Genebank Phenomics: A Strategic Approach to Enhance Value and Utilization of Crop Germplasm
Abstract
1. Introduction
2. Phenomics to Unlock the Genetic Potential of Genebank Germplasm
2.1. Plant Phenomics and Its Potential Applications for Plant Genetic Resources Research
2.2. Why Genebank Phenomics?
3. Phenomic Characterization and Evaluation of Genebank Accessions
3.1. Morphology
3.2. Inflorescence and Fruit
3.3. Seed Characteristics
3.4. Phenology
3.5. Physiological and Agronomic Traits
4. Challenges
4.1. Lack of Resources
4.2. Technical Difficulties in Data Management and Analysis
4.3. Users’ Awareness
5. Future Perspectives
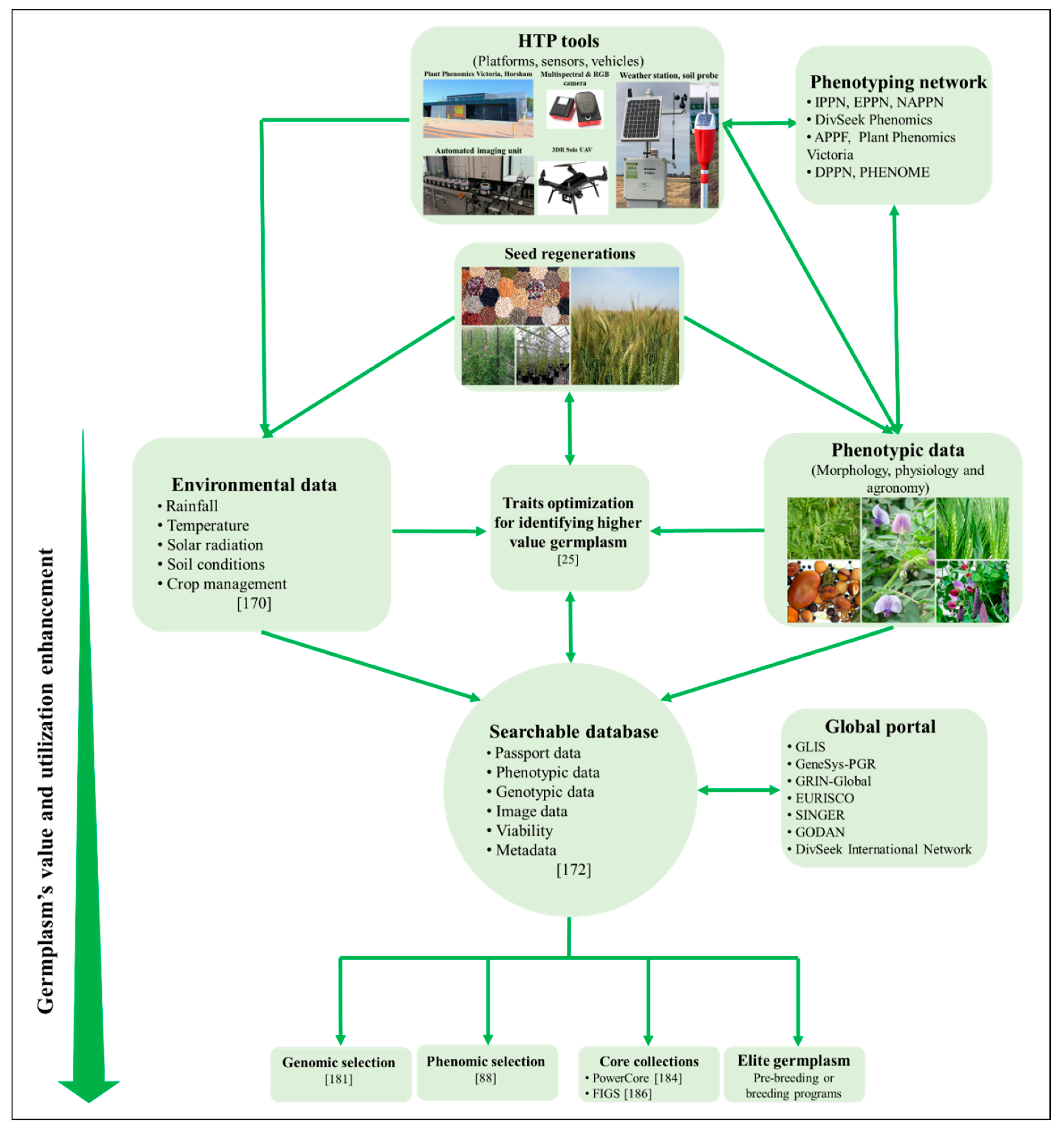
5.1. Systematic HTP Phenotyping of Routine Genebank Seed Regenerations
5.2. A Combination of Genebanks’ Data Mining Approaches
5.3. A Collaborative Network for Data Collection, Analyzing and Sharing
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Legrève, A.; Duveiller, E. Prevailing potential diseases and pest epidemics under a changing climate. In Climate Change and Crop Production; Reynolds, M.P., Ed.; CABI Press: Oxfordshire, UK, 2010; Volume 13, pp. 50–70. [Google Scholar]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, M.A.P.; Bebeli, P.J.; Bettencourt, E.; Costa, G.; Dias, S.; Dos Santos, T.M.; Slaski, J.J. Cereal landraces genetic resources in worldwide GeneBanks. A review. Agron. Sustain. Dev. 2013, 33, 177–203. [Google Scholar] [CrossRef]
- Roa, C.; Hamilton, R.S.; Wenzl, P.; Powell, W. Plant genetic resources: Needs, rights, and opportunities. Trends Plant Sci. 2016, 21, 633–636. [Google Scholar] [CrossRef] [PubMed]
- González, M.Y.; Weise, S.; Zhao, Y.; Philipp, N.; Arend, D.; Börner, A.; Oppermann, M.; Graner, A.; Reif, J.C.; Schulthess, A.W. Unbalanced historical phenotypic data from seed regeneration of a barley ex situ collection. Sci. Data 2018, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gotor, E.; Alercia, A.; Rao, V.R.; Watts, J.; Caracciolo, F. The scientific information activity of Bioversity International: The descriptor lists. Genet. Resour. Crop Evol. 2008, 55, 757–772. [Google Scholar] [CrossRef]
- Norton, S.L.; Khoury, C.K.; Sosa, C.C.; Castañeda-Álvarez, N.P.; Achicanoy, H.A.; Sotelo, S. Priorities for enhancing the ex situ conservation and use of Australian crop wild relatives. Aust. J. Bot. 2017, 65, 638–645. [Google Scholar] [CrossRef]
- FAO. The Second Report on the State of the World’s Plant Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2010. [Google Scholar]
- Tadesse, W.; Sanchez-Garcia, M.; Assefa, S.G.; Amri, A.; Bishaw, Z.; Ogbonnaya, F.C.; Baum, M. Genetic gains in wheat breeding and its role in feeding the world. CBGG 2019, 1, e190005. [Google Scholar] [CrossRef]
- Li, H.; Rasheed, A.; Hickey, L.T.; He, Z. Fast-forwarding genetic gain. Trends Plant Sci. 2018, 23, 184–186. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Ndjiondjop, M.-N.; Henry, R.J. Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief. Funct. Genom. 2018, 17, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Díez, M.J.; De la Rosa, L.; Martín, I.; Guasch, L.; Cartea, M.E.; Mallor, C.; Casals, J.; Simó, J.; Rivera, A.; Anastasio, G.; et al. Plant genebanks: Present situation and proposals for their improvement. the case of the Spanish network. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Anglin, N.L.; Amri, A.; Kehel, Z.; Ellis, D. A case of need: Linking traits to genebank accessions. Biopreservation Biobanking 2018, 16, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Finkers, R.; Chibon, P.-Y.; Van Treuren, R.; Visser, R.; Van Hintum, T. Genebanks and genomics: How to interconnect data from both communities? Plant Genet. Resour. 2014, 13, 90–93. [Google Scholar] [CrossRef]
- McCouch, S.; Baute, G.J.; Bradeen, J.; Bramel, P.; Bretting, P.K.; Buckler, E.; Burke, J.M.; Charest, D.; Cloutier, S.; Cole, G.; et al. Feeding the future. Nature 2013, 499, 23–24. Available online: https://www.nature.com/articles/499023a#supplementary-information (accessed on 24 April 2020). [CrossRef] [PubMed]
- McCouch, S.R.; McNally, K.L.; Wang, W.; Sackville, H.R. Genomics of gene banks: A case study in rice. Am. J. Bot. 2012, 99, 407–423. [Google Scholar] [CrossRef]
- Yu, X.; Li, X.; Guo, T.; Zhu, C.; Wu, Y.; Mitchell, S.E.; Roozeboom, K.L.; Wang, D.; Wang, M.L.; Pederson, G.A.; et al. Genomic prediction contributing to a promising global strategy to turbocharge gene banks. Nat. Plants 2016, 2, 1–7. [Google Scholar] [CrossRef]
- Kilian, B.; Graner, A. NGS technologies for analyzing germplasm diversity in genebanks. Brief. Funct. Genom. 2012, 11, 38–50. [Google Scholar] [CrossRef]
- Philipp, N.; Weise, S.; Oppermann, M.; Börner, A.; Graner, A.; Keilwagen, J.; Kilian, B.; Zhao, Y.; Reif, J.C.; Schulthess, A.W.; et al. Leveraging the use of historical data gathered during seed regeneration of an ex-situ genebank collection of wheat. Front. Plant Sci. 2018, 9, 609. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics—Technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Krajewski, P.; Chen, D.; Ćwiek, H.; Van Dijk, A.D.J.; Fiorani, F.; Kersey, P.; Klukas, C.; Lange, M.; Markiewicz, A.; Nap, J.P.; et al. Towards recommendations for metadata and data handling in plant phenotyping. J. Exp. Bot. 2015, 66, 5417–5427. [Google Scholar] [CrossRef] [PubMed]
- González, M.Y.; Philipp, N.; Schulthess, A.W.; Weise, S.; Zhao, Y.; Börner, A.; Oppermann, M.; Graner, A.; Reif, J.C. Unlocking historical phenotypic data from an ex situ collection to enhance the informed utilization of genetic resources of barley (Hordeum sp.). Theor. Appl. Genet. 2018, 131, 2009–2019. [Google Scholar] [CrossRef]
- Keilwagen, J.; Kilian, B.; Özkan, H.; Babben, S.; Perovic, D.; Mayer, K.F.X.; Walther, A.; Poskar, C.H.; Ordon, F.; Eversole, K.; et al. Separating the wheat from the chaff—A strategy to utilize plant genetic resources from ex situ genebanks. Sci. Rep. 2014, 4, 5231. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Kefauver, S.C. Breeding to adapt agriculture to climate change: Affordable phenotyping solutions. Curr. Opin. Plant Biol. 2018, 45, 237–247. [Google Scholar] [CrossRef]
- White, J.W.; Andrade-Sanchez, P.; Gore, M.A.; Bronson, K.F.; Coffelt, T.A.; Conley, M.M.; Feldmann, K.A.; French, A.N.; Heun, J.T.; Hunsaker, D.J.; et al. Field-based phenomics for plant genetics research. Field Crops Res. 2012, 133, 101–112. [Google Scholar] [CrossRef]
- Rutkoski, J.; Poland, J.; Mondal, S.; Autrique, E.; Pérez, L.G.; Crossa, J.; Reynolds, M.; Singh, R. Canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat. G3 Genes Genom. Genet. 2016, 6, 2799–2808. [Google Scholar] [CrossRef]
