucleolar Stress Response, a New Face in the NAC-Dependent Cellular Stress Responses. Front. Plant Sci. 2018, 8, 2247. [Google Scholar] [CrossRef]
- Correll, C.C.; Bartek, J.; Dundr, M. The Nucleolus: A Multiphase Condensate Balancing Ribosome Synthesis and Translational Capacity in Health, Aging and Ribosomopathies. Cells 2019, 8, 869. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.W.; Green, R. Ribosomopathies: There’s strength in numbers. Science 2017, 358, eaan2755. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Thomas, G.; Volarević, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Farley-Barnes, K.I.; Ogawa, L.M.; Baserga, S.J. Ribosomopathies: Old Concepts, New Controversies. Trends Genet. 2019, 35, 754–767. [Google Scholar] [CrossRef]
- Catez, F.; Dalla Venezia, N.; Marcel, V.; Zorbas, C.; Lafontaine, D.L.J.; Diaz, J.J. Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem. Pharmacol. 2019, 159, 74–81. [Google Scholar] [CrossRef]
- Cerezo, E.; Plisson-Chastang, C.; Henras, A.K.; Lebaron, S.; Gleizes, P.-E.; O’Donohue, M.-F.; Romeo, Y.; Henry, Y. Maturation of pre-40S particles in yeast and humans. Wiley Interdiscip. Rev. RNA 2019, 10, e1516. [Google Scholar] [CrossRef]
- Joazeiro, C.A.P. Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol. Cell Biol. 2019, 20, 368–383. [Google Scholar] [CrossRef]
- Beese, C.J.; Brynjólfsdóttir, S.H.; Frankel, L.B. Selective Autophagy of the Protein Homeostasis Machinery: Ribophagy, Proteaphagy and ER-Phagy. Front. Cell Dev. Biol. 2020, 7, 373. [Google Scholar] [CrossRef]
- Ojha, S.; Malla, S.; Lyons, S.M. snoRNPs: Functions in Ribosome Biogenesis. Biomolecules 2020, 10, 783. [Google Scholar] [CrossRef]
- Dupuis-Sandoval, F.; Poirier, M.; Scott, M.S. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip. Rev. RNA 2015, 6, 381–397. [Google Scholar] [CrossRef]
- Yu, G.; Zhao, Y.; Li, H. The multistructural forms of box C/D ribonucleoprotein particles. RNA 2018, 24, 1625–1633. [Google Scholar] [CrossRef]
- Yan, Q.; Zhu, C.; Guang, S.; Feng, X. The Functions of Non-coding RNAs in rRNA Regulation. Front. Genet. 2019, 10, 290. [Google Scholar] [CrossRef] [PubMed]
- Hutzinger, R.; Feederle, R.; Mrazek, J.; Schiefermeier, N.; Balwierz, P.J.; Zavolan, M.; Polacek, N.; Delecluse, H.J.; Hüttenhofer, A. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009, 5, e1000547. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.W.; Clark, G.P.; Leader, D.J.; Simpson, C.G.; Lowe, T. Multiple snoRNA gene clusters from Arabidopsis. RNA 2001, 7, 1817–1832. [Google Scholar] [PubMed]
- Zhu, P.; Wang, Y.; Qin, N.; Wang, F.; Wang, J.; Deng, X.W.; Zhu, D. Arabidopsis small nucleolar RNA monitors the efficient pre-rRNA processing during ribosome biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, 11967–11972. [Google Scholar] [CrossRef]
- Barneche, F.; Gaspin, C.; Guyot, R.; Echeverría, M. Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: Extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-O-methylation sites. J. Mol. Biol. 2001, 311, 57–73. [Google Scholar] [CrossRef]
