Stay in Touch—The Cortical ER of Moss Protonemata in Osmotic Stress Situations
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Culture
2.2. Fluorescence Markers and Staining
2.3. Reduced Turgor and Plasmolysis
2.4. Actin Depolymerisation
2.5. Confocal Laser Scanning Microscopy (CLSM)
2.6. Three-Dimensional Reconstruction
3. Results
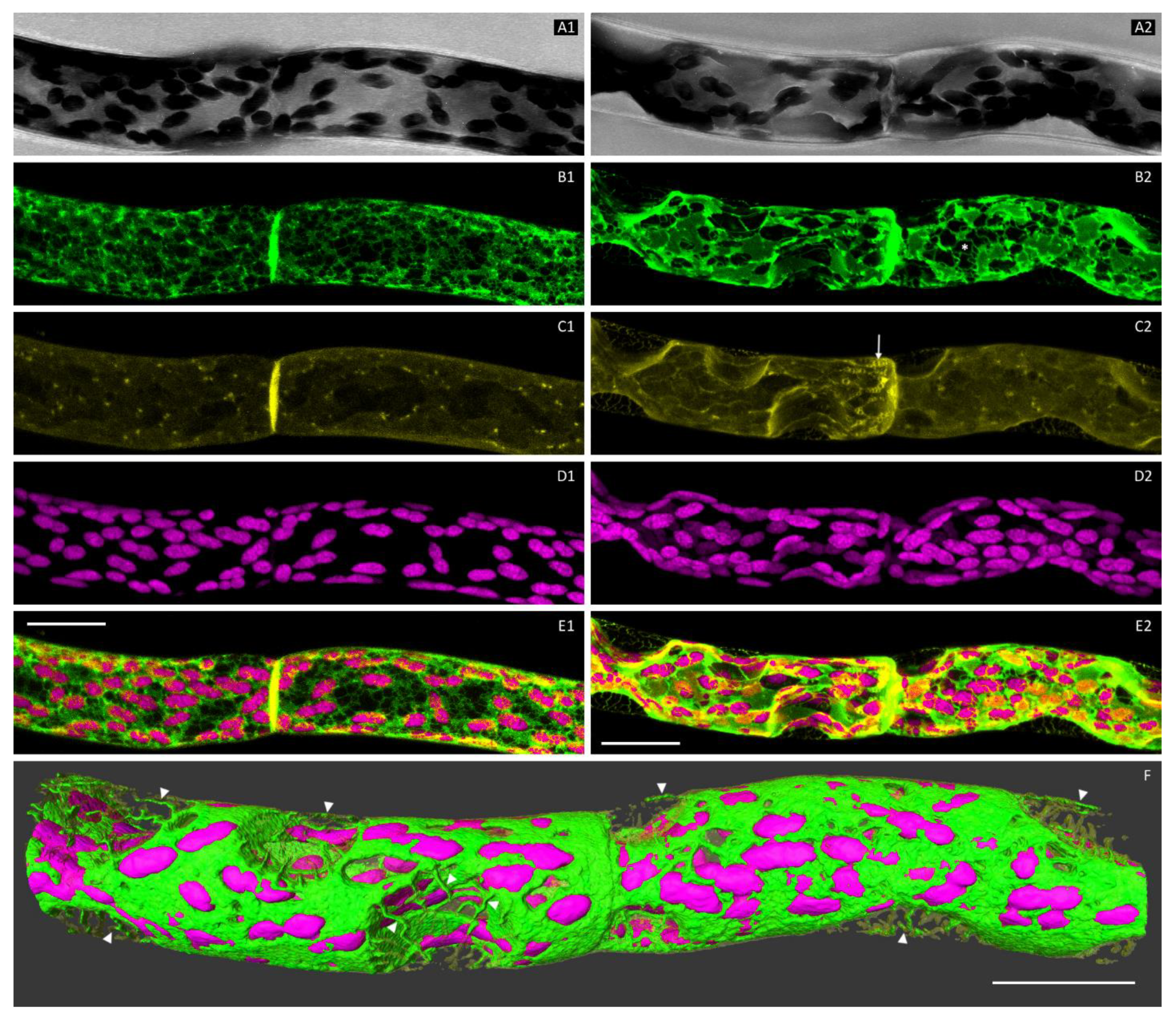
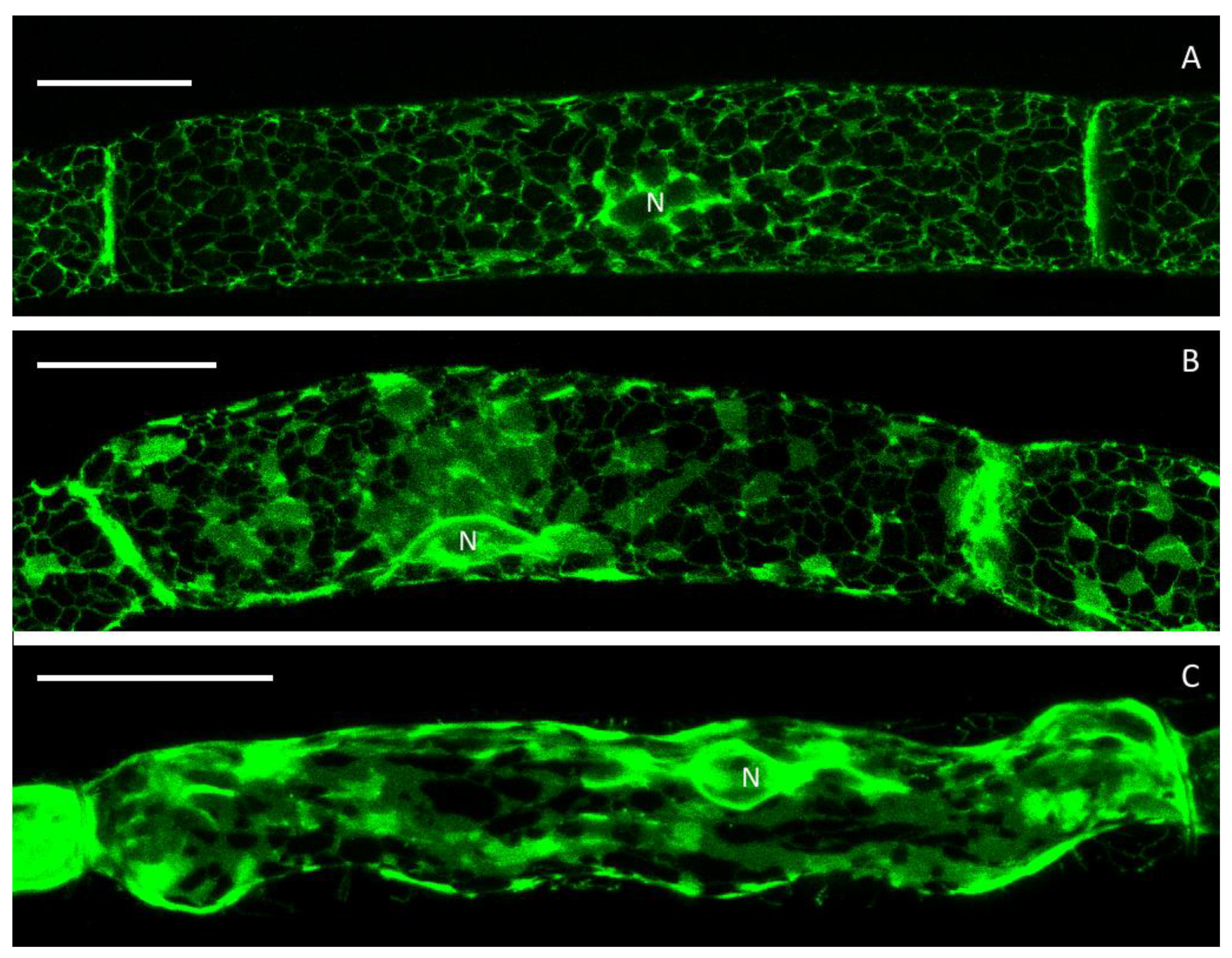
3.1. The Cortical ER of P. patens Before Plasmolysis
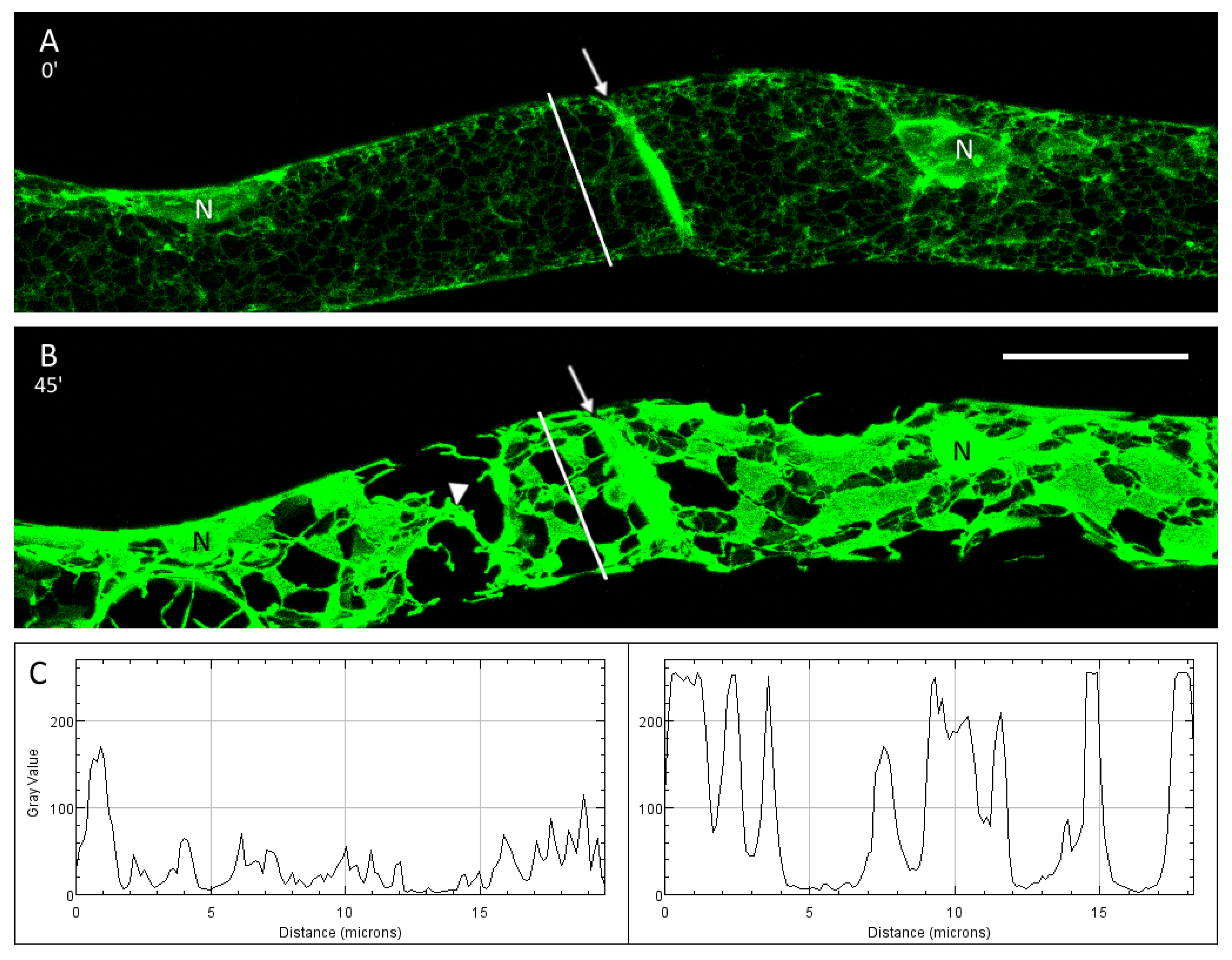
3.2. Plasmolysis of P. patens Protonema Cells
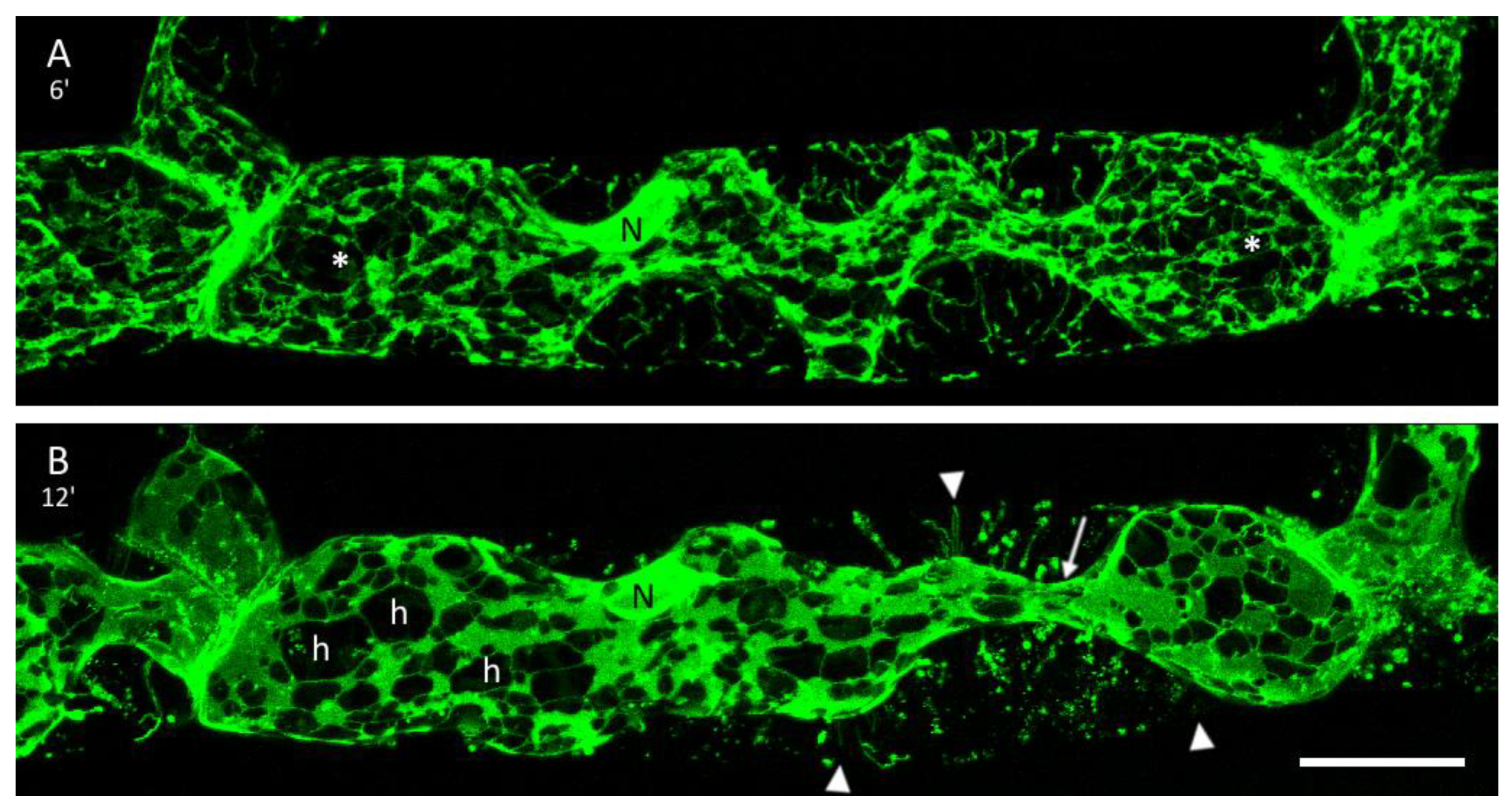
3.3. The ER and Plasmolysis
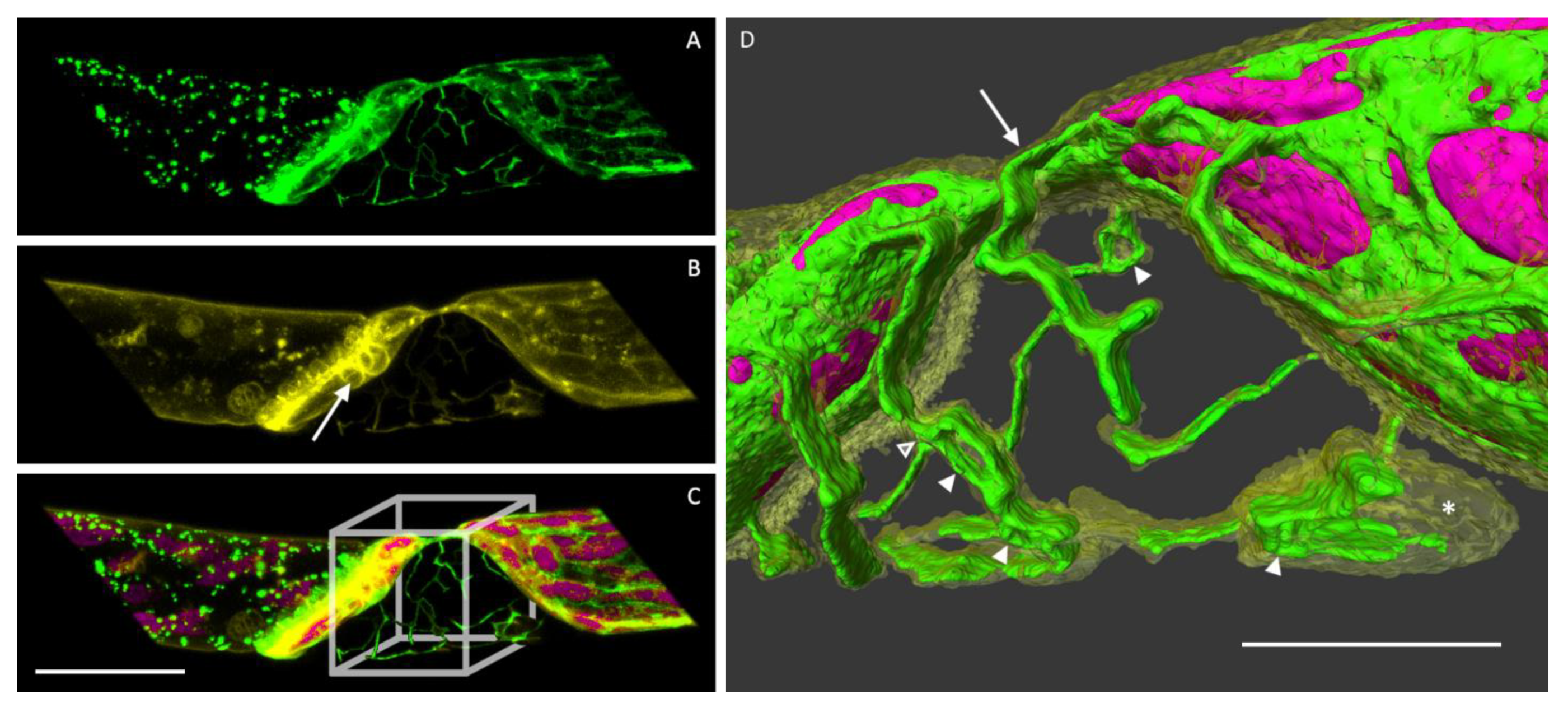
3.4. The ER in Hechtian Reticula
3.5. ER after Treatment with Latrunculin B
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reski, R. Enabling the water-to-land transition. Nat. Plants 2018, 4, 67–68. [Google Scholar] [CrossRef] [PubMed]
- Vidali, L.; Bezanilla, M. Physcomitrella patens: A model for tip cell growth and differentiation. Curr. Opin. Plant Biol. 2012, 15, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Pressel, S.; Ligrone, R.; Duckett, J.G. Cellular Differentiation in Moss Protonemata: A Morphological and Experimental Study. Ann. Bot. 2008, 102, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Furt, F.; Lemoi, K.; Tüzel, E.; Vidali, L. Quantitative analysis of organelle distribution and dynamics in Physcomitrella patens protonemal cells. BMC Plant Biol. 2012, 12, 70. [Google Scholar] [CrossRef]
