Abstract
Seed germination, one of the most important stages in a plant’s life cycle, can be affected by abiotic stresses, such as salinity. The plant hormone abscisic acid (ABA) and high concentrations of glucose are also known to inhibit germination. In contrast, nitrate is known to stimulate germination in many plants. However, this stimulatory effect has not yet been investigated in the presence of inhibitory effects caused by abiotic stresses, ABA, and glucose. In this study, we show that nitrate can alleviate the inhibitory effects of sodium chloride (NaCl) or high concentrations of glucose on seed germination in Arabidopsis, while it was not able to promote germination that was inhibited by exogenous ABA and mannitol (an inducer of osmotic stress). An analysis of the gene expression involved in the regulation of germination showed that GA20ox1, encoding the gibberellin (GA) synthesis enzyme, SPATULA (SPT), encoding a bHLH transcription factor, and CYP707A2, encoding an ABA catabolic enzyme, were significantly upregulated by the addition of KNO3 in the presence of NaCl or glucose. Our results suggest the possibility that these genes are involved in the nitrate-mediated control of seed germination in the presence of NaCl or glucose.
Keywords:
seed germination; Arabidopsis; nitrate; NaCl; glucose; abscisic acid (ABA); CYP707A2; SPATULA (SPT) 1. Introduction
Germination is a critical stage in the life of spermatophytes that is elaborately controlled by environmental factors such as light, temperature, water, and nutrients, in addition to endogenous signals such as the balance of phytohormones, gibberellin (GA), and abscisic acid (ABA) [1,2]. The influence of environmental factors on seed germination mostly occurs through the metabolism and signaling pathways of GA and ABA [3,4], where ABA promotes and maintains dormancy, while GA promotes germination [5].
In the GA metabolism, the enzymes involved in the final step of active GA synthesis, namely the GA 20-oxidase (GA20ox), GA 3-oxidase (GA3ox), and the enzyme involved in the deactivation of GAs, namely the GA 2-oxidase (GA2ox), are considered to be important in regulating seed germination. The major GA signaling components are the DELLA proteins, which belong to the GRAS family of transcription factors. These proteins inhibit plant growth and germination by negatively regulating the GA signaling (Table S1) [6,7].
Some enzymes of the ABA metabolism, for example the 9-cis-epoxy carotenoid dioxygenase (NCED), that catalyzes the synthesis of xanthoxine in plastids, and the cytochrome p450 type CYP707As, that catalyze ABA deactivation, resulting in phaseic acid (PA) and dihydrophaseic acid (DPA), are considered to be important for the regulation of seed germination [8]. Among the ABA signaling components, the ABSCISIC ACID INSENTIVE (ABI) 3 and 5 are well known to be involved in the regulation of seed germination (Table S1) [9,10].
The regulation mechanisms of seed germination have been extensively investigated in Arabidopsis. Arabidopsis seeds require after-ripening or low temperatures to break dormancy, and light to germinate. Recent studies identified many genes that are involved in the regulation of germination by light and dormancy-breaking (by cold stratification) also to be involved in the balance of GA and ABA [6,7,11]. Light induces the breakdown of PHYTOCHROME INTERACTING FACTOR 3 -LIKE5 (PIL5 or PIF1) proteins, which suppress germination in the dark by inducing the SOMNUS (SOM) and MOTHER OF FT and TFL1 (MFT) genes [12,13]. Cold stratification decreases the expression of SOM and MFT, but increases the expression of SPATULA (SPT) genes, resulting in the breakdown of dormancy. In addition, SPT promotes germination by repressing MFT under red light conditions [13]. These genes were shown to affect the expression of other genes involved in the GA or ABA metabolism and/or signaling, either directly or indirectly (Table S1) [13].
Nitrate is known to stimulate germination in a wide variety of plant species [14,15]; therefore, it is used as an agent for seed priming [16]. The effect of nitrate on germination does not depend on nitrate reductase (NR) [14,17], indicating that nitrate itself promotes seed germination. Recent research has shown that nitrate induced the expression of CYP707A2 gene in imbibed Arabidopsis seeds, and the cyp707a2 mutant was less sensitive to nitrate during both seed development and germination [18,19]. In addition, the NIN-like protein 8 (NLP8) was found to regulate CYP707A2 by directly binding to the CYP707A2 promoter region required for nitrate induction. As such, the nlp8 mutant was nonresponsive to nitrate. These results suggest that NLP8 and its downstream CYP707A2, are key genes in the nitrate-regulated germination in Arabidopsis [19] (Table S1).
Nitric oxide (NO) is another nitrogen compound that promotes germination and induces the expression of CYP707A2 [20,21]. Therefore, it was postulated that NO would act downstream of the nitrate signaling [22]. Recent research showed that in the presence of NO, group VII of the ethylene response factors (ERFs) is destroyed through the N-end rule pathway. Group VII ERF is involved in ABA signaling by regulating the expression and activity of ABI5 [23]. Mutants defective in the N-end rule pathway and abi5 displayed a NO-insensitive germination [23,24], however they were sensitive to nitrate. ABI5 expression was not altered in the nlp8 mutant [19]. Thus, all these results indicate that NO signaling seems to be more than just a simple, linear pathway, downstream of nitrate signaling.
It is well known that germination is also affected by abiotic stresses, such as salinity, drought, and unfavorable temperature [25] although the molecular mechanism by which these stresses inhibit germination has not been completely understood. In addition to these stresses, germination is also inhibited by high concentrations of glucose (or sucrose) and ABA, as mentioned above [3,26,27].