- Rodor, J.; Jobet, E.; Bizarro, J.; Vignols, F.; Carles, C.; Suzuki, T.; Nakamura, K.; Echeverría, M. AtNUFIP, an essential protein for plant development, reveals the impact of snoRNA gene organisation on the assembly of snoRNPs and rRNA methylation in Arabidopsis thaliana. Plant J. 2011, 65, 807–819. [Google Scholar] [CrossRef]
- Ballarino, M.; Morlando, M.; Pagano, F.; Fatica, A.; Bozzoni, I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 2005, 25, 5396–5403. [Google Scholar] [CrossRef]
- Liang, X.H.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef]
- Taoka, M.; Nobe, Y.; Yamaki, Y.; Sato, K.; Ishikawa, H.; Izumikawa, K.; Yamauchi, Y.; Hirota, K.; Nakayama, H.; Takahashi, N.; et al. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018, 46, 9289–9298. [Google Scholar] [CrossRef]
- Armanios, M.; Chen, J.L.; Chang, Y.P.; Brodsky, R.A.; Hawkins, A.; Griffin, C.A.; Eshleman, J.R.; Cohen, A.R.; Chakravarti, A.; Hamosh, A.; et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl. Acad. Sci. USA 2005, 102, 15960–15964. [Google Scholar] [CrossRef]
- Horiguchi, G.; Molla´-Morales, A.; Pe´rez-Pe´rez, J.M.; Kojima, K.; Robles, P.; Micol-Ponce, R.; Micol, J.L.; Tsukaya, H. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 2011, 65, 724–736. [Google Scholar] [CrossRef] [PubMed]
- Micol-Ponce, R.; Sarmiento-Mañús, R.; Ruiz-Bayón, A.; Montacié, C.; Sáez-Vasquez, J.; Ponce, M.R. Arabidopsis RIBOSOMAL RNA PROCESSING7 is Required for 18S rRNA Maturation. Plant Cell. 2018, 30, 2855–2872. [Google Scholar] [CrossRef] [PubMed]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2016, 113, E1625–E1634. [Google Scholar] [CrossRef] [PubMed]
- Holley, C.L.; Li, M.W.; Scruggs, B.S.; Matkovich, S.J.; Ory, D.S.; Schaffer, J.E. Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J. Biol. Chem. 2015, 290, 11741–11748. [Google Scholar] [CrossRef]
- Li, M.W.; Sletten, A.C.; Lee, J.; Pyles, K.D.; Matkovich, S.J.; Ory, D.S.; Schaffer, J.E. Nuclear export factor 3 regulates localization of small nucleolar RNAs. J. Biol. Chem. 2017, 292, 20228–20239. [Google Scholar] [CrossRef]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.F.; Chakraborty, A.; Gleizes, P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef]
- Kiss, T.; Marshallsay, C.; Filipowicz, W. Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell 1991, 65, 517–526. [Google Scholar] [CrossRef]
- Leader, D.J.; Sanders, J.F.; Waugh, R.; Shaw, P.; Brown, J.W. Molecular characterisation of plant U14 small nucleolar RNA genes: Closely linked genes are transcribed as polycistronic U14 transcripts. Nucleic Acids Res. 1994, 22, 5196–5203. [Google Scholar] [CrossRef]
- Leader, D.J.; Clark, G.P.; Watters, J.; Beven, A.F.; Shaw, P.J.; Brown, J.W. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997, 16, 5742–5751. [Google Scholar] [CrossRef]
- Brown, J.W.; Shaw, P.J. Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell. 1998, 10, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Qu, L.H.; Meng, Q.; Zhou, H.; Chen, Y.Q. Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 2001, 29, 1623–1630. [Google Scholar] [PubMed]
- Marker, C.; Zemann, A.; Terhörst, T.; Kiefmann, M.; Kastenmayer, J.P.; Green, P.; Bachellerie, J.P.; Brosius, J.; Hüttenhofer, A. Experimental RNomics: Identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol. 2002, 12, 2002–2013. [Google Scholar] [CrossRef]