- Roberts, A.W.; Roberts, E.M.; Haigler, C.H. Moss cell walls: Structure and biosynthesis. Front. Plant Sci. 2012, 3, 166. [Google Scholar] [CrossRef] [PubMed]
- Lang-Pauluzzi, I.; Gunning, B.E.S. A plasmolytic cycle: The fate of cytoskeletal elements. Protoplasma 2000, 212, 174–185. [Google Scholar] [CrossRef]
- Hecht, K. Studien über den Vorgang der Plasmolyse. Beiträge zur Biol. der Pflanz. 1912, 11, 133–189. [Google Scholar]
- Oparka, K.J.; Prior, D.A.M.; Crawford, J.W. Behaviour of plasma membrane, cortical ER and plasmodesmata during plasmolysis of onion epidermal cells. Plant. Cell Environ. 1994, 17, 163–171. [Google Scholar] [CrossRef]
- Lang, I.; Barton, D.A.; Overall, R.L. Membrane-wall attachments in plasmolysed plant cells. Protoplasma 2004, 224, 231–243. [Google Scholar] [CrossRef]
- Komis, G.; Apostolakos, P.; Galatis, B. Hyperosmotic stress-induced actin filament reorganization in leaf cells of Chlorophyton comosum. J. Exp. Bot. 2002, 53, 1699–1710. [Google Scholar] [CrossRef]
