Activation of Early Defense Signals in Seedlings of Nicotiana benthamiana Treated with Chitin Nanoparticles
Abstract
1. Introduction
2. Results
2.1. CNPs Can Be Synthesized by Emulsion W/O
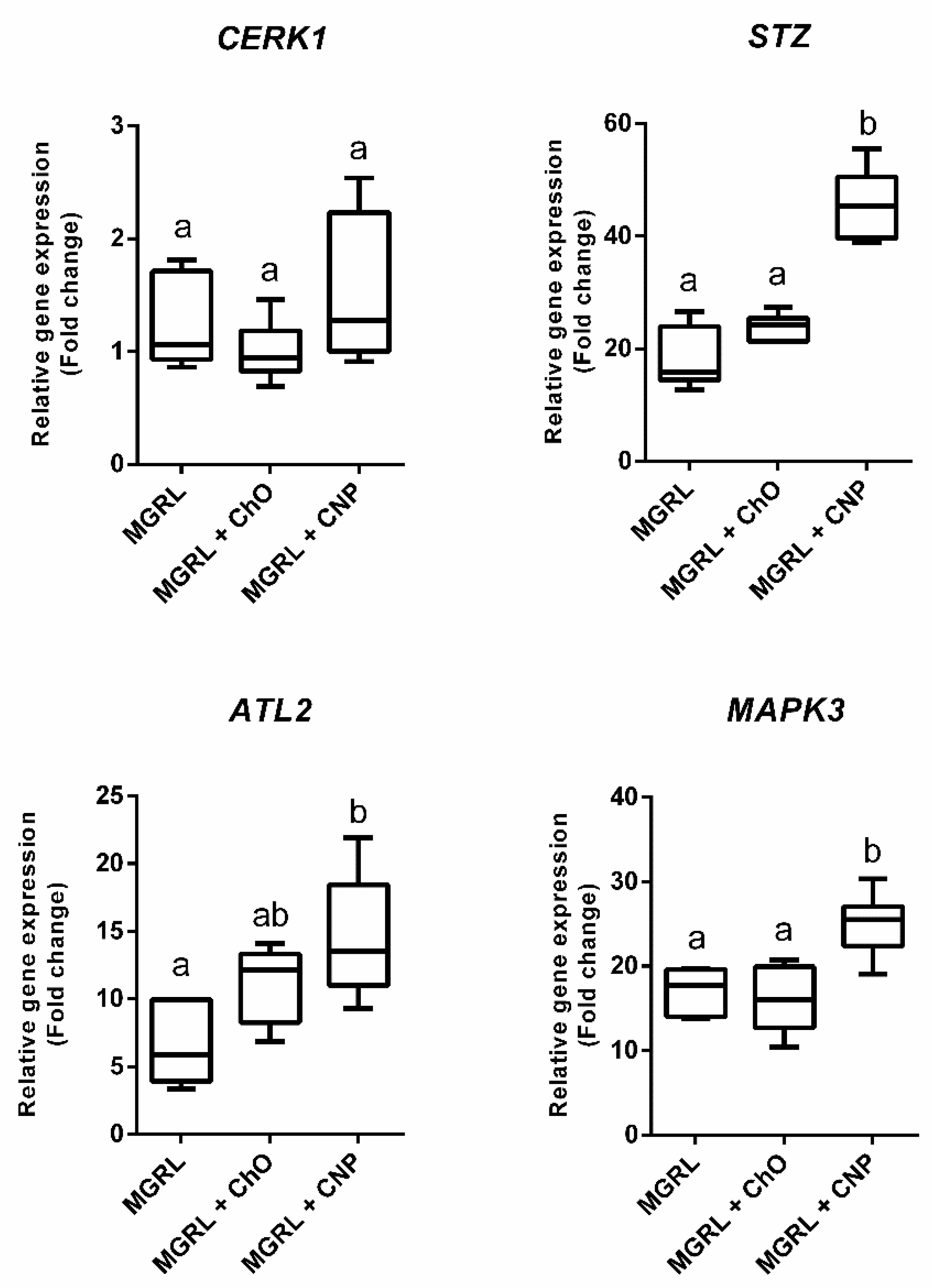
2.2. CNPs Induce Expression of Genes Activated by Chitin Recognition in Nicotiana benthamiana
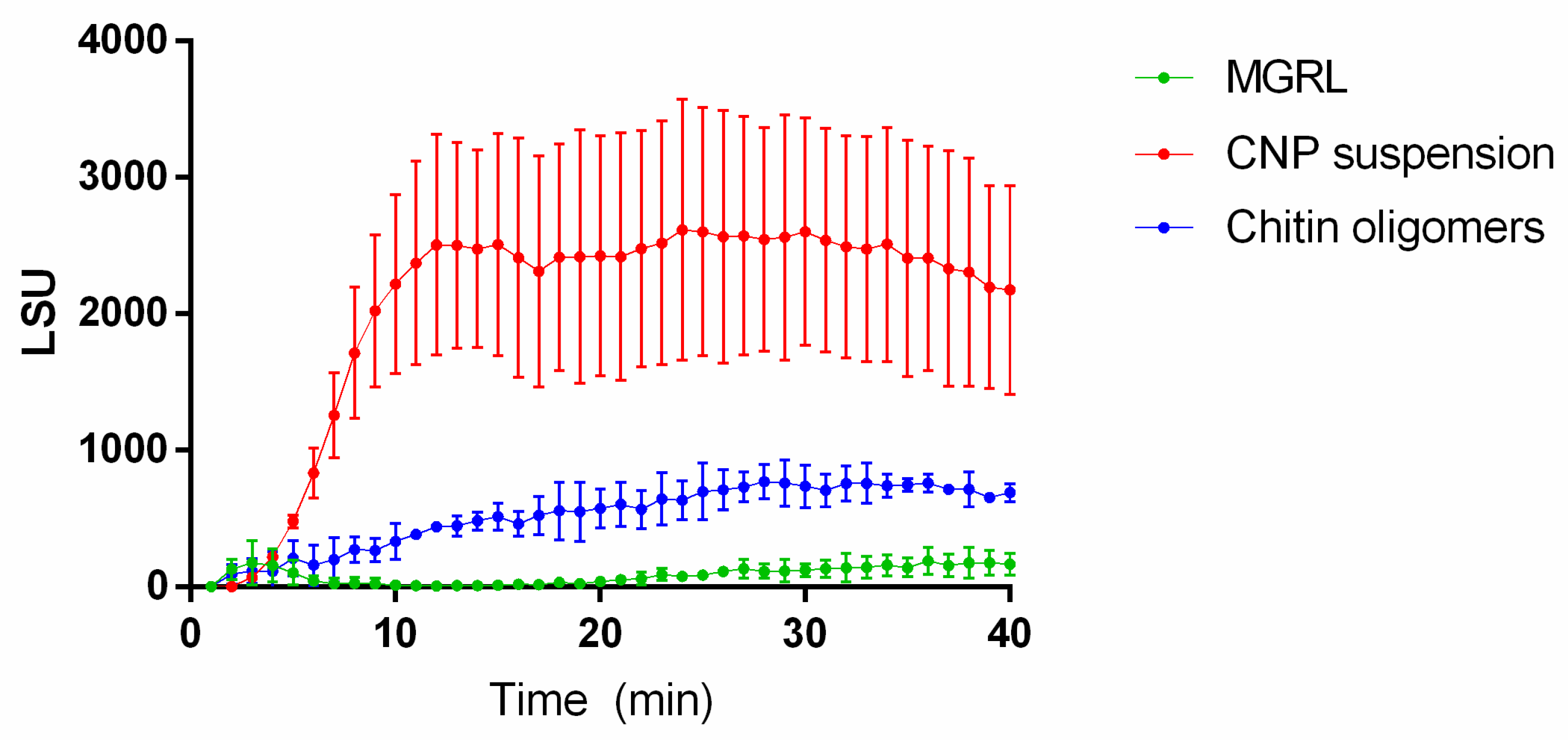
2.3. ROS Production Increases in Plants Treated with CNPs
3. Discussion
4. Materials and Methods
4.1. Dissolution of Chitin
4.2. Synthesis of Chitin Oligomers
4.3. Generation of CNP Using W/O Emulsion
4.4. Plant Material
4.5. Incubation of Seedlings with CNP, Chitin Oligomers, and RNA Extraction
4.6. Primer Design for qRT-PCR
4.7. qRT-PCR
4.8. ROS Measurement Using Chemiluminescence Assay
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CNP | chitin nanoparticles |
| W/O | water in oil |
| ROS | reactive oxygen species |
| MAMP | microbe-associated molecular pattern |
| LPS | lipopolysaccharide |
| dsRNA | double-stranded RNA |
| PTI | PAMP-triggered immunity |
| CS | chitin solution |
| qRT-PCR | quantitative reverse transcription polymerase chain reaction |
References
- Kaya, M.; Česoniene, L.; Daubaras, R.; Leskauskaite, D.; Zabulione, D. Chitosan coating of red kiwifruit (Actinidia melanandra) for extending of the shelf life. Int. J. Biol. Macromol. 2016, 85, 355–360. [Google Scholar] [CrossRef]
- Sharp, R. A Review of the Applications of Chitin and Its Derivatives in Agriculture to Modify Plant-Microbial Interactions and Improve Crop Yields. Agronomy 2013, 3, 757–793. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, X.-C.; Stacey, G. Chitin signaling and plant disease resistance. Plant Signal. Behav. 2008, 3, 831–833. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Desaki, Y.; Miyata, K.; Suzuki, M.; Shibuya, N.; Kaku, H. Plant immunity and symbiosis signaling mediated by LysM receptors. Innate Immun. 2018, 24, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Moya, F.; Suarez-Fernandez, M.; Lopez-Llorca, L.V. Molecular mechanisms of chitosan interactions with fungi and plants. Int. J. Mol. Sci. 2019, 20, 332. [Google Scholar] [CrossRef]