- Krause, M.R.; González-Pérez, L.; Crossa, J.; Pérez-Rodríguez, P.; Montesinos-López, O.; Singh, R.P.; Dreisigacker, S.; Poland, J.; Rutkoski, J.; Sorrells, M.; et al. Hyperspectral reflectance-derived relationship matrices for genomic prediction of grain yield in wheat. G3 Genes Genom. Genet. 2019, 9, 1231–1247. [Google Scholar] [CrossRef]
- Milner, S.G.; Jost, M.; Taketa, S.; Mazón, E.R.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea, M.; König, P.; et al. Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 2019, 51, 319–326. [Google Scholar] [CrossRef]
- Van, K.; Kim, D.H.; Shin, J.H.; Lee, S.-H. Genomics of plant genetic resources: Past, present and future. Plant Genet. Resour. 2011, 9, 155–158. [Google Scholar] [CrossRef]
- Mir, R.R.; Reynolds, M.; Pinto, F.; Khan, M.A.; Bhat, M.A. High-throughput phenotyping for crop improvement in the genomics era. Plant Sci. 2019, 282, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.; Borrell, A.; Braun, H.; Edmeades, G.; Flavell, R.; Gwyn, J.; Jordan, D.; Pixley, K.; Rebetzke, G. Translational research for climate resilient, higher yielding crops. CBGG 2019, 1, e190016. [Google Scholar] [CrossRef]
- Reynolds, M.; Tattaris, M.; Cossani, C.M.; Ellis, M.; Yamaguchi-Shinozaki, K.; Pierre, C.S. Exploring genetic resources to increase adaptation of wheat to climate change. In Advances in Wheat Genetics: From Genome to Field; Ogihara, Y., Takumi, S., Handa, H., Eds.; Springer: Tokyo, Japan, 2015; pp. 355–368. [Google Scholar]
- Tardieu, F.; Cabrera-Bosquet, L.; Pridmore, T.; Bennett, M. Plant phenomics, from sensors to knowledge. Curr. Biol. 2017, 27, R770–R783. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef]
- Homolová, L.; Malenovský, Z.; Clevers, J.G.P.W.; García-Santos, G.; Schaepman, M.E. Review of optical-based remote sensing for plant trait mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Jimenez-Berni, J.; Fischer, R.A.; Deery, D.M.; Smith, D.J. Review: High-throughput phenotyping to enhance the use of crop genetic resources. Plant Sci. 2019, 282, 40–48. [Google Scholar] [CrossRef]
- Nguyen, G.N.; Maharjan, P.; Maphosa, L.; Vakani, J.; Thoday-Kennedy, E.; Kant, S. A robust automated image-based phenotyping method for rapid vegetative screening of wheat germplasm for nitrogen use efficiency. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Nguyen, G.N.; Norton, S.L.; Rosewarne, G.M.; James, L.E.; Slater, A.T. Automated phenotyping for early vigour of field pea seedlings in controlled environment by colour imaging technology. PLoS ONE 2018, 13, e0207788. [Google Scholar] [CrossRef]
- Sadeghi-Tehran, P.; Sabermanesh, K.; Virlet, N.; Hawkesford, M.J. Automated method to determine two critical growth stages of wheat: Heading and flowering. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Andrade-Sanchez, P.; Gore, M.A.; Heun, J.T.; Thorp, K.R.; Carmo-Silva, A.E.; French, A.N.; Salvucci, M.E.; White, J.W. Development and evaluation of a field-based high-throughput phenotyping platform. Funct. Plant Biol. 2013, 41, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Sun, N.; Bai, H.; Wang, N.; Fan, Z.; Wang, Y.; Meng, Z.; Li, B.; Cong, Y. Field-based high-throughput phenotyping for maize plant using 3D LiDAR point cloud generated with a “Phenomobile”. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.C.; Merz, T.; Chan, A.; Jackway, P.; Hrabar, S.; Dreccer, M.F.; Holland, E.; Zheng, B.; Ling, T.J.; Jimenez-Berni, J.; et al. Pheno-Copter: A low-altitude, autonomous remote-sensing robotic helicopter for high-throughput field-based phenotyping. Agronomy 2014, 4, 279–301. [Google Scholar] [CrossRef]
- Hu, P.; Chapman, S.C.; Wang, X.; Potgieter, A.; Duan, T.; Jordan, D.; Guo, Y.; Zheng, B. Estimation of plant height using a high throughput phenotyping platform based on unmanned aerial vehicle and self-calibration: Example for sorghum breeding. Eur. J. Agron. 2018, 95, 24–32. [Google Scholar] [CrossRef]
- Silva-Perez, V.; Molero, G.; Serbin, S.P.; Condon, A.G.; Reynolds, M.P.; Furbank, R.T.; Evans, J.R. Hyperspectral reflectance as a tool to measure biochemical and physiological traits in wheat. J. Exp. Bot. 2017, 69, 483–496. [Google Scholar] [CrossRef]
- Dreccer, M.F.; Barnes, L.R.; Meder, R. Quantitative dynamics of stem water soluble carbohydrates in wheat can be monitored in the field using hyperspectral reflectance. Field Crops Res. 2014, 159, 70–80. [Google Scholar] [CrossRef]
- Duan, T.; Chapman, S.C.; Holland, E.; Rebetzke, G.J.; Guo, Y.; Zheng, B. Dynamic quantification of canopy structure to characterize early plant vigour in wheat genotypes. J. Exp. Bot. 2016, 67, 4523–4534. [Google Scholar] [CrossRef]
- Khan, Z.; Chopin, J.; Cai, J.; Eichi, V.-R.; Haefele, S.; Miklavcic, S. Quantitative estimation of wheat phenotyping traits using ground and aerial imagery. Remote Sens. 2018, 10, 950. [Google Scholar] [CrossRef]
- Hassan, M.; Yang, M.; Rasheed, A.; Jin, X.; Xia, X.; Xiao, Y.; He, Z. Time-series multispectral indices from unmanned aerial vehicle imagery reveal senescence rate in bread wheat. Remote Sens. 2018, 10, 809. [Google Scholar] [CrossRef]
- Shorten, P.R.; Leath, S.R.; Schmidt, J.; Ghamkhar, K. Predicting the quality of ryegrass using hyperspectral imaging. Plant Methods 2019, 15, 63. [Google Scholar] [CrossRef]
- Deery, D.M.; Rebetzke, G.J.; Jimenez-Berni, J.A.; Bovill, W.D.; James, R.A.; Condon, A.G.; Furbank, R.T.; Chapman, S.C.; Fischer, R.A. Evaluation of the phenotypic repeatability of canopy temperature in wheat using continuous-terrestrial and airborne measurements. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Berni, J.A.; Deery, D.M.; Rozas-Larraondo, P.; Condon, A.G.; Rebetzke, G.J.; James, R.A.; Bovill, W.D.; Furbank, R.T.; Sirault, X.R.R. High throughput determination of plant height, ground cover, and above-ground biomass in wheat with LiDAR. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.N.; Kant, S. Improving nitrogen use efficiency in plants: Effective phenotyping in conjunction with agronomic and genetic approaches. Funct. Plant Biol. 2018, 45, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.N.; Panozzo, J.; Spangenberg, G.; Kant, S. Phenotyping approaches to evaluate nitrogen-use efficiency related traits of diverse wheat varieties under field conditions. Crop Pasture Sci. 2016, 67, 1139–1148. [Google Scholar] [CrossRef]
- Guo, W.; Fukatsu, T.; Ninomiya, S. Automated characterization of flowering dynamics in rice using field-acquired time-series RGB images. Plant Methods 2015, 11, 7. [Google Scholar] [CrossRef]