- Hawes, C.; Kiviniemi, P.; Kriechbaumer, V. The endoplasmic reticulum: A dynamic and well-connected organelle. J. Integr. Plant Biol. 2015, 57, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.; Tolley, N.; Aller, I.; Svozil, J.; Osterrieder, A.; Botchway, S.; Mueller, C.; Frigerio, L.; Hawes, C. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 2010, 22, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Shemesh, T.; Prinz, W.A.; Palazzo, A.F.; Kozlov, M.M.; Rapoport, T.A. Mechanisms determining the morphology of the peripheral ER. Cell 2010, 143, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.G. The Plant Endoplasmic Reticulum; Plant Cell Monographs; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4, ISBN 978-3-540-32531-4. [Google Scholar]
- Lin, C.; White, R.R.; Sparkes, I.; Ashwin, P. Modeling Endoplasmic Reticulum Network Maintenance in a Plant Cell. Biophys. J. 2017, 113, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.; Runions, J.; Hawes, C.; Griffing, L. Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell 2009, 21, 3937–3949. [Google Scholar] [CrossRef]
- Wang, P.; Hawes, C.; Hussey, P.J. Plant Endoplasmic Reticulum-Plasma Membrane Contact Sites. Trends Plant Sci. 2017, 22, 289–297. [Google Scholar] [CrossRef]
- Bayer, E.M.; Sparkes, I.; Vanneste, S.; Rosado, A. From shaping organelles to signalling platforms: The emerging functions of plant ER–PM contact sites. Curr. Opin. Plant Biol. 2017, 40, 89–96. [Google Scholar] [CrossRef]