Although it is important to know whether nitrate can stimulate germination, even in the presence of inhibitors such as abiotic stresses, ABA, and high concentrations of glucose, it has not yet been investigated. In this study, we investigated the effects of nitrate on germination affected by salt, osmotic stress (mannitol), ABA and glucose, using Arabidopsis seeds. We found that nitrate promoted germination under inhibition by NaCl or high concentrations of glucose, but not under exogenously applied ABA and mannitol. Gene expression analysis showed that in addition to CYP707A2, the expression of GA20ox1 and SPT were increased by KNO3 in the presence of NaCl or glucose.
2. Materials and Methods
2.1. Plant Materials and Seed Germination Assays
Arabidopsis (Col-0) plants were grown in growth room at 23 °C under 16 h light/ 8 h dark cycle (100 μmol m−2 sec−1). Seeds were harvested when siliques turned dry on the plants. Seeds were stored in a dry box at 23 °C before use.
For the germination assay, seeds stored for one-two months were used. For the germination assay of freshly harvested seeds, seeds just after harvested were used.
These seeds were surface sterilized in 20% bleach for 5 min, rinsed five times with sterile water, and then plated on 0.8% agar plates. To examine the effect of NaCl and mannitol, half-strength Murashige and Skoog (1/2 MS) medium or distilled water (DW), containing 0, 102, 170, and 340 mM NaCl, or 0, 100, 200, 400, and 500 mM mannitol, respectively, were used. Since the 1/2 MS medium already contains nitrates as macronutrients, to examine the effect of these nitrogen compounds we prepared the DW+N medium, which contained the same concentration of nitrates (9 mM KNO3 and 10 mM NH4NO3) that are found in the 1/2 MS.
To examine the effect of KNO3 or NH4NO3, a medium containing 9 mM KNO3 and either 170 mM NaCl or 278 mM glucose (DW + KNO3 + NaCl or Glu), and a medium containing 10 mM NH4NO3 and either 170 mM NaCl or 278 mM glucose (DW + NH4NO3 + NaCl or Glu) were prepared. To examine the effect of NH4Cl, a DW + NH4Cl + NaCl or Glu medium that contained 10 mM NH4Cl and 170 mM NaCl or 278 mM glucose, respectively, were prepared. To examine the effect of KCl, a DW + KCl + NaCl or Glu medium that contained either 10 mM KCl and 170 mM NaCl or 278 mM glucose, respectively, were prepared.
Plates with seeds were first placed at 4 °C for 3 days (stratification) and then transferred to a growth chamber at 23 °C with continuous light (65 μmol m−2 sec−1). We used continuous light to avoid the effect of a light-dark cycle during the germination assay. Germination was scored by radicle emergence at 24, 32, 48, 56, 72, and 80 h after being transferred to 23 °C. In the experiments with glucose, germination scoring at 96 and 104 h was included. In the experiments with freshly harvested seeds, the germination was scored until 200 h (with additional scoring at 120, 128, 144, 152, 168, 176, 192, and 200 h). Each plate contained 50 seeds, and three plates were used for each experiment. The data on the final germination of each experiment were analyzed using an ANOVA followed by Tukey’s test.
2.2. RNA Extraction
Approximately 30 mg of seeds were sown on sterile filters in Petri dishes (60 × 15 mm), containing DW or 10 mM KNO3 solution (DW + KNO3) containing 170 mM NaCl, 278 mM glucose, 5 μM ABA, or 500 mM mannitol. Plates with seeds were first placed at 4 °C for 3 days and then transferred to a growth chamber at 23 °C with continuous light. After 6 or 24 h of incubation in a growth chamber, seeds were collected and stored at −80 °C for RNA extraction.
Total RNA from seeds was isolated using the ISOGEN II (Nippon Gene, Tokyo, Japan) and Fruit-mate (TAKARA, Ohtsu, Japan) reagents. cDNA was synthesized (from 0.5 μg RNA template) using the ReverTra Ace Kit (TOYOBO, Osaka, Japan) according to the manufacturer’s instructions.
2.3. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Real-time PCR amplification of cDNAs was conducted using a LightCycler 480 (Roche Diagnostics, Rotkreuz, Switzerland) in a 384-well PCR plate. The reactions were carried out in 10 μL reaction volumes, containing 5 μL of FastStart Essential DNA Green Master (Roche Diagnostics) with 0.2 μM of the forward and reverse primers and 1 μL cDNA (10-fold dilution). The primer sets used for real-time PCR are shown in Table S2. The primers used in this experiment were designed by the Universal ProbeLibrary Assay Design Center (Roche Diagnostics). The ELONGATION FACTOR1 αA4 (EF1αA4) gene was used for signal normalization in the real-time PCR.
Relative expression levels were calculated using the 2−ΔΔCT method [28]. All PCR reactions were performed at least five times, out of which at least three sets of consistent data were used for the analyses. In order to validate the reliability of data, we compared the amplification efficiencies of the target and reference genes for all PCR reactions, and examined the dissociation curves for all PCR products. Data were statistically analyzed by a Student’s t test.
2.4. Semi-Quantitative RT-PCR
These PCR reactions were performed with the Emeraldamp PCR master mix (Takara) using the following program: 98 °C for 2 min; 30–35 cycles of 15 s at 98 °C, 30 s at 55 °C, and 0.5–2 min at 72 °C; then hold at 72 °C for 5 min. The ACTIN2/8 gene was used as a reference gene. The primer sets used for the expression analysis are shown in Table S2.
2.5. Statistical Analysis
Statistical analysises were performed using the EZR software [29] which is based on R commander.