- Chen, C.L.; Liang, D.; Zhou, H.; Zhuo, M.; Chen, Y.Q.; Qu, L.H. The high diversity of snoRNAs in plants: Identification and comparative study of 120 snoRNA genes from Oryza sativa. Nucleic Acids Res. 2003, 31, 2601–2613. [Google Scholar] [CrossRef]
- Li, W.; Jiang, G.; Zeng, D.; Jin, Y. Identification of six new box C/D snoRNA gene clusters from rice. IUBMB Life 2007, 59, 664–674. [Google Scholar] [CrossRef]
- Chen, H.M.; Wu, S.H. Mining small RNA sequencing data: A new approach to identify small nucleolar RNAs in Arabidopsis. Nucleic Acids Res. 2009, 37, e69. [Google Scholar] [CrossRef]
- Hoeppner, M.P.; Poole, A.M. Comparative genomics of eukaryotic small nucleolar RNAs reveals deep evolutionary ancestry amidst ongoing intragenomic mobility. BMC Evol. Biol. 2012, 12, 183. [Google Scholar] [CrossRef]
- Patra Bhattacharya, D.; Canzler, S.; Kehr, S.; Hertel, J.; Grosse, I.; Stadler, P.F. Phylogenetic distribution of plant snoRNA families. BMC Genom. 2016, 17, 969. [Google Scholar] [CrossRef]
- Qu, G.; Kruszka, K.; Plewka, P.; Yang, S.Y.; Chiou, T.J.; Jarmolowski, A.; Szweykowska-Kulinska, Z.; Echeverria, M.; Karlowski, W.M. Promoter-based identification of novel non-coding RNAs reveals the presence of dicistronic snoRNA-miRNA genes in Arabidopsis thaliana. BMC Genom. 2015, 16, 1009. [Google Scholar] [CrossRef]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs derived from snoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.T.; Zhu, D.; Chen, W.; Deng, W.; He, H.; He, G.; Bai, B.; Qi, Y.; Chen, R.; Deng, X.W. A global identification and analysis of small nucleolar RNAs and possible intermediate-sized non-coding RNAs in Oryza sativa. Mol. Plant. 2013, 6, 830–846. [Google Scholar] [CrossRef]
- Zheng, J.; Zeng, E.; Du, Y.; He, C.; Hu, Y.; Jiao, Z.; Wang, K.; Li, W.; Ludens, M.; Fu, J.; et al. Temporal Small RNA Expression Profiling under Drought Reveals a Potential Regulatory Role of Small Nucleolar RNAs in the Drought Responses of Maize. Plant Genome 2019, 12, 1–15. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Sun, Q.; Yao, Y. Characterization of Small RNAs Derived from tRNAs, rRNAs and snoRNAs and Their Response to Heat Stress in Wheat Seedlings. PLoS ONE 2016, 11, e0150933. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.K.; Kumar, V.; Shukla, V.; Mattoo, R.; Liu, Y.; Chung, S.H.; Giovannoni, J.J.; Mattoo, A.K. Features of a unique intronless cluster of class I small heat shock protein genes in tandem with box C/D snoRNA genes on chromosome 6 in tomato (Solanum lycopersicum). Planta 2012, 235, 453–471. [Google Scholar] [CrossRef] [PubMed]
- Palm, D.; Streit, D.; Shanmugam, T.; Weis, B.L.; Ruprecht, M.; Simm, S.; Schleiff, E. Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing. Nucleic Acids Res. 2019, 47, 1880–1895. [Google Scholar] [CrossRef]
- Weis, B.L.; Missbach, S.; Marzi, J.; Bohnsack, M.T.; Schleiff, E. The 60S associated ribosome biogenesis factor LSG1–2 is required for 40S maturation in Arabidopsis thaliana. Plant J. 2014, 80, 1043–1056. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef]
- Michaud, M.; Cognat, V.; Duchêne, A.M.; Maréchal-Drouard, L. A global picture of tRNA genes in plant genomes. Plant J. 2011, 66, 80–93. [Google Scholar] [CrossRef]