- Taj, G.; Agarwal, P.; Grant, M.; Kumar, A. MAPK machinery in plants: Recognition and response to different stresses through multiple signal transduction pathways. Plant Signal. Behav. 2010, 5, 1370–1378. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Zhang, C.; Zhu, P.; Dai, H.; Yu, N.; He, Z.; Xu, L.; Wang, E. OsCERK1-Mediated Chitin Perception and Immune Signaling Requires Receptor-like Cytoplasmic Kinase 185 to Activate an MAPK Cascade in Rice. Mol. Plant 2017, 10, 619–633. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Shinya, T.; Yamaguchi, K.; Desaki, Y.; Yamada, K.; Narisawa, T.; Kobayashi, Y.; Maeda, K.; Suzuki, M.; Tanimoto, T.; Takeda, J.; et al. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 2014, 79, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The Arabidopsis CERK 1-associated kinase PBL 27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, G.V.; Shimoda, Y.; Nielsen, M.W.; Jorgensen, F.G.; Grossmann, C.; Sandal, N.; Sørensen, K.; Thirup, S.; Madsen, L.H.; Tabata, S.; et al. Evolution and regulation of the lotus japonicus LysM receptor gene family. Mol. Plant Microbe Interact. 2010, 23, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Bozsoki, Z.; Cheng, J.; Feng, F.; Gysel, K.; Vinther, M.; Andersen, K.R.; Oldroyd, G.; Blaise, M.; Radutoiu, S.; Stougaard, J. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc. Natl. Acad. Sci. USA 2017, 114, E8118–E8127. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Yoshimura, S.; Terauchi, A.; Kawasaki, T. Conservation of chitin-induced MAPK signaling pathways in rice and arabidopsis. Plant Cell Physiol. 2017, 58, 993–1002. [Google Scholar] [CrossRef]
- Zargar, V.; Asghari, M.; Dashti, A. A Review on Chitin and Chitosan Polymers: Structure, Chemistry, Solubility, Derivatives, and Applications. ChemBioEng Rev. 2015, 2, 204–226. [Google Scholar] [CrossRef]
- Dietz, K.J.; Herth, S. Plant nanotoxicology. Trends Plant Sci. 2011, 16, 582–589. [Google Scholar] [CrossRef]