- Bendig, J.; Willkomm, M.; Tilly, N.; Gnyp, M.; Bennertz, S.; Qiang, C.; Miao, Y.; Lenz-Wiedemann, V.; Bareth, G. Very high resolution crop surface models (CSMs) from UAV-based stereo images for rice growth monitoring in Northeast China. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2013, 40, 45–50. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- LeMasurier, L.S.; Panozzo, J.F.; Walker, C.K. A digital image analysis method for assessment of lentil size traits. J. Food Eng. 2014, 128, 72–78. [Google Scholar] [CrossRef]
- Hansen, M.A.E.; Hay, F.R.; Carstensen, J.M. A virtual seed file: The use of multispectral image analysis in the management of genebank seed accessions. Plant Genet. Resour. 2015, 14, 238–241. [Google Scholar] [CrossRef]
- McDonald, L.S.; Salisbury, P.A.; Ford, R.; Panozzo, J.F. Quantifying the colour loss of green field pea (Pisum sativum L.) due to bleaching. PLoS ONE 2019, 14, e0221523. [Google Scholar] [CrossRef]
- Coupel-Ledru, A.; Pallas, B.; Delalande, M.; Boudon, F.; Carrié, E.; Martinez, S.; Regnard, J.-L.; Costes, E. Multi-scale high-throughput phenotyping of apple architectural and functional traits in orchard reveals genotypic variability under contrasted watering regimes. Hortic. Res. 2019, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Patrick, A.; Li, C. High throughput phenotyping of blueberry bush morphological traits using unmanned aerial systems. Remote Sens. 2017, 9, 1250. [Google Scholar] [CrossRef]
- Ludovisi, R.; Tauro, F.; Salvati, R.; Khoury, S.; Mugnozza Scarascia, G.; Harfouche, A. UAV-based thermal imaging for high-throughput field phenotyping of black poplar response to drought. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Rist, F.; Gabriel, D.; Mack, J.; Steinhage, V.; Töpfer, R.; Herzog, K. Combination of an automated 3D field phenotyping workflow and predictive modelling for high-throughput and non-invasive phenotyping of grape bunches. Remote Sens. 2019, 11, 2953. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Wendel, A.; Underwood, J. Spectral filter design based on in-field hyperspectral imaging and machine learning for mango ripeness estimation. Comput. Electron. Agric. 2019, 164, 104890. [Google Scholar] [CrossRef]
- Xiong, J.; Liu, Z.; Chen, S.; Liu, B.; Zheng, Z.; Zhong, Z.; Yang, Z.; Peng, H. Visual detection of green mangoes by an unmanned aerial vehicle in orchards based on a deep learning method. Biosyst. Eng. 2020, 194, 261–272. [Google Scholar] [CrossRef]
- Mochida, K.; Koda, S.; Inoue, K.; Hirayama, T.; Tanaka, S.; Nishii, R.; Melgani, F. Computer vision-based phenotyping for improvement of plant productivity: A machine learning perspective. GigaScience 2018. [Google Scholar] [CrossRef]
- Škrubej, U.; Rozman, Č.; Stajnko, D. Assessment of germination rate of the tomato seeds using image processing and machine learning. Eur. J. Hortic. Sci. 2015, 80, 68–75. [Google Scholar] [CrossRef]
- Hasan, M.M.; Chopin, J.P.; Laga, H.; Miklavcic, S.J. Detection and analysis of wheat spikes using Convolutional Neural Networks. Plant Methods 2018, 14, 100. [Google Scholar] [CrossRef]
- Sadeghi-Tehran, P.; Virlet, N.; Ampe, E.M.; Reyns, P.; Hawkesford, M.J. DeepCount: In-field automatic quantification of wheat spikes using simple linear iterative clustering and deep convolutional neural networks. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Pantazi, X.E.; Moshou, D.; Alexandridis, T.; Whetton, R.L.; Mouazen, A.M. Wheat yield prediction using machine learning and advanced sensing techniques. Comput. Electron. Agric. 2016, 121, 57–65. [Google Scholar] [CrossRef]
- Nagel, M.; Holstein, K.; Willner, E.; Börner, A. Machine learning links seed composition, glucosinolates and viability of oilseed rape after 31 years of long-term storage. Seed Sci. Res. 2018, 28, 340–348. [Google Scholar] [CrossRef]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: Current status and perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed]
- Furbank, R.T.; Jimenez-Berni, J.A.; George-Jaeggli, B.; Potgieter, A.B.; Deery, D.M. Field crop phenomics: Enabling breeding for radiation use efficiency and biomass in cereal crops. New Phytol. 2019, 223, 1714–1727. [Google Scholar] [CrossRef]
- Lobet, G.; Draye, X.; Périlleux, C. An online database for plant image analysis software tools. Plant Methods 2013, 9, 38. [Google Scholar] [CrossRef]
- Brown, T.B.; Cheng, R.; Sirault, X.R.R.; Rungrat, T.; Murray, K.D.; Trtilek, M.; Furbank, R.T.; Badger, M.; Pogson, B.J.; Borevitz, J.O. TraitCapture: Genomic and environment modelling of plant phenomic data. Curr. Opin. Plant Biol. 2014, 18, 73–79. [Google Scholar] [CrossRef]
- Cobb, J.N.; DeClerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef]
- FAO. Genebank Standards for Plant Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014. [Google Scholar]
- Longin, C.F.H.; Reif, J.C. Redesigning the exploitation of wheat genetic resources. Trends Plant Sci. 2014, 19, 631–636. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; De los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Hu, Z.; Xu, C. Genomic selection methods for crop improvement: Current status and prospects. Crop J. 2018, 6, 330–340. [Google Scholar] [CrossRef]
- Jannink, J.L. Dynamics of long-term genomic selection. Genet. Sel. Evol. 2010, 42, 35. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.; Langridge, P. Physiological breeding. Curr. Opin. Plant Biol. 2016, 31, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Rutkoski, J.; Singh, R.P.; Huerta-Espino, J.; Bhavani, S.; Poland, J.; Jannink, J.L.; Sorrells, M.E. Genetic gain from phenotypic and genomic selection for quantitative resistance to stem rust of wheat. Plant Genome 2015, 8, 1–10. [Google Scholar] [CrossRef]
- Rincent, R.; Charpentier, J.-P.; Faivre-Rampant, P.; Paux, E.; Le Gouis, J.; Bastien, C.; Segura, V. Phenomic selection is a low-cost and high-throughput method based on indirect predictions: Proof of concept on wheat and poplar. G3 Genes Genom. Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, D.; Wilson, C.; McCullum, C.; Huang, R.; Dwen, P.; Flack, J.; Tran, Q.; Saltman, T.; Cliff, B. Economic and environmental benefits of biodiversity. BioScience 1997, 47, 747–757. [Google Scholar] [CrossRef]
- Van De Wouw, M.; Kik, C.; Van Hintum, T.; Van Treuren, R.; Visser, B. Genetic erosion in crops: Concept, research results and challenges. Plant Genet. Resour. 2010, 8, 1–15. [Google Scholar] [CrossRef]
- Schoen, D.J.; Brown, A.H.D. The conservation of wild plant species in seed banks: Attention to both taxonomic coverage and population biology will improve the role of seed banks as conservation tools. BioScience 2001, 51, 960–966. [Google Scholar] [CrossRef]
- Solberg, S.O.; Yndgaard, F.; Palmè, A. Morphological and phenological consequences of ex situ conservation of natural populations of red clover (Trifolium pratense L.). Plant Genet. Resour. 2015, 15, 97–108. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef]