- Hildebrandt, T.M.; Nunes Nesi, A.; Araújo, W.L.; Braun, H.P. Amino Acid Catabolism in Plants. Mol. Plant 2015, 8, 1563–1579. [Google Scholar] [CrossRef]
- Varenne, S.; Buc, J.; Lloubes, R.; Lazdunski, C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 1984, 180, 549–576. [Google Scholar] [CrossRef]
- Gingold, H.; Tehler, D.; Christoffersen, N.R.; Nielsen, M.M.; Asmar, F.; Kooistra, S.M.; Christophersen, N.S.; Christensen, L.L.; Borre, M.; Sørensen, K.D.; et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 2014, 158, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Nguyen, H.C.B.; Zhang, S.; Dill, B.D.; Molina, H.; Tavazoie, S.F. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell 2016, 165, 1416–1427. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Kawamata, T.; Horie, T.; Tsugawa, H.; Nakayama, Y.; Ohsumi, Y.; Fukusaki, E. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 2015, 34, 154–168. [Google Scholar] [CrossRef]
- Liu, Y.; Zou, W.; Yang, P.; Wang, L.; Ma, Y.; Zhang, H.; Wang, X. Autophagy-dependent ribosomal RNA degradation is essential for maintaining nucleotide homeostasis during C. elegans development. Elife 2018, 7, e36588. [Google Scholar] [CrossRef]
- Yoshihama, M.; Nakao, A.; Kenmochi, N. snOPY: A small nucleolar RNA orthological gene database. BMC Res. Notes 2013, 6, 426. [Google Scholar] [CrossRef]
- Brown, J.W.; Echeverria, M.; Qu, L.H.; Lowe, T.M.; Bachellerie, J.P.; Hüttenhofer, A.; Kastenmayer, J.P.; Green, P.J.; Shaw, P.; Marshall, D.F. Plant snoRNA database. Nucleic Acids Res. 2003, 31, 432–435. [Google Scholar] [CrossRef]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, J.; Ye, K. Structural and functional analysis of the U3 snoRNA binding protein Rrp9. RNA 2013, 19, 701–711. [Google Scholar] [CrossRef]
- Ebersberger, I.; Simm, S.; Leisegang, M.S.; Schmitzberger, P.; Mirus, O.; von Haeseler, A.; Bohnsack, M.T.; Schleiff, E. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 2014, 42, 1509–1523. [Google Scholar] [CrossRef]
- Simm, S.; Fragkostefanakis, S.; Paul, P.; Keller, M.; Einloft, J.; Scharf, K.D.; Schleiff, E. Identification and Expression Analysis of Ribosome Biogenesis Factor Co-orthologs in Solanum lycopersicum. Bioinform. Biol. Insights 2015, 4, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Marz, M.; Gruber, A.R.; Höner Zu Siederdissen, C.; Amman, F.; Badelt, S.; Bartschat, S.; Bernhart, S.H.; Beyer, W.; Kehr, S.; Lorenz, R.; et al. Animal snoRNAs and scaRNAs with exceptional structures. RNA Biol. 2011, 8, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xu, Y.; Bai, S.; Bai, X.; Zhu, H.; Dong, H.; Wang, W.; Zhu, X.; Hao, F.; Song, C.P. Transcriptome-wide analysis of pseudouridylation of mRNA and non-coding RNAs in Arabidopsis. J. Exp. Bot. 2019, 70, 5089–5600. [Google Scholar] [CrossRef] [PubMed]
- Meier, U.T. RNA modification in Cajal bodies. RNA Biol. 2017, 14, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Nostramo, R.T.; Hopper, A.K. Beyond rRNA and snRNA: tRNA as a 2′-O-methylation target for nucleolar and Cajal body box C/D RNPs. Genes Dev. 2019, 33, 739–740. [Google Scholar] [CrossRef]
- Yin, Q.F.; Yang, L.; Zhang, Y.; Xiang, J.F.; Wu, Y.W.; Carmichael, G.G.; Chen, L.L. Long noncoding RNAs with snoRNA ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef]