3. Results
3.1. Germination in the Presence of NaCl or Mannitol
We examined the inhibitory effect of NaCl and mannitol (inducer of osmotic stress) on the germination of Arabidopsis seeds, and found that germination was reduced on 1/2 MS containing 170 mM (1%) NaCl, and was completely inhibited on 340 mM NaCl (Figure 1A). Interestingly, germination was already reduced at 102 mM NaCl and 170 mM NaCl could inhibit germination fully in DW. The same concentration of KCl (170 mM) delayed germination only slightly, indicating that the inhibition was caused mainly by sodium and not chloride. Mannitol delayed germination in a concentration-dependent manner, and the germination was significantly reduced by 500 mM mannitol in both 1/2 MS and DW. The effect of mannitol on germination was similar between treatment using 1/2 MS and DW as the medium (Figure 1B).
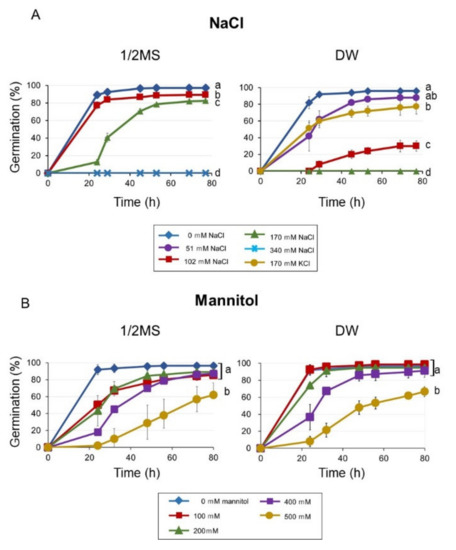
Figure 1.
Seed germination on 1/2 MS or distilled water (DW), in the presence of several concentrations of NaCl (A) and mannitol (B) Seeds were stratified for three days before incubation at 23 °C. Germination was scored after transferred to 23 °C. Data are means ± standard deviation (SD) of three replicates. Each replicate contained 50 seeds. Different letters indicate significant differences in germination at the final time point (ANOVA and Tukey’s test, p < 0.05).
Since the germination-promoting effects of nitrogen compounds, such as nitrate and nitrogen oxide (NO) are known [14,19,22,30], we speculated that nitrogen (N) components in 1/2 MS medium would promote germination in the presence of 170 mM NaCl. Therefore, we examined the germination on both 1/2 MS and DW in the presence of N compounds (DW + N: 9 mM KNO3 and 10 mM NH4NO3) containing 170 mM NaCl (Figure 2). The germination on DW + N containing 170 mM NaCl (DW + N + NaCl) was almost the same as on 1/2 MS containing 170 mM NaCl (1/2 MS + NaCl) (Figure 2A). The results suggested that the reason why the germination on 1/2 MS + NaCl was better than that on DW + NaCl was mainly due to the presence of N compounds in the 1/2 MS medium. We also examined the germination on DW + N containing 500 mM mannitol (DW + N + Man), which was almost the same as that on DW containing 500 mM mannitol (DW + Man) (Figure 2D).
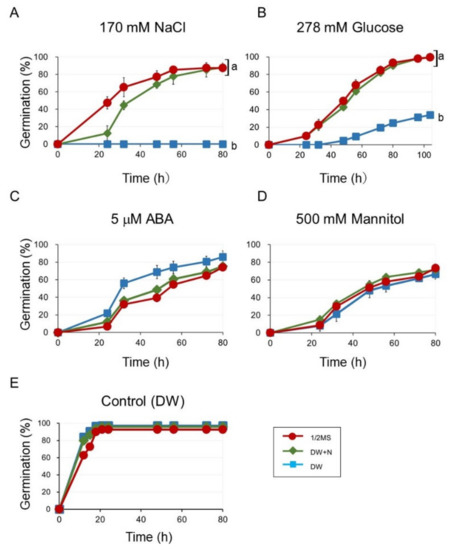
Figure 2.
Seed germination on 1/2 MS, nitrogen components of 1/2 MS (DW+N), and DW, in the presence of (A) 170 mM NaCl, (B) 278 mM glucose, (C) 5 μM ABA, and (D) 500 mM mannitol. (E) controls (no inhibitory compounds) Seeds were stratified for three days before incubation at 23 °C. Germination was scored after transferred to 23 °C. Data are means ± SD of three replicates with each replicate containing 50 seeds. Different letters indicate significant differences in germination at the final time point (ANOVA and Tukey’s test, p < 0.05).
The differences observed in the effect of N compounds on the germination inhibited by NaCl or mannitol prompted us to analyze other germination inhibitors, such as ABA and high concentrations of glucose. Interestingly, germination in DW containing 5 μM ABA (DW + ABA) was inhibited to a similar extent as that on 1/2 MS containing 5 μM ABA (1/2 MS + ABA) and DW+N containing 5 μM ABA (DW + N + ABA) (Figure 2C), whereas germinations on 1/2 MS containing 278 mM (5%) glucose (1/2 MS + Glu) and DW + N containing 278 mM glucose (DW + N + Glu) were much higher than that on DW containing 278 mM glucose (DW + Glu) (Figure 2B).
Higher germination in the presence of NaCl or glucose on 1/2 MS or DW + N were observed when the germination assay was carried out using freshly harvested seeds without stratification (Figure S1).
3.2. Nitrate Promotes Germination Inhibited by NaCl or High Concentrations of Glucose
As we used a mixture of 9 mM KNO3 and 10 mM NH4NO3 as nitrates in the media, we had to examine the effect of each nitrogen compound on germination individually. Our experimental setup consisted of germinations inhibited by NaCl or Glu (DW + NaCl or DW + Glu) in the presence of either 9 mM KNO3 (DW + KNO3 + NaCl or DW + KNO3 + Glu) or 10 mM NH4NO3 (DW + NH4KO3 + NaCl or DW + NH4NO3 + Glu). For comparison, we also included germination with DW + N + NaCl and DW + N + Glu. The germination for DW + KNO3 + NaCl and DW + NH4NO3 + NaCl were similar but slower than that on DW + N + NaCl (Figure 3A). However, when we reduced KNO3 concentration to 1 mM, the germination was also reduced (Figure 3A).