- Cvrčková, F.; Oulehlová, D.; Žárský, V. Formins: Linking cytoskeleton and endomembranes in plant cells. Int. J. Mol. Sci. 2014, 16, 1–18. [Google Scholar] [CrossRef]
- Finka, A.; Schaefer, D.G.; Saidi, Y.; Goloubinoff, P.; Zrӱd, J.-P. In vivo visualization of F-actin structures during the development of the moss Physcomitrella patens. New Phytol. 2007, 174, 63–76. [Google Scholar] [CrossRef]
- Bascom, C.S.; Winship, L.J.; Bezanilla, M. Simultaneous imaging and functional studies reveal a tight correlation between calcium and actin networks. Proc. Natl. Acad. Sci. USA. 2018, 115, E2869–E2878. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- McCauley, M.M.; Hepler, P.K. Visualization of the endoplasmic reticulum in living buds and branches of the moss Funaria hygrometrica by confocal laser scanning microscopy. Development 1990, 109. [Google Scholar]
- Cheng, X.; Lang, I.; Adeniji, O.S.; Griffing, L. Plasmolysis-deplasmolysis causes changes in endoplasmic reticulum form, movement, flow, and cytoskeletal association. J. Exp. Bot. 2017, 68, 4075–4087. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Nishii, I.; Okada, S.; Noguchi, T. 3D reconstruction of endoplasmic reticulum in a hydrocarbon-secreting green alga, Botryococcus braunii (Race B). Planta 2018, 247, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Hepler, P.K.; Palevitz, B.A.; Lancelle, S.A.; MCCauley, M.M.; Lichtscheidl, I. Cortical endoplasmic reticulum in plants. J. Cell Sci. 1990, 96. [Google Scholar]
