Insights into the Root Invasion by the Plant Pathogenic Bacterium Ralstonia solanacearum
Abstract
:1. Introduction
2. Plant Defence Responses Encountered by R. solanacearum
3. The Establishment of R. solanacearum In Vitro Inoculation Assays Paved the Way to the Study of the Early Stages of the Infection
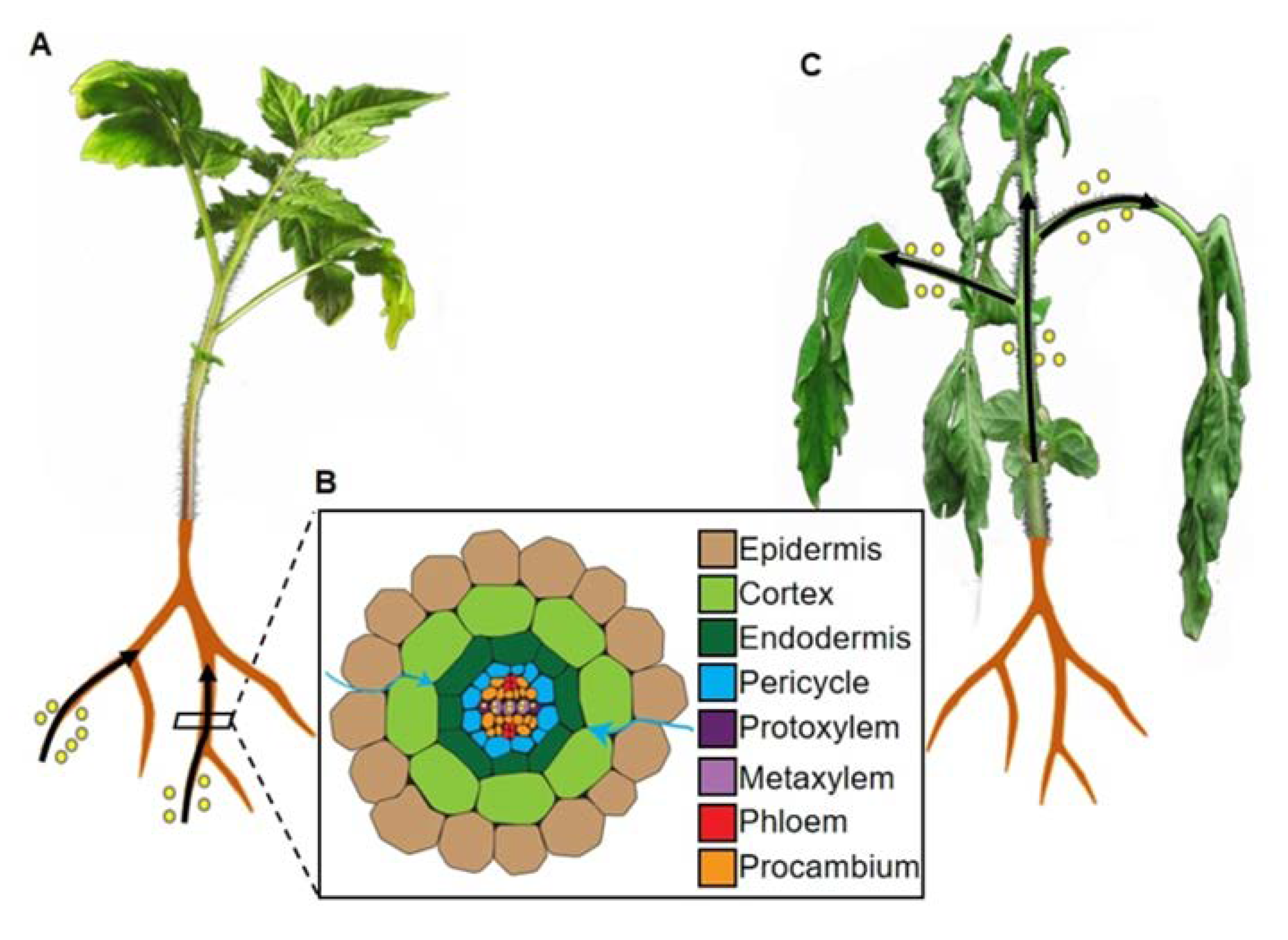
3.1. Classic Soil-Drenching Assay
3.2. In Vitro Inoculation Assays and their Application to Observe Root Development during the Infection by R. solanacearum
4. Root Morphological Alterations upon R. solanacearum Inoculation
5. Root Transcriptome Reprogramming during R. solanacearum Infection May Contribute to Root Morphological Alterations
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genin, S. Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 2010, 187, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Vasse, J.; Pascal, F.; Trigalet, A. Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 1995, 8, 241–251. [Google Scholar] [CrossRef]
- Vailleau, F.; Sartorel, E.; Jardinaud, M.-F.; Chardon, F.; Genin, S.; Huguet, T.; Gentzbittel, L.; Petitprez, M. Characterization of the interaction between the bacterial wilt pathogen Ralstonia solanacearum and the model legume plant Medicago truncatula. Mol. Plant-Microbe Interact. 2007, 20, 159–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Digonnet, C.; Martinez, Y.; Denancé, N.; Chasseray, M.; Dabos, P.; Ranocha, P.; Marco, Y.; Jauneau, A.; Goffner, D. Deciphering the route of Ralstonia solanacearum colonization in Arabidopsis thaliana roots during a compatible interaction: Focus at the plant cell wall. Planta 2012, 236, 1419–1431. [Google Scholar] [CrossRef]
- Turner, M.; Jauneau, A.; Genin, S.; Tavella, M.-J.; Vailleau, F.; Gentzbittel, L.; Jardinaud, M.-F. Dissection of bacterial wilt on Medicago truncatula revealed two type III secretion system effectors acting on root infection process and disease development. Plant Physiol. 2009, 150, 1713–1722. [Google Scholar] [CrossRef] [Green Version]
- Saile, E.; McGarvey, J.A.; Schell, M.A.; Denny, T.P. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 1997, 87, 1264–1271. [Google Scholar] [CrossRef] [Green Version]
- Hawes, M.; Allen, C.; Turgeon, B.G.; Curlango-Rivera, G.; Tran, T.M.; Huskey, D.A.; Xiong, Z. Root border cells and their role in plant defense. Annu. Rev. Phytopathol. 2016, 54, 143–161. [Google Scholar] [CrossRef]
- Tran, T.M.; MacIntyre, A.; Hawes, M.; Allen, C. Escaping underground nets: Extracellular DNases degrade plant extracellular traps and contribute to virulence of the plant pathogenic bacterium Ralstonia solanacearum. PLOS Pathog. 2016, 12, e1005686. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P.-I.; Yasuda, S. Pattern recognition receptors and signaling in plant–Microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-Associated molecular patterns and danger signals by pattern-Recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR receptor–Like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 2004, 16, 3496–3507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.G.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-Mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Pfund, C.; Tans-Kersten, J.; Dunning, F.M.; Alonso, J.M.; Ecker, J.R.; Allen, C.; Bent, A.F. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2004, 17, 696–706. [Google Scholar] [CrossRef] [Green Version]