- Parisi, C.; Vigani, M.; Rodriguez-Cerezo, E. Agricultural nanotechnologies: What are the current possibilities? Curr. Sci. 2015, 108, 1210–1213. [Google Scholar] [CrossRef]
- Shibuya, N.; Minami, E. Oligosaccharide signalling for defence responses in plant. Physiol. Mol. Plant Pathol. 2001, 59, 223–233. [Google Scholar] [CrossRef]
- Kurita, K. Chitin and chitosan: Functional biopolymers from marine crustaceans. Mar. Biotechnol. 2006, 8, 203–226. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Du, Y.; Tang, Y.; Wang, Q.; Feng, T.; Yang, J.; Kennedy, J.F. Solubility and property of chitin in NaOH/urea aqueous solution. Carbohydr. Polym. 2007, 70, 451–458. [Google Scholar] [CrossRef]
- Eckerson, S. The Number and Size of the Stomata. Bot. Gaz. 1908, 46, 221–224. [Google Scholar] [CrossRef]
- Judy, J.D.; Unrine, J.M.; Rao, W.; Wirick, S.; Bertsch, P.M. Bioavailability of gold nanomaterials to plants: Importance of particle size and surface coating. Environ. Sci. Technol. 2012, 46, 8467–8474. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.C.; Wells, B.; Roberts, K. Direct visualization of cross-links in the primary plant cell wall. J. Cell Sci. 1990, 96, 323–334. [Google Scholar]
- Hamid, R.; Khan, M.A.; Ahmad, M.; Ahmad, M.M.; Abdin, M.Z.; Musarrat, J.; Javed, S. Chitinases: An update. J. Pharm. Bioallied Sci. 2013, 5, 21–29. [Google Scholar] [PubMed]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad, K. Plant chitinases. Plant J. 1993, 3, 31–40. [Google Scholar] [CrossRef]
- Kasprzewska, A. Plant chitinases—Regulation and function. Cell. Mol. Biol. Lett. 2003, 8, 809–824. [Google Scholar]
- Egusa, M.; Matsui, H.; Urakami, T.; Okuda, S.; Ifuku, S.; Nakagami, H.; Kaminaka, H. Chitin Nanofiber Elucidates the Elicitor Activity of Polymeric Chitin in Plants. Front. Plant Sci. 2015, 6, 1098. [Google Scholar] [CrossRef]