- Nixon-Abell, J.; Obara, C.J.; Weigel, A.V.; Li, D.; Legant, W.R.; Xu, C.S.; Pasolli, H.A.; Harvey, K.; Hess, H.F.; Betzig, E.; et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 2016, 354, aaf3928. [Google Scholar] [CrossRef]
- Hawes, C.; Osterrieder, A.; Sparkes, I.A.; Ketelaar, T. Optical tweezers for the micromanipulation of plant cytoplasm and organelles. Curr. Opin. Plant Biol. 2010, 13, 731–735. [Google Scholar] [CrossRef]
- West, M.; Zurek, N.; Hoenger, A.; Voeltz, G.K. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 2011, 193, 333–346. [Google Scholar] [CrossRef]
- Friedman, J.R.; Voeltz, G.K. The ER in 3D: A multifunctional dynamic membrane network. Trends Cell Biol. 2011, 21, 709–717. [Google Scholar] [CrossRef]
- Schroeder, L.K.; Barentine, A.E.S.; Merta, H.; Schweighofer, S.; Zhang, Y.; Baddeley, D.; Bewersdorf, J.; Bahmanyar, S. Dynamic nanoscale morphology of the ER surveyed by STED microscopy. J. Cell Biol. 2019, 218, 83–96. [Google Scholar] [CrossRef]
- Wang, P.; Hussey, P.J. Interactions between plant endomembrane systems and the actin cytoskeleton. Front. Plant Sci. 2015, 6, 422. [Google Scholar] [CrossRef] [PubMed]