- Mueller, K.; Bittel, P.; Chinchilla, D.; Jehle, A.K.; Albert, M.; Boller, T.; Felix, G. Chimeric FLS2 receptors reveal the basis for differential flagellin perception in Arabidopsis and tomato. Plant Cell 2012, 24, 2213–2224. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Caceres-Moreno, C.; Jimenez-Gongora, T.; Wang, K.; Sang, Y.; Lozano-Duran, R.; Macho, A.P. The Ralstonia solanacearum csp22 peptide, but not flagellin-Derived peptides, is perceived by plants from the Solanaceae family. Plant Biotechnol. J. 2018, 16, 1349–1362. [Google Scholar] [CrossRef] [Green Version]
- Clarke, C.R.; Chinchilla, D.; Hind, S.R.; Taguchi, F.; Miki, R.; Ichinose, Y.; Martin, G.B.; Leman, S.; Felix, G.; Vinatzer, B.A. Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol. 2013, 200, 847–860. [Google Scholar] [CrossRef] [Green Version]
- Lacombe, S.; Rougon-Cardoso, A.; Sherwood, E.; Peeters, N.; Dahlbeck, D.; van Esse, H.P.; Smoker, M.; Rallapalli, G.; Thomma, B.P.H.J.; Staskawicz, B.; et al. Interfamily transfer of a plant pattern-Recognition receptor confers broad-Spectrum bacterial resistance. Nature Biotechnol. 2010, 28, 365–369. [Google Scholar] [CrossRef]
- Wang, L.; Albert, M.; Einig, E.; Fürst, U.; Krust, D.; Felix, G. The pattern-Recognition receptor CORE of Solanaceae detects bacterial cold-Shock protein. Nat. Plants 2016, 2, 16185. [Google Scholar] [CrossRef]
- Pfeilmeier, S.; George, J.; Morel, A.; Roy, S.; Smoker, M.; Stransfeld, L.; Downie, J.A.; Peeters, N.; Malone, J.G.; Zipfel, C. Expression of the Arabidopsis thaliana immune receptor EFR in Medicago truncatula reduces infection by a root pathogenic bacterium, but not nitrogen-fixing rhizobial symbiosis. Plant Biotechnol. J. 2019, 17, 569–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Emonet, A.; Dénervaud Tendon, V.; Marhavy, P.; Wu, D.; Lahaye, T.; Geldner, N. Co-Incidence of damage and microbial patterns controls localized immune responses in roots. Cell 2020, 180, 440.e418–453.e418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunnac, S.; Boucher, C.; Genin, S. Characterization of the cis-Acting regulatory element controlling HrpB-Mediated activation of the Type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 2004, 186, 2309–2318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deslandes, L.; Rivas, S. Catch me if you can: Bacterial effectors and plant targets. Trends Plant Sci. 2012, 17, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Macho, A.P. Subversion of plant cellular functions by bacterial type-III effectors: Beyond suppression of immunity. New Phytol. 2016, 210, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Subramaniam, R.; Desveaux, D. Of guards, decoys, baits and traps: Pathogen perception in plants by type III effector sensors. Curr. Opin. Microbiol. 2016, 29, 49–55. [Google Scholar] [CrossRef]
- Gassmann, W.; Hinsch, M.E.; Staskawicz, B.J. The Arabidopsis RPS4 bacterial-Resistance gene is a member of the TIR-NBS-LRR family of disease-Resistance genes. Plant J. 1999, 20, 265–277. [Google Scholar] [CrossRef]