- Ramonell, K.M.; Zhang, B.; Ewing, R.M.; Chen, Y.; Xu, D.; Stacey, G.; Somerville, S. Microarray analysis of chitin elicitation in Arabidopsis thaliana. Mol. Plant Pathol. 2002, 3, 301–311. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. CRC Crit. Rev. Plant Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Mezaki, H.; Fujiwara, M.; Hara, Y.; Kawasaki, T. Arabidopsis ubiquitin ligase PUB12 interacts with and negatively regulates Chitin Elicitor Receptor Kinase 1 (CERK1). PLoS ONE 2017, 12, e0188886. [Google Scholar] [CrossRef] [PubMed]
- El Hadrami, A.; Adam, L.R.; El Hadrami, I.; Daayf, F. Chitosan in plant protection. Mar. Drugs 2010, 8, 968–987. [Google Scholar] [CrossRef] [PubMed]
- El Gueddari, N.E.; Rauchhaus, U.; Moerschbacher, B.M.; Deising, H.B. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 2002, 156, 103–112. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, B.; Zhao, J.H.; Huang, J.F.; Jia, P.S.; Wang, S.; Zhang, J.; Zhou, J.M.; Guo, H.S. Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 2019, 5, 1167–1176. [Google Scholar] [CrossRef]
- Revol, J.F.; Marchessault, R.H. In vitro chiral nematic ordering of chitin crystallites. Int. J. Biol. Macromol. 1993, 15, 329–335. [Google Scholar] [CrossRef]
- Naito, S.; Hirai, M.Y.; Chino, M.; Komeda, Y. Expression of a Soybean (Glycine max [L.] Merr.) Seed Storage Protein Gene in Transgenic Arabidopsis thaliana and Its Response to Nutritional Stress and to Abscisic Acid Mutations. Plant Physiol. 1994, 104, 497–503. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034-1. [Google Scholar] [CrossRef]
- Liu, D.; Shi, L.; Han, C.; Yu, J.; Li, D.; Zhang, Y. Validation of Reference Genes for Gene Expression Studies in Virus-Infected Nicotiana benthamiana Using Quantitative Real-Time PCR. PLoS ONE 2012, 7, e46451. [Google Scholar] [CrossRef]

| Name | Function | Arabidopsis thaliana ID | N. benthamiana ID |
|---|---|---|---|
| CERK1 | Perception and transduction of chitin signaling | AT3G21630 | Niben101Scf01037g04009.1 |
| MAPK3 | Kinase activated under specific biotic and abiotic stress | AT3G45640 | Niben101Scf36191g00003.1 |