- Kleist, T.J.; Cartwright, H.N.; Perera, A.M.; Christianson, M.L.; Lemaux, P.G.; Luan, S. Genetically encoded calcium indicators for fluorescence imaging in the moss Physcomitrella: GCaMP3 provides a bright new look. Plant Biotechnol. J. 2017, 15, 1235–1237. [Google Scholar] [CrossRef] [PubMed]
- Bonza, M.C.; Loro, G.; Behera, S.; Wong, A.; Kudla, J.; Costa, A. Analyses of Ca2+ accumulation and dynamics in the endoplasmic reticulum of Arabidopsis root cells using a genetically encoded Cameleon sensor. Plant Physiol. 2013, 163, 1230–1241. [Google Scholar] [CrossRef]
- Storti, M.; Costa, A.; Golin, S.; Zottini, M.; Morosinotto, T.; Alboresi, A. Systemic Calcium Wave Propagation in Physcomitrella patens. Plant Cell Physiol. 2018, 59, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Knox, K.; Wang, P.; Kriechbaumer, V.; Tilsner, J.; Frigerio, L.; Sparkes, I.; Hawes, C.; Oparka, K. Putting the Squeeze on Plasmodesmata: A Role for Reticulons in Primary Plasmodesmata Formation. Plant Physiol. 2015, 168, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Saheki, Y. Endoplasmic reticulum—Plasma membrane crosstalk mediated by the extended synaptotagmins. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2017; Volume 997, pp. 83–93. [Google Scholar]