- Deslandes, L.; Olivier, J.; Theulières, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. 2002, 99, 2404–2409. [Google Scholar] [CrossRef] [Green Version]
- Poueymiro, M.; Cunnac, S.; Barberis, P.; Deslandes, L.; Peeters, N.; Cazale-Noel, A.-C.; Boucher, C.; Genin, S. Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host-range specificity on tobacco. Mol. Plant-Microbe Interact. 2009, 22, 538–550. [Google Scholar] [CrossRef] [Green Version]
- Solé, M.; Popa, C.; Mith, O.; Sohn, K.H.; Jones, J.D.G.; Deslandes, L.; Valls, M. The awr gene family encodes a novel class of Ralstonia solanacearum type III effectors displaying virulence and avirulence activities. Mol. Plant-Microbe Interact. 2012, 25, 941–953. [Google Scholar] [CrossRef] [Green Version]
- Nahar, K.; Matsumoto, I.; Taguchi, F.; Inagaki, Y.; Yamamoto, M.; Toyoda, K.; Shiraishi, T.; Ichinose, Y.; Mukaihara, T. Ralstonia solanacearum type III secretion system effector Rip36 induces a hypersensitive response in the nonhost wild eggplant Solanum torvum. Mol. Plant Pathol. 2014, 15, 297–303. [Google Scholar] [CrossRef]
- Morel, A.; Guinard, J.; Lonjon, F.; Sujeeun, L.; Barberis, P.; Genin, S.; Vailleau, F.; Daunay, M.-C.; Dintinger, J.; Poussier, S.; et al. The eggplant AG91-25 recognizes the Type III-secreted effector RipAX2 to trigger resistance to bacterial wilt (Ralstonia solanacearum species complex). Mol. Plant Pathol. 2018, 19, 2459–2472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, M.; Mukaihara, T. The type III effector RipB from Ralstonia solanacearum RS1000 acts as a major avirulence factor in Nicotiana benthamiana and other Nicotiana species. Mol. Plant Pathol. 2019, 20, 1237–1251. [Google Scholar] [CrossRef] [Green Version]
- Jeon, H.; Kim, W.; Kim, B.; Lee, S.; Jayaraman, J.; Jung, G.; Choi, S.; Sohn, K.H.; Segonzac, C. Ralstonia solanacearum type III effectors with predicted nuclear localization signal localize to various cell compartments and modulate immune responses in Nicotiana spp. Plant Pathol. J. 2020, 36, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Yu, W.; Zhuang, H.; Wei, Y.; Derevnina, L.; Yu, G.; Luo, J.; Macho, A.P. Intra-Strain elicitation and suppression of plant Immunity by Ralstonia solanacearum Type-III effectors in Nicotiana benthamiana. Plant Commun. 2020. [Google Scholar] [CrossRef]
- Peeters, N.; Carrère, S.; Anisimova, M.; Plener, L.; Cazalé, A.-C.; Genin, S. Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genom. 2013, 14, 859. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, C.R.R.; Carrere, S.; Lonjon, F.; Vailleau, F.; Macho, A.P.; Genin, S.; Peeters, N. Pangenomic type III effector database of the plant pathogenic Ralstonia spp. PeerJ 2019, 7, e7346. [Google Scholar] [CrossRef] [Green Version]
- Morel, A.; Peeters, N.; Vailleau, F.; Barberis, P.; Jiang, G.; Berthomé, R.; Guidot, A. Plant pathogenicity phenotyping of Ralstonia solanacearum strains. In Host-Pathogen Interactions: Methods and Protocols; Medina, C., López-Baena, F.J., Eds.; Springer: New York, NY, USA, 2018; pp. 223–239. [Google Scholar] [CrossRef]