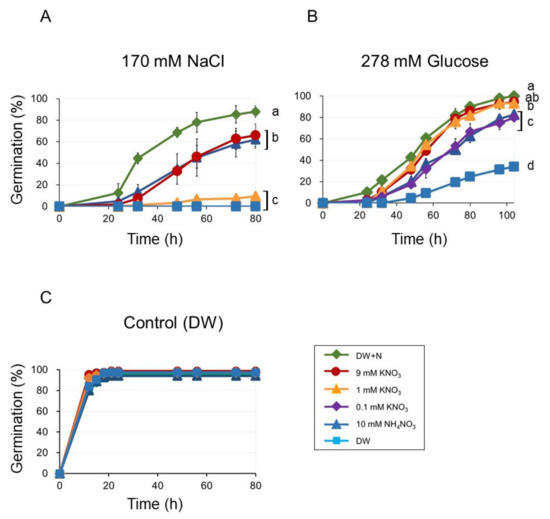
Figure 3.
Seed germination on KNO3 or NH4NO3 medium (DW, DW + N, 9 mM KNO3, 10 mM NH4NO3, 1 mM KNO3 and 0.1 mM KNO3) in the presence of inhibitory compounds (A) 170 mM NaCl, (B) 278 mM glucose, and (C) controls (no inhibitory compounds). Seeds were stratified for three days before incubation at 23 °C. Germination was scored after transferred to 23 °C. Data are means ± SD of three replicates. Each replicate contained 50 seeds. Different letters indicate significant differences in germination the final time point (ANOVA and Tukey’s test, p < 0.05).
Furthermore, we decided to analyze the effect of ammonium salt (NH4Cl) instead of NH4NO3, as the latter contains both NH4+ and NO3−. The germination rate in DW + NaCl containing 10 mM NH4Cl (DW + NH4Cl + NaCl) was better than in DW + 1 mM KNO3 + NaCl, but worse than in DW + KNO3 + NaCl (Figure S2A). Since KNO3 also contains potassium (K+), we examined the effect of potassium using KCl. The germination rate in DW + NaCl containing 10 mM KCl (DW + KCl + NaCl) was similar to that of DW + NH4Cl + NaCl, indicating that both NH4Cl and KCl could improve germination in the presence of NaCl, although the effects were weaker than for KNO3 (Figure S2A).
The germination rates were similar in DW + KNO3+ Glu and DW + 1 mM KNO3 + Glu (Figure 3B). Moreover, germination in DW + Glu was similarly improved by 0.1 mM KNO3 and 10 mM NH4NO3 (Figure 3B). Germination in DW + Glu containing 10 mM NH4Cl (DW + NH4Cl+Glu) or 10 mM KCl (DW + KCl + Glu) was only slightly lower than in DW + KNO3 + Glu (Figure S2B). Thus, a much lower KNO3 concentration was already effective at enhancing the germination inhibited by glucose than what we observed for NaCl.
In the control experiments (without any inhibitory compounds), no significant differences in germination were observed under both KNO3 and NH4NO3 (Figure 3C).
Furthermore, we decided to analyze the effect of ammonium salt (NH4Cl) instead of NH4NO3, as the latter contains both NH4+ and NO3−. The germination rate in DW + NaCl containing 10 mM NH4Cl (DW + NH4Cl + NaCl) was better than in DW + 1 mM KNO3 + NaCl, but worse than in DW + KNO3 + NaCl (Figure S2A). Since KNO3 also contains potassium (K+), we examined the effect of potassium using KCl. The germination rate in DW + NaCl containing 10 mM KCl (DW + KCl + NaCl) was similar to that of DW + NH4Cl + NaCl, indicating that both NH4Cl and KCl could improve germination in the presence of NaCl, although the effects were weaker than for KNO3 (Figure S2A).
The germination rates were similar in DW + KNO3 + Glu and DW + 1 mM KNO3 + Glu (Figure 3B). Moreover, germination in DW + Glu was similarly improved by 0.1 mM KNO3 and 10 mM NH4NO3 (Figure 3B). Germination in DW + Glu containing 10 mM NH4Cl (DW + NH4Cl+Glu) or 10 mM KCl (DW + KCl + Glu) was only slightly lower than in DW + KNO3 + Glu (Figure S2B). Thus, a much lower KNO3 concentration was already effective at enhancing the germination inhibited by glucose than what we observed for NaCl.
In the control experiments (without any inhibitory compounds), no significant differences in germination were observed under both KNO3 and NH4NO3 (Figure 3C).
3.3. Gene Expression in Seeds Imbibed with NaCl, Glucose, ABA, and Mannitol
Next, we analyzed the gene expression in seeds imbibed in the presence of several inhibitory compounds, namely, 170 mM NaCl, 278 mM glucose, 5 μM ABA, and 500 mM mannitol by qRT-PCR (Figure 4). The length of the imbibing period (6 and 24 h) was chosen to detect both rapid and slower changes in gene expression, as many of the genes could be detected within 24 h of imbibition, while genes that are rapidly induced by nitrate are detected within 6 h [31].