- Boyle, R.D.; Corke, F.M.K.; Doonan, J.H. Automated estimation of tiller number in wheat by ribbon detection. Mach. Vis. Appl. 2016, 27, 637–646. [Google Scholar] [CrossRef]
- Yamamoto, K.; Guo, W.; Ninomiya, S. Node detection and internode length estimation of tomato seedlings based on image analysis and machine learning. Sensors 2016, 16, 1044. [Google Scholar] [CrossRef]
- Conn, A.; Pedmale, U.V.; Chory, J.; Stevens, C.F.; Navlakha, S. A statistical description of plant shoot architecture. Curr. Biol. 2017, 27, 2078–2088.e2073. [Google Scholar] [CrossRef] [PubMed]
- Crowell, S.; Falcão, A.X.; Shah, A.; Wilson, Z.; Greenberg, A.J.; McCouch, S.R. High-resolution inflorescence phenotyping using a novel image-analysis pipeline, PANorama. Plant Physiol. 2014, 165, 479–495. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Singh, D.; Marla, S.; Morris, G.; Poland, J. Field-based high-throughput phenotyping of plant height in sorghum using different sensing technologies. Plant Methods 2018, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Holman, F.; Riche, A.; Michalski, A.; Castle, M.; Wooster, M.; Hawkesford, M. High throughput field phenotyping of wheat plant height and growth rate in field plot trials using UAV based remote sensing. Remote Sens. 2016, 8, 1031. [Google Scholar] [CrossRef]
- Han, L.; Yang, G.; Dai, H.; Yang, H.; Xu, B.; Feng, H.; Li, Z.; Yang, X. Fuzzy clustering of maize plant-height patterns using time series of UAV remote-sensing images and variety traits. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Bendig, J.; Bolten, A.; Bareth, G. UAV-based imaging for multi-temporal, very high resolution crop surface models to monitor crop growth variability. Photogramm. Fernerkund. Geoinf. 2013, 2013, 551–562. [Google Scholar] [CrossRef]
- Larese, M.G.; Granitto, P.M. Finding local leaf vein patterns for legume characterization and classification. Mach. Vis. Appl. 2016, 27, 709–720. [Google Scholar] [CrossRef]
- Wang, Y.; Wen, W.; Wu, S.; Wang, C.; Yu, Z.; Guo, X.; Zhao, C. Maize plant phenotyping: Comparing 3D laser scanning, multi-view stereo reconstruction, and 3D digitizing estimates. Remote Sens. 2018, 11, 63. [Google Scholar] [CrossRef]
- Brichet, N.; Fournier, C.; Turc, O.; Strauss, O.; Artzet, S.; Pradal, C.; Welcker, C.; Tardieu, F.; Cabrera-Bosquet, L. A robot-assisted imaging pipeline for tracking the growths of maize ear and silks in a high-throughput phenotyping platform. Plant Methods 2017, 13, 1–12. [Google Scholar] [CrossRef]
- AL-Tam, F.; Adam, H.; Anjos, A.d.; Lorieux, M.; Larmande, P.; Ghesquière, A.; Jouannic, S.; Shahbazkia, H.R. P-TRAP: A panicle trait phenotyping tool. BMC Plant Biol. 2013, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Rebolledo, M.C.; Peña, A.L.; Duitama, J.; Cruz, D.F.; Dingkuhn, M.; Grenier, C.; Tohme, J. Combining image analysis, genome wide association studies and different field trials to reveal stable genetic regions related to panicle architecture and the number of spikelets per panicle in rice. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Duan, L.; Liu, L.; Tu, H.; Yang, P.; Wu, D.; Chen, G.; Xiong, L.; Yang, W.; Liu, Q. Panicle-SEG: A robust image segmentation method for rice panicles in the field based on deep learning and superpixel optimization. Plant Methods 2017, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Cao, Z.; Xiao, Y.; Fang, Z.; Zhu, Y.; Xian, K. Fine-grained maize tassel trait characterization with multi-view representations. Comput. Electron. Agric. 2015, 118, 143–158. [Google Scholar] [CrossRef]
- Lu, H.; Cao, Z.; Xiao, Y.; Li, Y.; Zhu, Y. Region-based colour modelling for joint crop and maize tassel segmentation. Biosyst. Eng. 2016, 147, 139–150. [Google Scholar] [CrossRef]
- Boyle, R.; Corke, F.; Howarth, C. Image-based estimation of oat panicle development using local texture patterns. Funct. Plant Biol. 2015, 42, 433–443. [Google Scholar] [CrossRef]
- Song, Y.; Glasbey, C.; Horgan, G.; Polder, G.; Dieleman, J.; Van der Heijden, G. Automatic fruit recognition and counting from multiple images. Biosyst. Eng. 2014, 118, 203–215. [Google Scholar] [CrossRef]
- Xiong, B.; Wang, B.; Xiong, S.; Lin, C.; Yuan, X. 3D morphological processing for wheat spike phenotypes using computed tomography images. Remote Sens. 2019, 11, 1110. [Google Scholar] [CrossRef]
- Genaev, M.A.; Komyshev, E.G.; Smirnov, N.V.; Kruchinina, Y.V.; Goncharov, N.P.; Afonnikov, D.A. Morphometry of the wheat spike by analyzing 2D images. Agronomy 2019, 9, 390. [Google Scholar] [CrossRef]
- McDonald, L.S.; Panozzo, J.F.; Salisbury, P.A.; Ford, R. Discriminant analysis of defective and non-defective field pea (Pisum sativum L.) into broad market grades based on digital image features. PLoS ONE 2016, 11, e0155523. [Google Scholar] [CrossRef]
- Rolletschek, H.; Fuchs, J.; Friedel, S.; Börner, A.; Todt, H.; Jakob, P.M.; Borisjuk, L. A novel noninvasive procedure for high-throughput screening of major seed traits. Plant Biotechnol. J. 2015, 13, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Tanabata, T.; Shibaya, T.; Hori, K.; Ebana, K.; Yano, M. SmartGrain: High-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 2012, 160, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Miller, N.D.; Haase, N.J.; Lee, J.; Kaeppler, S.M.; De Leon, N.; Spalding, E.P. A robust, high-throughput method for computing maize ear, cob, and kernel attributes automatically from images. Plant J. 2017, 89, 169–178. [Google Scholar] [CrossRef]
- Makanza, R.; Zaman-Allah, M.; Cairns, J.E.; Eyre, J.; Burgueño, J.; Pacheco, Á.; Diepenbrock, C.; Magorokosho, C.; Tarekegne, A.; Olsen, M.; et al. High-throughput method for ear phenotyping and kernel weight estimation in maize using ear digital imaging. Plant Methods 2018, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Martín-Gómez, J.J.; Rewicz, A.; Goriewa-Duba, K.; Wiwart, M.; Tocino, Á.; Cervantes, E. Morphological description and classification of wheat kernels based on geometric models. Agronomy 2019, 9, 399. [Google Scholar] [CrossRef]
- Gentallan, R.P.; Altoveros, N.C.; Borromeo, T.H.; Endonela, L.E.; Hay, F.R.; Lalusin, A.G.; Reaño, C.E.; Yoshioka, Y. An objective method of shape descriptor state establishment using elliptic Fourier analysis (EFA). Plant Genet. Resour. 2019, 17, 480–487. [Google Scholar] [CrossRef]
- Jahnke, S.; Roussel, J.; Hombach, T.; Kochs, J.; Fischbach, A.; Huber, G.; Scharr, H. phenoSeeder-A robot system for automated handling and phenotyping of individual seeds. Plant Physiol. 2016, 172, 1358–1370. [Google Scholar] [CrossRef]