- Cruz, A.P.Z.; Ferreira, V.; Pianzzola, M.J.; Siri, M.I.; Coll, N.S.; Valls, M. A novel, sensitive method to evaluate potato germplasm for bacterial wilt resistance using a luminescent Ralstonia solanacearum reporter strain. Mol. Plant-Microbe Interact. 2014, 27, 277–285. [Google Scholar] [CrossRef] [Green Version]
- Zolobowska, L.; Van Gijsegem, F. Induction of lateral root structure formation on Petunia roots: A novel effect of GMI1000 Ralstonia solanacearum infection impaired in Hrp mutants. Mol. Plant-Microbe Interact. 2006, 19, 597–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Lema, A.S.; Planas-Marquès, M.; Alonso-Díaz, A.; Valls, M.; Coll, N.S. Type III secretion-Dependent and-independent phenotypes caused by Ralstonia solanacearum in Arabidopsis roots. Mol. Plant-Microbe Interact. 2018, 31, 175–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasse, J.; Genin, S.; Frey, P.; Boucher, C.; Brito, B. The hrpB and hrpG regulatory genes of Ralstonia solanacearum are required for different stages of the tomato root infection process. Mol. Plant-Microbe Interact. 2000, 13, 259–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Wang, H.; Lu, Y.; Hu, J.; Qu, L.; Li, Z.; Wang, D.; He, Y.; Valls, M.; Coll, N.S.; et al. Deep sequencing reveals early reprogramming of Arabidopsis root transcriptomes upon Ralstonia solanacearum infection. Mol. Plant-Microbe Interact. 2019, 32, 813–827. [Google Scholar] [CrossRef] [Green Version]
- Boucher, C.A.; Barberis, P.A.; Demery, D.A. Transposon mutagenesis of Pseudomonas solanacearum: Isolation of Tn5-induced avirulent mutants. Microbiology 1985, 131, 2449–2457. [Google Scholar] [CrossRef] [Green Version]
- Valls, M.; Genin, S.; Boucher, C. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLOS Pathog. 2006, 2, e82. [Google Scholar] [CrossRef] [Green Version]
- French, E.; Kim, B.-S.; Rivera-Zuluaga, K.; Iyer-Pascuzzi, A.S. Whole root transcriptomic analysis suggests a role for auxin pathways in resistance to Ralstonia solanacearum in tomato. Mol. Plant-Microbe Interact. 2018, 31, 432–444. [Google Scholar] [CrossRef] [Green Version]
- Zuluaga, A.P.; Solé, M.; Lu, H.; Góngora-Castillo, E.; Vaillancourt, B.; Coll, N.; Buell, C.R.; Valls, M. Transcriptome responses to Ralstonia solanacearum infection in the roots of the wild potato Solanum commersonii. BMC Genom. 2015, 16, 246. [Google Scholar] [CrossRef] [Green Version]
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, H.; Lozano-Durán, R.; Macho, A.P. Insights into the Root Invasion by the Plant Pathogenic Bacterium Ralstonia solanacearum. Plants 2020, 9, 516. https://doi.org/10.3390/plants9040516
Xue H, Lozano-Durán R, Macho AP. Insights into the Root Invasion by the Plant Pathogenic Bacterium Ralstonia solanacearum. Plants. 2020; 9(4):516. https://doi.org/10.3390/plants9040516
Chicago/Turabian StyleXue, Hao, Rosa Lozano-Durán, and Alberto P. Macho. 2020. "Insights into the Root Invasion by the Plant Pathogenic Bacterium Ralstonia solanacearum" Plants 9, no. 4: 516. https://doi.org/10.3390/plants9040516
APA StyleXue, H., Lozano-Durán, R., & Macho, A. P. (2020). Insights into the Root Invasion by the Plant Pathogenic Bacterium Ralstonia solanacearum. Plants, 9(4), 516. https://doi.org/10.3390/plants9040516