| STZ | Transcriptional repressor, activated by chitin oligomers | AT1G27730 | Niben101Scf06467g01006.1 |
| ATL2 | RING-H2 transcription factor, activated by chitin | AT3G16720 | Niben101Scf06556g10007.1 |
| Name | Primer | Primer Sequence | Product Length (bp) |
|---|---|---|---|
| CERK1 | Forward | GGT TAG GAG TTT TCC TTA TCC TAG | 111 |
| Reverse | CCC GCC AAA CAT AGA ATG AAG CTA AAG C | ||
| MAPK3 | Forward | GTC TGC TCG GTG TTG AAT ACG G | 143 |
| Reverse | CCA ATT ACA TTT TCA TGG TCT AAA TGG CG | ||
| STZ | Forward | CCT CGT AGT ATG GAA CGA CAG | 145 |
| Reverse | GCG CGT GTA TTT TCA TCA CTG GAA C | ||
| ATL2 | Forward | GGG AAG ATA ATG CTA AGC GCT ATT G | 145 |
| Reverse | GGG TAC GGC GAT GGT GGA TCC TAC | ||
| 18S a | Forward | GCA AGA CCG AAA CTC AAA GG | 107 |
| Reverse | TGT TCA TAT GTC AAG GGC TGG | ||
| EF1A a | Forward | AGC TTT ACC TCC CAA GTC ATC | 116 |
| Reverse | AGA ACG CCT GTC AAT CTT GG | ||
| L23 a | Forward | AAG GAT GCC GTG AAG AAG ATG T | 110 |
| Reverse | GCA TCG TAG TCA GGA GTC AAC C |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López, M.; Miranda, E.; Ramos, C.; García, H.; Neira-Carrillo, A. Activation of Early Defense Signals in Seedlings of Nicotiana benthamiana Treated with Chitin Nanoparticles. Plants 2020, 9, 607. https://doi.org/10.3390/plants9050607
López M, Miranda E, Ramos C, García H, Neira-Carrillo A. Activation of Early Defense Signals in Seedlings of Nicotiana benthamiana Treated with Chitin Nanoparticles. Plants. 2020; 9(5):607. https://doi.org/10.3390/plants9050607
Chicago/Turabian StyleLópez, Miguel, Elisa Miranda, Cecilia Ramos, Héctor García, and Andrónico Neira-Carrillo. 2020. "Activation of Early Defense Signals in Seedlings of Nicotiana benthamiana Treated with Chitin Nanoparticles" Plants 9, no. 5: 607. https://doi.org/10.3390/plants9050607
APA StyleLópez, M., Miranda, E., Ramos, C., García, H., & Neira-Carrillo, A. (2020). Activation of Early Defense Signals in Seedlings of Nicotiana benthamiana Treated with Chitin Nanoparticles. Plants, 9(5), 607. https://doi.org/10.3390/plants9050607