- Liu, Z.; Persson, S.; Sanchez-Rodriguez, C. At the border: The plasma membrane-cell wall continuum. J. Exp. Bot. 2015, 66, 1553–1563. [Google Scholar] [CrossRef]
- Wartenberg, M.; Hamann, J.; Pratsch, I.; Donath, E. Osmotically induced fluid-phase uptake of fluorescent markers by protoplasts of Chenopodium album. Protoplasma 1992, 166, 61–66. [Google Scholar] [CrossRef]
- Oparka, K.J. Plasmolysis: New insights into an old process. New Phytol. 1994, 126, 571–591. [Google Scholar] [CrossRef]
- Oparka, K.J.; Hawes, C. Vacuolar sequestration of fluorescent probes in plant cells: A review. J. Microsc. 1992, 166, 15–27. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harant, D.; Lang, I. Stay in Touch—The Cortical ER of Moss Protonemata in Osmotic Stress Situations. Plants 2020, 9, 421. https://doi.org/10.3390/plants9040421
Harant D, Lang I. Stay in Touch—The Cortical ER of Moss Protonemata in Osmotic Stress Situations. Plants. 2020; 9(4):421. https://doi.org/10.3390/plants9040421
Chicago/Turabian StyleHarant, Dominik, and Ingeborg Lang. 2020. "Stay in Touch—The Cortical ER of Moss Protonemata in Osmotic Stress Situations" Plants 9, no. 4: 421. https://doi.org/10.3390/plants9040421
APA StyleHarant, D., & Lang, I. (2020). Stay in Touch—The Cortical ER of Moss Protonemata in Osmotic Stress Situations. Plants, 9(4), 421. https://doi.org/10.3390/plants9040421