- Clohessy, J.W.; Pauli, D.; Kreher, K.M.; Buckler, E.S.; Armstrong, P.R.; Wu, T.; Hoekenga, O.A.; Jannink, J.-L.; Sorrells, M.E.; Gore, M.A. A low-cost automated system for high-throughput phenotyping of single oat seeds. Plant Phenome J. 2018, 1. [Google Scholar] [CrossRef]
- Wu, J.; Yang, G.; Yang, X.; Xu, B.; Han, L.; Zhu, Y. Automatic counting of in situ rice seedlings from UAV images based on a deep fully convolutional neural network. Remote Sens. 2019, 11, 691. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Paterson, A.H.; Robertson, J.S. DeepSeedling: Deep convolutional network and Kalman filter for plant seedling detection and counting in the field. Plant Methods 2019, 15, 141. [Google Scholar] [CrossRef]
- Jin, X.; Liu, S.; Baret, F.; Hemerlé, M.; Comar, A. Estimates of plant density of wheat crops at emergence from very low altitude UAV imagery. Remote Sens. Environ. 2017, 198, 105–114. [Google Scholar] [CrossRef]
- Duan, T.; Zheng, B.; Guo, W.; Ninomiya, S.; Guo, Y.; Chapman, S.C. Comparison of ground cover estimates from experiment plots in cotton, sorghum and sugarcane based on images and ortho-mosaics captured by UAV. Funct. Plant Biol. 2017, 44, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Cao, Z.; Lu, H.; Li, Y.; Xiao, Y. In-field automatic observation of wheat heading stage using computer vision. Biosyst. Eng. 2016, 143, 28–41. [Google Scholar] [CrossRef]
- Desai, S.V.; Balasubramanian, V.N.; Fukatsu, T.; Ninomiya, S.; Guo, W. Automatic estimation of heading date of paddy rice using deep learning. Plant Methods 2019, 15, 76. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Miller, N.D.; De Leon, N.; Kaeppler, S.M.; Spalding, E.P. Predicting Zea mays flowering time, yield, and kernel dimensions by analyzing aerial images. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Liu, S.; Martre, P.; Buis, S.; Abichou, M.; Andrieu, B.; Baret, F. Estimation of plant and canopy architectural traits using the digital plant phenotyping platform. Plant Physiol. 2019, 181, 881–890. [Google Scholar] [CrossRef]
- Wilke, N.; Siegmann, B.; Klingbeil, L.; Burkart, A.; Kraska, T.; Muller, O.; Van Doorn, A.; Heinemann, S.; Rascher, U. Quantifying lodging percentage and lodging severity using a UAV-based canopy height model combined with an objective threshold approach. Remote Sens. 2019, 11, 515. [Google Scholar] [CrossRef]
- Singh, D.; Wang, X.; Kumar, U.; Gao, L.; Noor, M.; Imtiaz, M.; Singh, R.P.; Poland, J. High-throughput phenotyping enabled genetic dissection of crop lodging in wheat. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, Y.; Song, M.; Ding, Y.; Lin, F.; Liang, D.; Zhang, D. Use of unmanned aerial vehicle imagery and deep learning UNet to extract rice lodging. Sensors 2019, 19, 3859. [Google Scholar] [CrossRef]
- Fu, P.; Meacham-Hensold, K.; Guan, K.; Bernacchi, C.J. Hyperspectral leaf reflectance as proxy for photosynthetic capacities: An ensemble approach based on multiple machine learning algorithms. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Yendrek, C.R.; Tomaz, T.; Montes, C.M.; Cao, Y.; Morse, A.M.; Brown, P.J.; McIntyre, L.M.; Leakey, A.D.B.; Ainsworth, E.A. High-throughput phenotyping of maize leaf physiological and biochemical traits using hyperspectral reflectance. Plant Physiol. 2017, 173, 614–626. [Google Scholar] [CrossRef]
- Zhang, L.; Niu, Y.; Zhang, H.; Han, W.; Li, G.; Tang, J.; Peng, X. Maize canopy temperature extracted from UAV thermal and RGB imagery and its application in water stress monitoring. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Sadras, V.O.; Mahadevan, M.; Zwer, P.K. Stay-green associates with low water soluble carbohydrates at flowering in oat. Field Crops Res. 2019, 230, 132–138. [Google Scholar] [CrossRef]
- Blancon, J.; Dutartre, D.; Tixier, M.-H.; Weiss, M.; Comar, A.; Praud, S.; Baret, F. A High-throughput model-assisted method for phenotyping maize green leaf area index dynamics using unmanned aerial vehicle imagery. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.D.C.; Edwards, J.; McDonald, G.; Kuchel, H. Estimating biomass and canopy height with LiDAR for field crop breeding. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Golzarian, M.R.; Frick, R.A.; Rajendran, K.; Berger, B.; Roy, S.; Tester, M.; Lun, D.S. Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 2011, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; De los Campos, G.; Alvarado, G.; Suchismita, M.; Rutkoski, J.; González-Pérez, L.; Burgueño, J. Predicting grain yield using canopy hyperspectral reflectance in wheat breeding data. Plant Methods 2017, 13, 4. [Google Scholar] [CrossRef]
- Fernandez-Gallego, J.; Kefauver, S.; Gutiérrez, N.A.; Nieto-Taladriz, M.T.; Araus, J. Automatic Wheat Ear Counting In-Field Conditions: Simulation and Implication of Lower Resolution Images; Proc. SPIE, Remote Sensing for Agriculture, Ecosystems, and Hydrology XX, 107830M: Berlin, Germany, 2018; Volume 10783. [Google Scholar] [CrossRef]
- Guo, W.; Zheng, B.; Potgieter, A.B.; Diot, J.; Watanabe, K.; Noshita, K.; Jordan, D.R.; Wang, X.; Watson, J.; Ninomiya, S.; et al. Aerial imagery analysis-quantifying appearance and number of sorghum heads for applications in breeding and agronomy. Front. Plant Sci. 2018, 9, 1544. [Google Scholar] [CrossRef]
- Madec, S.; Jin, X.; Lu, H.; De Solan, B.; Liu, S.; Duyme, F.; Heritier, E.; Baret, F. Ear density estimation from high resolution RGB imagery using deep learning technique. Agric. For. Meteorol. 2019, 264, 225–234. [Google Scholar] [CrossRef]
- Pérez-Harguindeguy, N.; Díaz, S.; Garnier, E.; Lavorel, S.; Poorter, H.; Jaureguiberry, P.; Bret-Harte, M.S.; Cornwell, W.K.; Craine, J.M.; Gurvich, D.E.; et al. New handbook for standardised measurement of plant functional traits worldwide. Aust. J. Bot. 2013, 61, 167–234. [Google Scholar] [CrossRef]
- Reynolds, M.; Foulkes, M.J.; Slafer, G.A.; Berry, P.; Parry, M.A.J.; Snape, J.W.; Angus, W.J. Raising yield potential in wheat. J. Exp. Bot. 2009, 60, 1899–1918. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Li, J.; Bhatta, M.; Shi, Y.; Baenziger, P.; Ge, Y. Wheat height estimation using LiDAR in comparison to ultrasonic sensor and UAS. Sensors 2018, 18, 3731. [Google Scholar] [CrossRef] [PubMed]
- Grillo, O.; Blangiforti, S.; Venora, G. Wheat landraces identification through glumes image analysis. Comput. Electron. Agric. 2017, 141, 223–231. [Google Scholar] [CrossRef]
- Cervantes, E.; Martín, J.J.; Saadaoui, E. Updated methods for seed shape analysis. Scientifica 2016, 2016. [Google Scholar] [CrossRef]
- ElMasry, G.; Mandour, N.; Al-Rejaie, S.; Belin, E.; Rousseau, D. Recent applications of multispectral imaging in seed phenotyping and quality monitoring—An overview. Sensors 2019, 19, 1090. [Google Scholar] [CrossRef]