- Falaleeva, M.; Stamm, S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays 2013, 35, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Mleczko, A.M.; Machtel, P.; Walkowiak, M.; Wasilewska, A.; Pietras, P.J.; Bąkowska-Żywicka, K. Levels of sdRNAs in cytoplasm and their association with ribosomes are dependent upon stress conditions but independent from snoRNA expression. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Bratkovič, T.; Božič, J.; Rogelj, B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 2020, 48, 1627–1651. [Google Scholar] [CrossRef]
- Palm, D.; Simm, S.; Darm, K.; Weis, B.L.; Ruprecht, M.; Schleiff, E.; Scharf, C. Proteome distribution between nucleoplasm and nucleolus and its relation to ribosome biogenesis in Arabidopsis thaliana. RNA Biol. 2016, 13, 441–454. [Google Scholar] [CrossRef]
- McKeown, P.; Pendle, A.F.; Shaw, P.J. Preparation of Arabidopsis nuclei and nucleoli. Methods Mol. Biol. 2008, 463, 67–75. [Google Scholar] [PubMed]
- Sedlazeck, F.J.; Rescheneder, P.; von Haeseler, A. NextGenMap: Fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 2013, 29, 2790–2791. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Cognat, V.; Pawlak, G.; Duchêne, A.M.; Daujat, M.; Gigant, A.; Salinas, T.; Michaud, M.; Gutmann, B.; Giegé, P.; Gobert, A.; et al. PlantRNA, a database for tRNAs of photosynthetic eukaryotes. Nucleic Acids Res. 2013, 41, D273–D279. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Missbach, S.; Weis, B.L.; Martin, R.; Simm, S.; Bohnsack, M.T.; Schleiff, E. 40S ribosome biogenesis co-factors are essential for gametophyte and embryo development. PLoS ONE 2013, 8, e54084. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. A computational screen for methylation guide snoRNAs in yeast. Science 1999, 283, 1168–1171. [Google Scholar] [CrossRef]
- Tafer, H.; Kehr, S.; Hertel, J.; Hofacker, I.L.; Stadler, P.F. RNAsnoop: Efficient target prediction for H/ACA snoRNAs. Bioinformatics 2010, 26, 610–616. [Google Scholar] [CrossRef]
- Duncan, S.; Olsson, T.S.; Hartley, M.; Dean, C.; Rosa, S. Single molecule RNA FISH in Arabidopsis root cells. Bio-Protocol 2017, 7, e2240. [Google Scholar] [CrossRef]
- Abràmo, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Streit, D.; Shanmugam, T.; Garbelyanski, A.; Simm, S.; Schleiff, E. The Existence and Localization of Nuclear snoRNAs in Arabidopsis thaliana Revisited. Plants 2020, 9, 1016. https://doi.org/10.3390/plants9081016
Streit D, Shanmugam T, Garbelyanski A, Simm S, Schleiff E. The Existence and Localization of Nuclear snoRNAs in Arabidopsis thaliana Revisited. Plants. 2020; 9(8):1016. https://doi.org/10.3390/plants9081016
Chicago/Turabian StyleStreit, Deniz, Thiruvenkadam Shanmugam, Asen Garbelyanski, Stefan Simm, and Enrico Schleiff. 2020. "The Existence and Localization of Nuclear snoRNAs in Arabidopsis thaliana Revisited" Plants 9, no. 8: 1016. https://doi.org/10.3390/plants9081016
APA StyleStreit, D., Shanmugam, T., Garbelyanski, A., Simm, S., & Schleiff, E. (2020). The Existence and Localization of Nuclear snoRNAs in Arabidopsis thaliana Revisited. Plants, 9(8), 1016. https://doi.org/10.3390/plants9081016