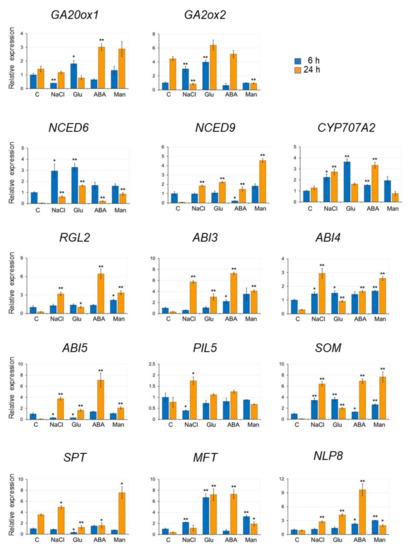
Figure 4.
Expression of genes involved in the regulation of seed germination in the presence of NaCl, glucose, ABA, and mannitol. Seeds were imbibed in the presence of 170 mM NaCl, 278 mM glucose (Glu), 5 μM ABA, and 500 mM mannitol (Man) for 6 h and 24 h. C: control (no inhibitory compounds). Gene expression was quantified by quantitative RT-PCR (qRT-PCR) and presented as relative to the levels in the control seeds (imbibed for 6 h). The expression levels were normalized against the expression of ELONGATION FACTOR1 αA4 (EF1αA4). Data are presented as the mean ± SD of three replicates. Asterisks indicate a significant difference between control and each treatment at p < 0.01 (**) and p < 0.05 (*). (Student’s t-test).
The selected genes are involved in the regulation of seed germination, specifically genes encoding enzymes involved in GA synthesis (GA20 ox1), GA catabolism (GA2ox1), ABA synthesis (NCED6 and NCED9), ABA catabolism (CYP707A2), DELLA (RGL2), factors involved in ABA signaling (ABI3, ABI4, and ABI5) and seed germination by light and temperature (PIL5, SOM, SPT, and MFT), plus NLP8, which was identified as an important factor in promoting CYP707A2 expression by the addition of nitrate (Table S1) [19].
Figure 4 shows the relative expression of each gene in seeds imbibed with NaCl, glucose, ABA, and mannitol for 6 or 24 h, compared with their expression in seeds imbibed for 6 h in DW. The expression pattern of each gene differed depending on the added inhibitory compounds. Since the expression of NCED6, NCED9, RGL2, ABI3, ABI4, ABI5, SOM, and MFT decreased after 24 h imbibition in DW, the relative expression of these genes in seeds imbibed in the presence of the inhibitory compounds was much higher than in seeds imbibed in DW. Overall, we found these inhibitory compounds to induce the expression of genes that repress germination or that are involved in ABA synthesis and ABA signaling.
3.4. Gene Expression in Seeds Imbibed With or Without KNO3 in the Presence of NaCl, Glucose, ABA, or Mannitol
We compared the gene expression in seeds imbibed with or without 10 mM KNO3 in the presence of NaCl, glucose, ABA, or mannitol, by semi-quantitative RT-PCR (Figure S3). We used KNO3 because of its widespread use in studies that examined the effect of nitrates on germination [14,18,19]. In addition to the genes shown on Figure 4, we also analyzed other genes by semi-quantitative RT-PCR: namely, genes for GA synthesis (GA3ox1 and GA3ox2), DELLAs (GAI, RGA, RGL1, and RGL3), and EXPANSINs (EXP1 and EXP2). The semi-quantitative RT-PCR analysis showed that the expression of several genes was altered by the addition of nitrate (Figure S3). The change in gene expression by the addition of KNO3 was different, depending on the added inhibitory compounds, so we could not find any common effect of KNO3 on gene expression. Since we could not detect significant changes in the expression of GA3oxes, DELLAs, and EXPANSINs by semi-quantitative analysis in the presence or absence of KNO3, we decided to examine the genes from Figure 4 by qRT-PCR.
Because the addition of KNO3 increased the germination in the presence of 170 mM NaCl or 278 mM Glu, we compared the gene expression in seeds imbibed with or without KNO3 in the presence of NaCl or glucose (Figure 5).
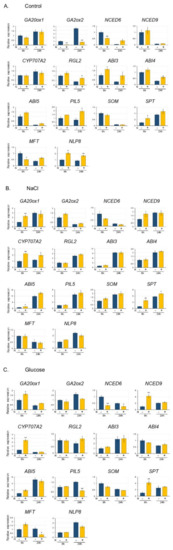
Figure 5.
Expression of genes involved in the regulation of seed germination in the presence (+) or absence (−) of 10 mM KNO3. Seeds were imbibed in (A) control: no additional compounds, (B) in the presence of 170 mM NaCl, (C) in the presence of 278 mM glucose, for 6 and 24 h, respectively. Gene expression was quantified by qRT-PCR and values are presented as relative to the control seeds (imbibed for 6 h). The expression levels were normalized against the expression of ELONGATION FACTOR1 αA4 (EF1αA4). Data are presented as the mean ± SD for three replicates. Asterisks indicate a significant difference between the absence (−) and the presence (+) of KNO3 at p < 0.01 (**) and p < 0.05 (*) (Student’s t-test).
In the absence of NaCl or Glu, GA2ox2 expression was significantly reduced by KNO3 in both 6 h and 24 h imbibition (Figure 5A). Although NLP8 expression was increased by the addition of KNO3, the expression of CYP707A2 did not change after either 6 or 24 h of imbibition. SPT expression however, increased, while MFT expression was decreased by KNO3 after 6 h of imbibition (Figure 5A).
In the presence of 170 mM NaCl, the expression of GA20ox1 was higher after 6 h of imbibition with KNO3, while the expression of GA2ox2 did not change. Interestingly, the expression of CYP707A2 was increased after both 6 and 24 h imbibition with KNO3, although the expression of NLP8 did not change. The expression of SPT was increased in both 6 h and 24 h imbibition with KNO3, while the expression of MFT did not change (Figure 5B).