- Feng, L.; Zhu, S.; Liu, F.; He, Y.; Bao, Y.; Zhang, C. Hyperspectral imaging for seed quality and safety inspection: A review. Plant Methods 2019, 15, 91. [Google Scholar] [CrossRef]
- Gentallan, R.P.; Altoveros, N.C.; Borromeo, T.H.; Endonela, L.E.; Hay, F.R.; Lalusin, A.G.; Reaño, C.E.; Yoshioka, Y. Systematic establishment of colour descriptor states through image-based phenotyping. Plant Genet. Resour. 2018, 17, 91–94. [Google Scholar] [CrossRef]
- Nielsen, N.H.; Jahoor, A.; Jensen, J.D.; Orabi, J.; Cericola, F.; Edriss, V.; Jensen, J. Genomic prediction of seed quality traits using advanced barley breeding lines. PLoS ONE 2016, 11, e0164494. [Google Scholar] [CrossRef]
- Kumar, J.; Choudhary, A.K.; Gupta, D.S.; Kumar, S. Towards exploitation of adaptive traits for climate-resilient smart pulses. Int. J. Mol. Sci. 2019, 20, 2971. [Google Scholar] [CrossRef]
- Van Hintum, T.J.; Brown, A.H.D.; Spillane, C. Core Collections of Plant Genetic Resources; International Plant Genetic Resources Institute: Rome, Italy, 2000. [Google Scholar]
- Reynolds, M.; Foulkes, J.; Furbank, R.; Griffiths, S.; King, J.; Murchie, E.; Parry, M.; Slafer, G. Achieving yield gains in wheat. Plant Cell Environ. 2012, 35, 1799–1823. [Google Scholar] [CrossRef]
- Khush, G.S. Strategies for increasing the yield potential of cereals: Case of rice as an example. Plant Breed. 2013, 132, 433–436. [Google Scholar] [CrossRef]
- Shunmugam, A.S.K.; Kannan, U.; Jiang, Y.; Daba, K.A.; Gorim, L.Y. Physiology based approaches for breeding of next-generation food legumes. Plants 2018, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Borrell, A.K.; Mullet, J.E.; George-Jaeggli, B.; Van Oosterom, E.J.; Hammer, G.L.; Klein, P.E.; Jordan, D.R. Drought adaptation of stay-green sorghum is associated with canopy development, leaf anatomy, root growth, and water uptake. J. Exp. Bot. 2014, 65, 6251–6263. [Google Scholar] [CrossRef] [PubMed]
- Kamal, N.M.; Gorafi, Y.S.A.; Abdelrahman, M.; Abdellatef, E.; Tsujimoto, H. Stay-green trait: A prospective approach for yield potential, and drought and heat stress adaptation in globally important cereals. Int. J. Mol. Sci. 2019, 20, 5837. [Google Scholar] [CrossRef]
- Kumari, M.; Pudake, R.N.; Singh, V.P.; Joshi, A.K. Association of staygreen trait with canopy temperature depression and yield traits under terminal heat stress in wheat (Triticum aestivum L.). Euphytica 2013, 190, 87–97. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Jimenez-Berni, J.A.; Bovill, W.D.; Deery, D.M.; James, R.A. High-throughput phenotyping technologies allow accurate selection of stay-green. J. Exp. Bot. 2016, 67, 4919–4924. [Google Scholar] [CrossRef]
- Li, X.; Ingvordsen, C.H.; Weiss, M.; Rebetzke, G.J.; Condon, A.G.; James, R.A.; Richards, R.A. Deeper roots associated with cooler canopies, higher normalized difference vegetation index, and greater yield in three wheat populations grown on stored soil water. J. Exp. Bot. 2019, 70, 4963–4974. [Google Scholar] [CrossRef]
- Reynolds, D.; Baret, F.; Welcker, C.; Bostrom, A.; Ball, J.; Cellini, F.; Lorence, A.; Chawade, A.; Khafif, M.; Noshita, K.; et al. What is cost-efficient phenotyping? Optimizing costs for different scenarios. Plant Sci. 2019, 282, 14–22. [Google Scholar] [CrossRef]
- Roitsch, T.; Cabrera-Bosquet, L.; Fournier, A.; Ghamkhar, K.; Jiménez-Berni, J.; Pinto, F.; Ober, E.S. Review: New sensors and data-driven approaches—A path to next generation phenomics. Plant Sci. 2019, 282, 2–10. [Google Scholar] [CrossRef]
- Czedik-Eysenberg, A.; Seitner, S.; Güldener, U.; Koemeda, S.; Jez, J.; Colombini, M.; Djamei, A. The ‘PhenoBox’, a flexible, automated, open-source plant phenotyping solution. New Phytol. 2018, 219, 808–823. [Google Scholar] [CrossRef]
- Coppens, F.; Wuyts, N.; Inzé, D.; Dhondt, S. Unlocking the potential of plant phenotyping data through integration and data-driven approaches. Curr. Opin. Syst. Biol. 2017, 4, 58–63. [Google Scholar] [CrossRef]
- Bolger, A.M.; Poorter, H.; Dumschott, K.; Bolger, M.E.; Arend, D.; Osorio, S.; Gundlach, H.; Mayer, K.F.X.; Lange, M.; Scholz, U.; et al. Computational aspects underlying genome to phenome analysis in plants. Plant J. 2019, 97, 182–198. [Google Scholar] [CrossRef]
- Poorter, H.; Fiorani, F.; Pieruschka, R.; Wojciechowski, T.; Van der Putten, W.H.; Kleyer, M.; Schurr, U.; Postma, J. Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytol. 2016, 212, 838–855. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Envirotyping for deciphering environmental impacts on crop plants. Theor. Appl. Genet. 2016, 129, 653–673. [Google Scholar] [CrossRef] [PubMed]
- Pieruschka, R.; Schurr, U. Plant phenotyping: Past, present, and future. Plant Phenomics 2019, 2019, 7507131. [Google Scholar] [CrossRef]
- Oppermann, M.; Weise, S.; Dittmann, C.; Knüpffer, H. GBIS: The information system of the German genebank. Database 2015, 2015. [Google Scholar] [CrossRef]
- Ćwiek-Kupczyńska, H.; Altmann, T.; Arend, D.; Arnaud, E.; Chen, D.; Cornut, G.; Fiorani, F.; Frohmberg, W.; Junker, A.; Klukas, C.; et al. Measures for interoperability of phenotypic data: Minimum information requirements and formatting. Plant Methods 2016, 12, 44. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 2016, 3. [Google Scholar] [CrossRef]
- Colmsee, C.; Joachim Keller, E.R.; Zanke, C.; Senula, A.; Funke, T.; Oppermann, M.; Weise, S.; Scholz, U. The garlic and shallot core collection image database of IPK presenting two vegetatively maintained crops in the Federal ex situ genebank for agricultural and horticultural crops at Gatersleben, Germany. Genet. Resour. Crop Evol. 2012, 59, 1407–1415. [Google Scholar] [CrossRef]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the rate of genetic gain in public-sector plant breeding programs: Lessons from the breeder’s equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef]
- Wang, C.; Hu, S.; Gardner, C.; Lübberstedt, T. Emerging avenues for utilization of exotic germplasm. Trends Plant Sci. 2017, 22, 624–637. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.M.; Lockwood, D.R.; Volk, G.M.; Walters, C. Modeling demographics and genetic diversity in ex situ collections during seed storage and regeneration. Crop Sci. 2010, 50, 2440–2447. [Google Scholar] [CrossRef]
- Hoban, S. New guidance for ex situ gene conservation: Sampling realistic population systems and accounting for collection attrition. Biol. Conserv. 2019, 235, 199–208. [Google Scholar] [CrossRef]
- Bustos-Korts, D.; Boer, M.P.; Malosetti, M.; Chapman, S.; Chenu, K.; Zheng, B.; Van Eeuwijk, F.A. Combining crop growth modeling and statistical genetic modeling to evaluate phenotyping strategies. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Crossa, J.; Jarquín, D.; Franco, J.; Pérez-Rodríguez, P.; Burgueño, J.; Saint-Pierre, C.; Vikram, P.; Sansaloni, C.; Petroli, C.; Akdemir, D.; et al. Genomic prediction of gene bank wheat landraces. G3 Genes Genom. Genet. 2016, 6, 1819–1834. [Google Scholar] [CrossRef]
- Gorjanc, G.; Jenko, J.; Hearne, S.J.; Hickey, J.M. Initiating maize pre-breeding programs using genomic selection to harness polygenic variation from landrace populations. BMC Genom. 2016, 17, 30. [Google Scholar] [CrossRef]
- Girma, G.; Bhattacharjee, R.; Lopez-Montes, A.; Gueye, B.; Ofodile, S.; Franco, J.; Abberton, M. Re-defining the yam (Dioscorea spp.) core collection using morphological traits. Plant Genet. Resour. 2017, 16, 193–200. [Google Scholar] [CrossRef]
- Chung, H.K.; Kim, K.W.; Chung, J.W.; Lee, J.R.; Lee, S.Y.; Dixit, A.; Kang, H.K.; Zhao, W.; McNally, K.L.; Hamilton, R.S.; et al. Development of a core set from a large rice collection using a modified heuristic algorithm to retain maximum diversity. J. Integr. Plant Biol. 2009, 51, 1116–1125. [Google Scholar] [CrossRef]
- Dutta, M.; Phogat, B.S.; Kumar, S.; Kumar, N.; Kumari, J.; Pandey, A.C.; Singh, T.P.; Tyagi, R.K.; Jacob, S.R.; Srinivasan, K.; et al. Development of core set of wheat (Triticum spp.) germplasm conserved in the national genebank in India. In Advances in Wheat Genetics: From Genome to Field; Ogihara, Y., Takumi, S., Handa, H., Eds.; Springer: Tokyo, Japan, 2015; pp. 33–45. [Google Scholar]
- Sanders, R. A New Approach to Mining Agricultural Gene Banks–To Speed the Pace of Research Innovation for Food Security; International Center for Agricultural Research in the Dry Areas: Beirut, Lebanon, 2013. [Google Scholar]
- Khazaei, H.; Street, K.; Bari, A.; Mackay, M.; Stoddard, F.L. The FIGS (Focused Identification of Germplasm Strategy) approach identifies traits related to drought adaptation in Vicia faba genetic resources. PLoS ONE 2013, 8, e63107. [Google Scholar] [CrossRef]
- Endresen, D.T.F.; Street, K.; Mackay, M.; Bari, A.; Amri, A.; De Pauw, E.; Nazari, K.; Yahyaoui, A. Sources of resistance to stem rust (Ug99) in bread wheat and Durum wheat identified using focused identification of germplasm strategy. Crop Sci. 2012, 52, 764–773. [Google Scholar] [CrossRef]
- Bhullar, N.K.; Street, K.; Mackay, M.; Yahiaoui, N.; Keller, B. Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc. Natl. Acad. Sci. USA 2009, 106, 9519–9524. [Google Scholar] [CrossRef] [PubMed]
- El Bouhssini, M.; Street, K.; Amri, A.; Mackay, M.; Ogbonnaya, F.C.; Omran, A.; Abdalla, O.; Baum, M.; Dabbous, A.; Rihawi, F.; et al. Sources of resistance in bread wheat to Russian wheat aphid (Diuraphis noxia) in Syria identified using the Focused Identification of Germplasm Strategy (FIGS). Plant Breed. 2011, 130, 96–97. [Google Scholar] [CrossRef]
- Haupt, M.; Schmid, K. Combining focused identification of germplasm and core collection strategies to identify genebank accessions for central European soybean breeding. BioRxiv 2019. [Google Scholar] [CrossRef] [PubMed]
- Halewood, M.; Chiurugwi, T.; Sackville Hamilton, R.; Kurtz, B.; Marden, E.; Welch, E.; Michiels, F.; Mozafari, J.; Sabran, M.; Patron, N.; et al. Plant genetic resources for food and agriculture: Opportunities and challenges emerging from the science and information technology revolution. New Phytol. 2018, 217, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.-B. The vulnerability of plant genetic resources conserved ex situ. Crop Sci. 2017, 57, 2314–2328. [Google Scholar] [CrossRef]
- Gourdji, S.M.; Mathews, K.L.; Reynolds, M.; Crossa, J.; Lobell, D.B. An assessment of wheat yield sensitivity and breeding gains in hot environments. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122190. [Google Scholar] [CrossRef]
- Walters, C.; Volk, G.M.; Richards, C.M. Genebanks in the post-genomic age: Emerging roles and anticipated uses. Biodiversity 2008, 9, 68–71. [Google Scholar] [CrossRef]

| Traits | Description | Sensors and Capture mode | Species | Environment | References |
|---|---|---|---|---|---|
| Morphology | |||||
| Plant architecture | Number of tillers of wheat plants | Automated RGB1 imaging platform, LemnaTec 3D Scanalyzer | Triticum aestivum | Greenhouse | [94] |
| Node and internode length of tomato seedlings | RGB imagery | Solanum lycopersicum | Greenhouse | [95] | |
| Characterization of plant architecture by 3D scanning reconstruction | Blue-laser scanner | Solanum lycopersicum, Nicotiana benthamiana, Sorghum bicolor | Greenhouse | [96] | |
| Canopy structure (tiller and leaf number, leaf length and angle, leaf elongation rate) | RGB imagery | Triticum aestivum | Greenhouse | [50] | |
| A phenotyping platform, PANorama, measuring architectural properties (panicle, branch, leaf) of various crop species. | RGB imaging unit | Oryza sativa Zea mays Solanum lycopersicum | Laboratory | [97] | |
| Plant height | Sorghum plant height estimates | Ultrasonic, LiDAR2-Lite, Kinect camera, imaging array, UAV3 RGB imagery | Sorghum bicolor | Field | [98] |
| Wheat plant height estimation | UGV4 and UAV RGB imagery | Triticum aestivum | Field | [51,99] | |
| Rice plant height estimation | UAV RGB imagery | Oryza sativa | Field | [59] | |
| Barley plant height measurement | UAV RGB imagery | Hordeum vulgare | Field | [60] | |
| Maize plant height estimates | UAV RGB imagery | Zea mays | Field | [100,101] | |
| Leaf properties | Characterization of local leaf vein in legume leaves | Color scanner | Vigna angularis Phaseolus vulgaris Glycine max | Laboratory | [102] |
| Leaf morphological properties and height of maize, measured by various digital phenotyping methods | 3D scanner, Multi-view stereo cameras, FastTrack 3D digitizer | Zea mays | Laboratory | [103] | |
| Inflorescence and fruit | |||||
| An automated imaging system for monitoring the growths of maize ear and silks | RGB imaging platform | Zea mays | Greenhouse | [104] | |
| Analysis of panicle architecture and spikelet numbers in rice by an imaging tool, P-TRAP5 | RGB imagery | Oryza sativa | Field, laboratory | [105,106] | |
| Rice panicle phenotyping using Panicle-SEG6 algorithm | RGB imagery | Oryza sativa | Field | [107] | |
| Characterization of maize tassel traits by RGB imaging and machine vision | RGB imaging sensors | Zea mays | Field | [108,109] | |
| Analysis of oat panicle development | RGB imaging platform | Avena sativa | Greenhouse | [110] | |
| Fruit recognition and counting by an imaging robot, SPYSEE | RGB imaging robot | Capsicum annuum | Greenhouse | [111] | |
| Morphological characterization of wheat spike (grain number, size and angle, stem node) | Computed Tomography imagery | Triticum aestivum | Laboratory | [112] | |
| Automatic quantification of wheat heads | Automated RGB imagery platform, Field Scanalyzer | Triticum aestivum | Field | [73] | |