In the presence of 278 mM Glu, the expression of GA20ox1, NCED9, and CYP707A2 increased after 6 h of imbibition with KNO3, while the expression of NCED6 decreased following both 6 and 24 h of imbibition with KNO3. Moreover, SPT expression also increased after 6 h of imbibition with KNO3, while that of MFT decreased after 24 h of imbibition (Figure 5C).
4. Discussion
Nitrate has been shown to be one of the signals that relieve seed dormancy in many species, including Arabidopsis [14,15]. However, the effect of nitrate on germinations affected by abiotic stresses, ABA, and glucose has not been investigated. Our study showed that nitrate was able to enhance germinations that were inhibited by NaCl and high concentrations of glucose, but was unsuccessful in promoting germinations inhibited by exogenous ABA and mannitol (Figure 2).
It is well known that salt affects seed germination [32,33,34], however, the mechanisms by which salt inhibits seed germination remain largely unknown. Salinity stress by NaCl consists of a primarily osmotic stress plus the toxic effect of Na+. Since our results indicated that the germinations inhibited by osmotic stress were not improved by the addition of nitrate (Figure 2), it seems that the toxicity of Na+ could have been mitigated by nitrate. Ethylene signaling has been reported to modulate salt response, including germination inhibition [35] and recent research revealed the roles of ethylene signaling in salt stress. Yu et al. (2016) showed that salt treatment caused the CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1) protein to be retained in the cytosol and inhibited the interaction between COP1 and LONG HYPOCOTYL 5 (HY5) [36]. COP1, a ring finger E3 ligase, is translocated to the nucleus in the dark and is responsible for the proteasome-mediated degradation of photomorphogenesis-promoting factors, such as HY5 [37,38]. HY5 has been reported to mediate ABA response through ABI5 during seed germination and early seedling growth [35]. Therefore, HY5 activates ABA signaling in the presence of salt stress to inhibit germination. In contrast, ethylene enhances the localization of COP1 to the nucleus to promote germination [36]. In addition, Li et al. (2015) reported that NO managed to alleviate germination inhibition induced by salt stress, by enhancing ethylene signaling [39]. In many plants, NO can be produced from nitrate by nitrate reductase (NR) [31,40]. Therefore, it could be possible that the effect of nitrate on germination inhibited by NaCl is attributed to the effect of NO, through the activation of ethylene signaling. However, our expression analysis showed that the expression of ABI5 was not significantly different in the presence or the absence of nitrate (Figure 5), suggesting that nitrate did not attenuate ABA signaling through ABI5 in our experimental setup. However, further research is required to elucidate whether the effect of nitrate is related to NO and/or ethylene signaling.
In general, the expression of all examined genes was affected by the addition of NaCl (Figure 5B), but only the expression of GA20ox1, CYP707A2, and SPT was upregulated by the addition of KNO3 in the presence of NaCl. CYP707A2 has been reported to be a key gene for the enhancement of seed germination by nitrate in Arabidopsis [8,18], so the observed increase in the expression of CYP707A2 could have contributed to the improvement of germination in the presence of NaCl.
By modulating gene expression and influencing a variety of processes such as germination, early seedling development, flowering, or senescence, glucose plays an important regulatory role as a central signaling molecule [40].
High concentrations of glucose are known to delay germination in several plants [26,27,41], but the mechanisms by which this happens have not been completely clarified. Exogenously applied high concentrations of glucose during germination lead to enhancement of ABA biosynthesis [42] and the repression of genes associated with ABA catabolism [27]. It has been reported that the delay of germination by glucose is not caused by increased cellular ABA concentration, but rather by the fact that glucose appears to slow down the decline of endogenous ABA. Moreover, the glucose-induced delay in germination seems to be independent from hexokinase (HXK) [26,42,43].
Our expression analysis showed that genes of the ABA biosynthesis (NCED6 and NCED9) and ABA signaling (ABI3, 4, and 5) were increased by the addition of 278 mM glucose (Figure 4). CYP707A2, however, involved in ABA degradation, was also increased by the addition of glucose (Figure 4). This result contradicts other reports according to which the expression of CYP707A2 was reduced by the addition of glucose [27,43]. It is not clear why these expression data were different. In rice seeds the expressions of OsABA8ox2 and OsABA8ox3 (rice CYP707As orthologues) were decreased by 1% glucose, but their expression increased to a level that was similar in control (0% glucose) under higher concentrations (5%) of glucose [27]. The glucose concentration used in our experiment was 5%, which is higher than the one used by Zhu et al. (1% and 3%) in Arabidopsis [43]. Hence, it is possible that the response of CYP707A2 expression to glucose might be concentration-dependent.
With the addition of KNO3, the expression of CYP707A2 was higher than in control at 6 h, while the expression of NCED6 was lower at 6 and 24 h in the presence of 278 mM glucose. In contrast, NCED9 expression was higher than in control at 6 h, and dropped to a similar level at 24 h (Figure 5C). These results indicated that the expression of genes involved both in ABA biosynthesis and catabolism were affected by KNO3. In contrast, the expression of RGL2 and genes for ABA signal transduction were not changed by KNO3. Interestingly, SPT expression was significantly reduced by the addition of glucose (Figure 4), and it was increased by KNO3 application (Figure 5C). On the other hand, MFT expression was increased by the addition of glucose (Figure 4), but it was suppressed by KNO3 (Figure 5C). Therefore, the assumption that MFT and SPT would be directly involved in the regulation of germination by glucose and KNO3 needs further clarification.
The expression of GA20ox1, CYP707A2, and SPT were increased by KNO3 both in the presence of NaCl or glucose. The increase of CYP707A2 and GA20ox1 by KNO3 may partly explain the reason why KNO3 enhanced germination in the presence of NaCl or glucose. However, it is not yet clear how increased SPT expression could contribute to enhanced germination in the presence of NaCl or glucose.
The function of SPT in the physiology of germination are complicated. For example, SPT induces the expression of ABI5 and RGL3, while it represses that of MFT, ABI4, and RGA in freshly matured Arabidopsis seeds. As a result, SPT promotes dormancy in Columbia ecotype, but represses it in the Landsberg erecta ecotype [44]. However, SPT promotes germination in imbibed seeds, by repressing MFT under red light conditions [13]. It will be interesting to elucidate whether SPT is involved in the enhancement of germination by KNO3 in the presence of NaCl or glucose.
The fact that KNO3 did not alleviate the effects of inhibition by exogenously supplied ABA on the germination (Figure 2) was an unexpected finding, although the expression of CYP707A2 was induced by the addition of KNO3 both in our current and previous studies [18,19]. The addition of KNO3 triggered an increase in the expression of CYP707A2 in the presence of 5μM ABA (Figure 4), however it is possible that this increase was not enough to reduce the exogenous ABA level. However, KNO3 application did not enhance germination even when this was inhibited by a much lower concentration of ABA (1 μM) (Figure S4). The mapping of genes involved in the regulation of germination that code for enzymes involved in ABA synthesis and ABA degradation revealed overlapping expression patterns (during germination) in the root cap, epidermis, and vascular cells of the radicle in Arabidopsis embryos. On the other hand, the expression of ABA response-genes has been mainly localized to the outer cell layers of the embryo radicle, principally the root cap and epidermis [45]. These results indicate that, while endogenously-produced ABA is degraded efficiently, the degradation of exogenously-applied ABA may not be efficient, despite an increase in the CYP702A2 expression.
Finally, our germination analysis showed that both NH4Cl and KCl could alleviate the NaCl—or glucose-induced inhibition of germination, although they were less effective than KNO3 (Figure S2). Ammonium salts and other nitrogenous compounds have been reported to promote germination in some plants, but the mechanisms behind this have not been clarified [46]. These nitrogenous compounds yield NO under strong oxidation, and the produced NO might promote germination [30]. However, it is not clear whether the effect of NH4Cl on germination in our experiment is through the production of NO. Although KCl is sometimes used for seed priming [30], the mechanism by which KCl stimulates germination has also not been clarified. Thus, the elucidation of how these compounds can improve germination under inhibition by NaCl or glucose is of great importance.
Further research will be needed, with a comprehensive temporal and spatial gene expression analysis, to elucidate how the inhibition of germination under unfavorable conditions (by high salinity and glucose) are alleviated by nitrate.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/6/707/s1. Figure S1: Seed germination on 1/2 MS, nitrogen components of 1/2 MS (DW+N), and DW in the presence of (A) 170 mM NaCl, (B) 278 mM glucose, and (C) no additional compounds (control)., Figure S2: The effect of KCl and NH4Cl on seed germination in the presence of (A) 170 mM NaCl and (B) 278 mM Glucose., Figure S3: Expression of genes involved in the regulation of seed germination in the presence in the presence (+N) or absence (−N) of 10 mM KNO3., Figure. S4. Seed germination on 1/2 MS, nitrogen components of 1/2 MS (DW + N), and DW in the presence of 1 μM ABA. Table S1: List of genes used in this study., Table S2: Primers used in this study.
Author Contributions
The design of the study was made by A.K.; Experiments were performed by S.I., T.A., I.I., and A.T.; A.K. drafted the manuscript. All authors read and approved the final manuscript.
Funding
This research received no external funding.
Acknowledgments
We thank Global Science Campus supported by Japan Science and Technology Agency for supporting this study. We also thank Mano, F., and Yaguchi, M. for help in carrying out experiments.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Footitt, S. Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 2017, 68, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Liu, X.-D.; Xie, Q.; He, Z.-H. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef]
- Tuan, P.A.; Kumar, R.; Rehal, P.K.; Toora, P.K.; Ayele, B.T. Molecular Mechanisms Underlying Abscisic Acid/Gibberellin Balance in the Control of Seed Dormancy and Germination in Cereals. Front Plant Sci. 2018, 9, 668. [Google Scholar] [CrossRef]
- Gazzarrini, S.; Tsai, A.Y.-L. Hormone cross-talk during seed germination. Essays Biochem. 2015, 58, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Chen, Z. The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Rep. 2017, 36, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.-H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Piskurewicz, U.; Jikumaru, Y.; Kinoshita, N.; Nambara, E.; Kamiya, Y.; Lopez-Molina, L. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 2008, 20, 2729–2745. [Google Scholar] [CrossRef]
- Seo, M.; Nambara, E.; Choi, G.; Yamaguchi, S. Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 2009, 69, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Yamaguchi, S.; Lim, S.; Oh, E.; Park, J.; Hanada, A.; Kamiya, Y.; Choi, G. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 2008, 20, 1260–1277. [Google Scholar] [CrossRef] [PubMed]
- Vaistij, F.E.; Barros-Galvão, T.; Cole, A.F.; Gilday, A.D.; He, Z.; Li, Y.; Harvey, D.; Larson, T.R.; Graham, I.A. MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, 8442–8447. [Google Scholar] [CrossRef] [PubMed]