| Morphometric properties of wheat spikes | RGB imagery | Triticum aestivum | Laboratory | [113] | |
| Seed characteristics | |||||
| Seed quality of field pea (color, shape, and size) analyzed by multi-spectral imaging | Built-in multi-spectral camera, EyeFoss | Pisum sativum | Laboratory | [114] | |
| Evaluating of lentil seed size by multi-spectral imaging | Built-in multi-spectral camera, EyeFoss | Lens culinaris | Laboratory | [61] | |
| Screening method to evaluate seed properties | Nuclear magnetic resonance | Avena spp. | Laboratory | [115] | |
| Rice seed shape analyzed by an image processing pipeline | Color scanner | Oryza sativa | Laboratory | [116] | |
| Analysis of maize ear, cob and kernel properties | Color scanner | Zea mays | Laboratory | [117] | |
| Estimation of ear characteristics and kernel weight in maize | RGB imagery | Zea mays | Field | [118] | |
| Morphological characteristics of wheat kernels | Color scanner and RGB imagery | Triticum aestivum | Laboratory | [119] | |
| Phenotypic classification of rice seed accessions | Multispectral imagery, VideometerLab | Oryza sativa | Laboratory | [62] | |
| Shape description of pili seed by imaging technology | Multispectral imagery, VideometerLab | Canarium ovatum | Laboratory | [120] | |
| Automated morphological characterization of rapeseed and barley seeds | Automated RGB imaging unit, phenoSeeder | Brassica napus Hordeum vulgare | Laboratory | [121] | |
| Automated phenotyping of oat seed properties | NIR spectroscopy, Single-Seed Analyzer | Avena sativa | Laboratory | [122] | |
| Phenology | |||||
| Emergence count | Rice seedling counts | High resolution UAV RGB imagery | Oryza sativa | Field | [123] |
| Cotton seedling detection and count | Ground-based video recording | Gossypium hirsutum | Field | [124] | |
| Germination rate estimation in tomato by color imagery | RGB imagery | Solanum lycopersicum | Laboratory | [71] | |
| Determination of plant density at emergence in wheat | High resolution UAV RGB imagery | Triticum aestivum | Field | [125] | |
| Ground cover | Ground cover estimates | High resolution UAV RGB imagery | Sorghum bicolor Gossypium hirsutum Saccharum spp. | Field | [126] |
| Flowering | Automated observations of wheat flowering | CCD7 digital camera | Triticum aestivum | Field | [127] |
| Heading and flowering detection in wheat | Automated RGB imaging platform | Triticum aestivum | Field | [43] | |
| Automated flowering observation in rice from a time-series RGB images | Automated RGB imaging platform | Oryza sativa | Field | [58,128] | |
| Estimation of flowering time in maize | UAV RGB imagery | Zea mays | Field | [129] | |
| Physiology | |||||
| Early vigor | Early vigor of field pea seedlings | Automated RGB imaging platform, LemnaTec 3D Scanalyzer and handheld active sensor, crop circle | Pisum sativum | Greenhouse, field | [42] |
| Wheat vigor and canopy height quantification | UGV and UAV RGB imagery | Triticum aestivum | Field | [51] | |
| Monitoring plant and canopy growth dynamics | RGB or multispectral imagery, D3P8 | Triticum aestivum | Greenhouse | [130] | |
| Lodging | Lodging score estimation in barley by aerial imagery | High resolution UAV RGB imagery | Hordeum vulgare | Field | [131] |
| Estimation of crop lodging in wheat by aerial imagery | High resolution UAV RGB imagery | Triticum aestivum | Field | [132] | |
| Rice lodging scores estimated by UNet model derived from aerial imagery | High resolution UAV RGB and multispectral imagery | Oryza sativa | Field | [133] | |
| Photosynthesis and respiration | Photosynthetic capacities in tobacco | Handheld hyperspectral sensor, FieldSpec. | Nicotiana tabacum | Field | [134] |
| Leaf photosynthesis in maize | Handheld hyperspectral sensor, FieldSpec. | Zea mays | Field | [135] | |
| Leaf photosynthesis and relevant physiological parameters in wheat | Handheld hyperspectral sensor, FieldSpec. | Triticum aestivum | Field | [48] | |
| Water soluble carbohydrates | Estimates of stem water soluble carbohydrates at different growth stages in wheat | Handheld hyperspectral sensor, FieldSpec. | Triticum aestivum | Field | [49] |
| Predicting the quality of ryegrass (sugar) | Hyperspectral imaging platform | Lolium perenne | Field | [53] | |
| Canopy temperature | Wheat canopy temperature measurement | Airborne thermography and wireless infra-red thermometers | Triticum aestivum | Field | [54] |
| Maize canopy temperature measurement | UAV thermal and RGB imagery | Zea mays | Field | [136] | |
| Canopy temperature and vegetation indices of wheat | Airborne thermal and hyperspectral imagery | Triticum aestivum | Field | [29] | |
| Stay green | Stay-green associates with low water-soluble carbohydrates in oat | Handheld active sensor GreenSeeker | Avena sativa | Field | [137] |
| Characterization of maize green leaf area dynamics | UAV multispectral imagery | Zea mays | Field | [138] | |
| Senescence rate in wheat | UAV multispectral imagery | Triticum aestivum | Field | [52] | |
| Biomass and yield | Biomass, ground cover and canopy height estimates | UGV LiDAR | Triticum aestivum | Field | [55,139] |
| Estimation of shoot biomass by color imagery | RGB imaging platform, LemnaTec 3D Scanalyzer | Triticum aestivum | Greenhouse | [140] | |
| Grain yield prediction by canopy hyperspectral reflectance | Airborne hyperspectral imagery | Triticum aestivum | Field | [30,141] | |
| Wheat ear counts | Handheld thermal imagery | Triticum aestivum | Field | [142] | |
| Head counts in sorghum | High resolution UAV RGB imagery | Sorghum bicolor | Field | [143] | |
| Counting of wheat spikes | Handheld and UGV RGB imagery | Triticum aestivum | Field | [72,144] | |
| Wheat biomass and yield, nitrogen related traits | Automated RGB imaging platform, LemnaTec 3D Scanalyzer | Triticum aestivum | Greenhouse | [41] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, G.N.; Norton, S.L. Genebank Phenomics: A Strategic Approach to Enhance Value and Utilization of Crop Germplasm. Plants 2020, 9, 817. https://doi.org/10.3390/plants9070817
Nguyen GN, Norton SL. Genebank Phenomics: A Strategic Approach to Enhance Value and Utilization of Crop Germplasm. Plants. 2020; 9(7):817. https://doi.org/10.3390/plants9070817
Chicago/Turabian StyleNguyen, Giao N., and Sally L. Norton. 2020. "Genebank Phenomics: A Strategic Approach to Enhance Value and Utilization of Crop Germplasm" Plants 9, no. 7: 817. https://doi.org/10.3390/plants9070817
APA StyleNguyen, G. N., & Norton, S. L. (2020). Genebank Phenomics: A Strategic Approach to Enhance Value and Utilization of Crop Germplasm. Plants, 9(7), 817. https://doi.org/10.3390/plants9070817