- Alboresi, A.; Gestin, C.; Leydecker, M.T.; Bedu, M.; Meyer, C.; Truong, H.N. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant, Cell Environ. 2005, 28, 500–512. [Google Scholar] [CrossRef]
- Mei Wang, R.M.v.d.M.; Visser, R.M.v.d.M.; Mei Wang, K.; Van Schaik, H.-K.; Van Duijn, B.; De Boer, A.H. Effects of dormancy-breaking chemicals on ABA levels in barley grain embryos. Seed Sci. Res. 1998, 8, 128–137. [Google Scholar]
- Pill, W.G.; Fret, J.J.; Morneau, D.C. Germination and seedling emergence of primed tomato and asparagus seeds under adverse conditions. HortScience 1991, 26, 1160–1162. [Google Scholar] [CrossRef]
- Henk, W.M.; Hilhorst, C.M.K. Nitrate Reductase Independent Stimulation of Seed Germination in Sisymbrium officinale L. (Hedge Mustard) by Light and Nitrate. Ann. Bot. 1989, 63, 131–137. [Google Scholar]
- Matakiadis, T.; Alboresi, A.; Jikumaru, Y.; Tatematsu, K.; Pichon, O.; Renou, J.P.; Kamiya, Y.; Nambara, E.; Truong, H.N. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 2009, 149, 949–960. [Google Scholar] [CrossRef]
- Yan, D.; Easwaran, V.; Chau, V.; Okamoto, M.; Ierullo, M.; Kimura, M.; Endo, A.; Yano, R.; Pasha, A.; Gong, Y.; et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 2016, 7, 13179. [Google Scholar] [CrossRef]
- Arc, E.; Galland, M.; Godin, B.; Cueff, G.; Rajjou, L. Nitric oxide implication in the control of seed dormancy and germination. Front. Plant Sci. 2013, 4, 346. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, L.; Ye, N.; Liu, R.; Jia, W.; Zhang, J. Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 2009, 183, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Bethke, P.C.; Libourel, I.G.; Jones, R.L. Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 2006, 57, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Md Isa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marín-de la Rosa, N.; Vicente Conde, J.; Sousa Correia, C.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 2015, 6, 8669. [Google Scholar] [CrossRef] [PubMed]
- Vishal, B.; Kumar, P.P. Regulation of Seed Germination and Abiotic Stresses by Gibberellins and Abscisic Acid. Front. Plant Sci. 2018, 9, 838. [Google Scholar] [CrossRef]
- Dekkers, B.J.W.; Schuurmans, J.A.M.J.; Smeekens, S.C.M. Glucose delays seed germination in Arabidopsis thaliana. Planta 2004, 218, 579–588. [Google Scholar] [CrossRef]
- Zhu, G.; Ye, N.; Zhang, J. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 2009, 50, 644–651. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 2013, 48, 452–458. [Google Scholar] [CrossRef]
- Osuna, D.; Prieto, P.; Aguilar, M. Control of Seed Germination and Plant Development by Carbon and Nitrogen Availability. Front. Plant Sci. 2015, 6, 1023. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.; Neff, M.M.; Hong, S.-W.; Zhang, H.; Deng, X.-W.; Xiong, L. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. USA 2008, 105, 4495–4500. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Cao, Y.-R.; Chen, S.-Y.; Zhang, J.-S. Ethylene signaling regulates salt stress response: An overview. Plant Signal. Behav. 2008, 3, 761–763. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, J.; Shi, H.; Gu, J.; Dong, J.; Deng, X.W.; Huang, R. Salt Stress and Ethylene Antagonistically Regulate Nucleocytoplasmic Partitioning of COP1 to Control Seed Germination. Plant Physiol. 2016, 170, 2340–2350. [Google Scholar] [CrossRef]
- Osterlund, M.T.; Ang, L.H.; Deng, X.W. The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 1999, 9, 113–118. [Google Scholar] [CrossRef]
- Ang, L.H.; Chattopadhyay, S.; Wei, N.; Oyama, T.; Okada, K.; Batschauer, A.; Deng, X.W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1998, 1, 213–222. [Google Scholar] [CrossRef]
- Li, X.; Pan, Y.; Chang, B.; Wang, Y.; Tang, Z. NO Promotes Seed Germination and Seedling Growth Under High Salt May Depend on EIN3 Protein in Arabidopsis. Front. Plant Sci. 2015, 6, 1203. [Google Scholar] [CrossRef]
- Yamasaki, H.; Sakihama, Y. Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: In vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 2000, 468, 89–92. [Google Scholar] [CrossRef]
- Zhao, M.-G.; Liu, R.-J.; Chen, L.; Tian, Q.-Y.; Zhang, W.-H. Glucose-induced inhibition of seed germination in Lotus japonicus is alleviated by nitric oxide and spermine. J. Plant Physiol. 2009, 166, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Price, J.; Li, T.-C.; Kang, S.G.; Na, J.K.; Jang, J.-C. Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol. 2003, 132, 1424–1438. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Liu, Y.; Ye, N.; Liu, R.; Zhang, J. Involvement of the abscisic acid catabolic gene CYP707A2 in the glucose-induced delay in seed germination and post-germination growth of Arabidopsis. Physiologia Plantarum 2011, 143, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Vaistij, F.E.; Gan, Y.; Penfield, S.; Gilday, A.D.; Dave, A.; He, Z.; Josse, E.-M.; Choi, G.; Halliday, K.J.; Graham, I.A. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc. Natl. Acad. Sci USA 2013, 110, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- Topham, A.T.; Taylor, R.E.; Yan, D.; Nambara, E.; Johnston, I.G.; Bassel, G.W. Temperature variability is integrated by a spatially embedded decision-making center to break dormancy in Arabidopsis seeds. Proc. Natl. Acad. Sci USA 2017, 114, 6629–6634. [Google Scholar] [CrossRef]
- Hendricks, S.B.; Taylorson, R.B. Promotion of seed germination by nitrate, nitrite, hydroxylamine, and ammonium salts. Plant Physiol. 1974, 54, 304–309. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).