First Phylogeny of Bitterbush Family, Picramniaceae (Picramniales)
Abstract
1. Introduction
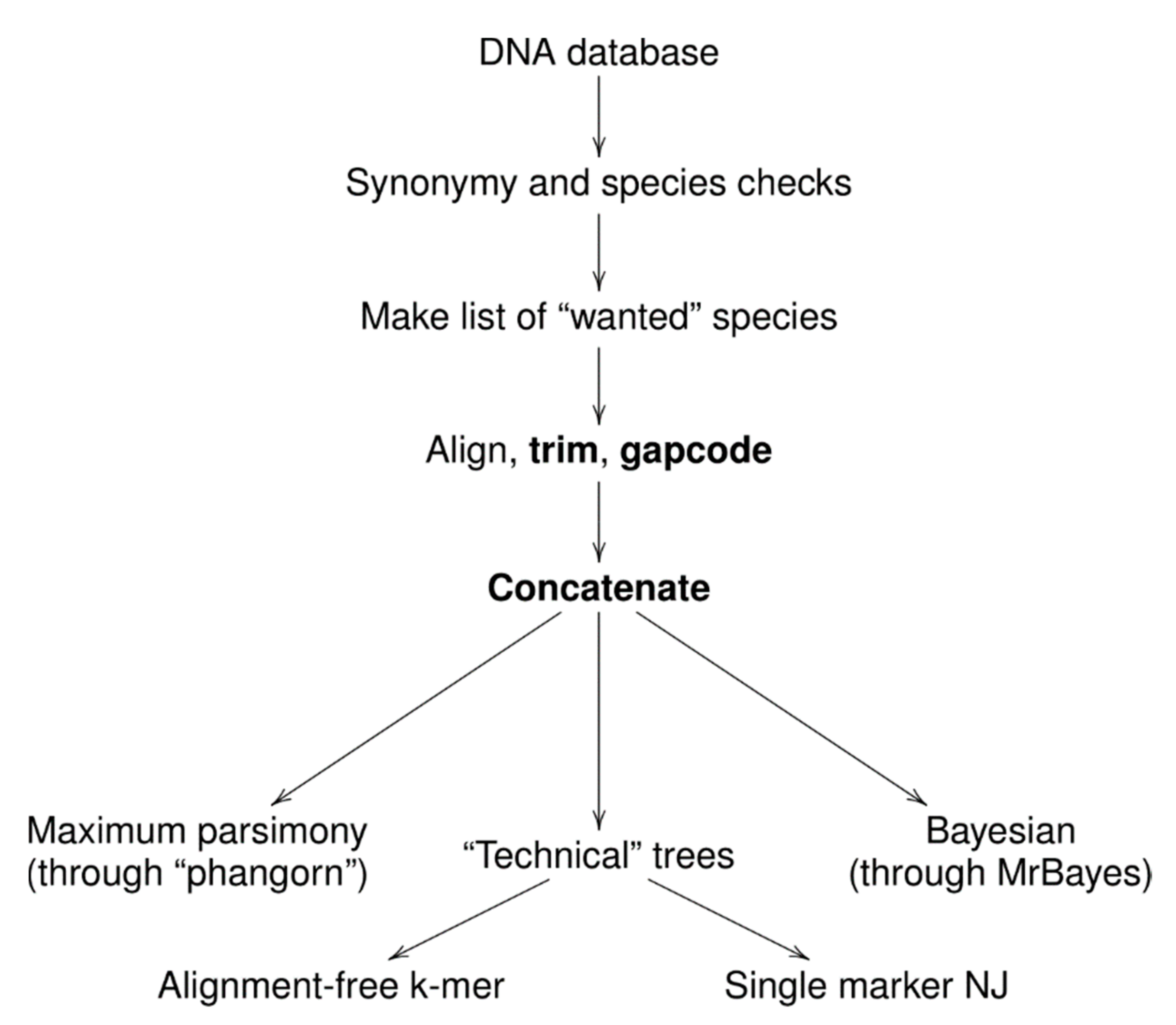
2. Material and Methods
3. Results
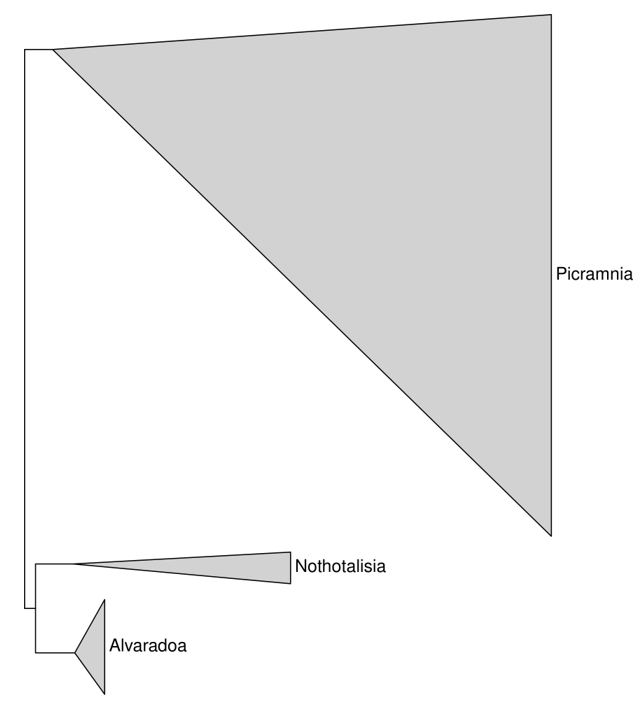
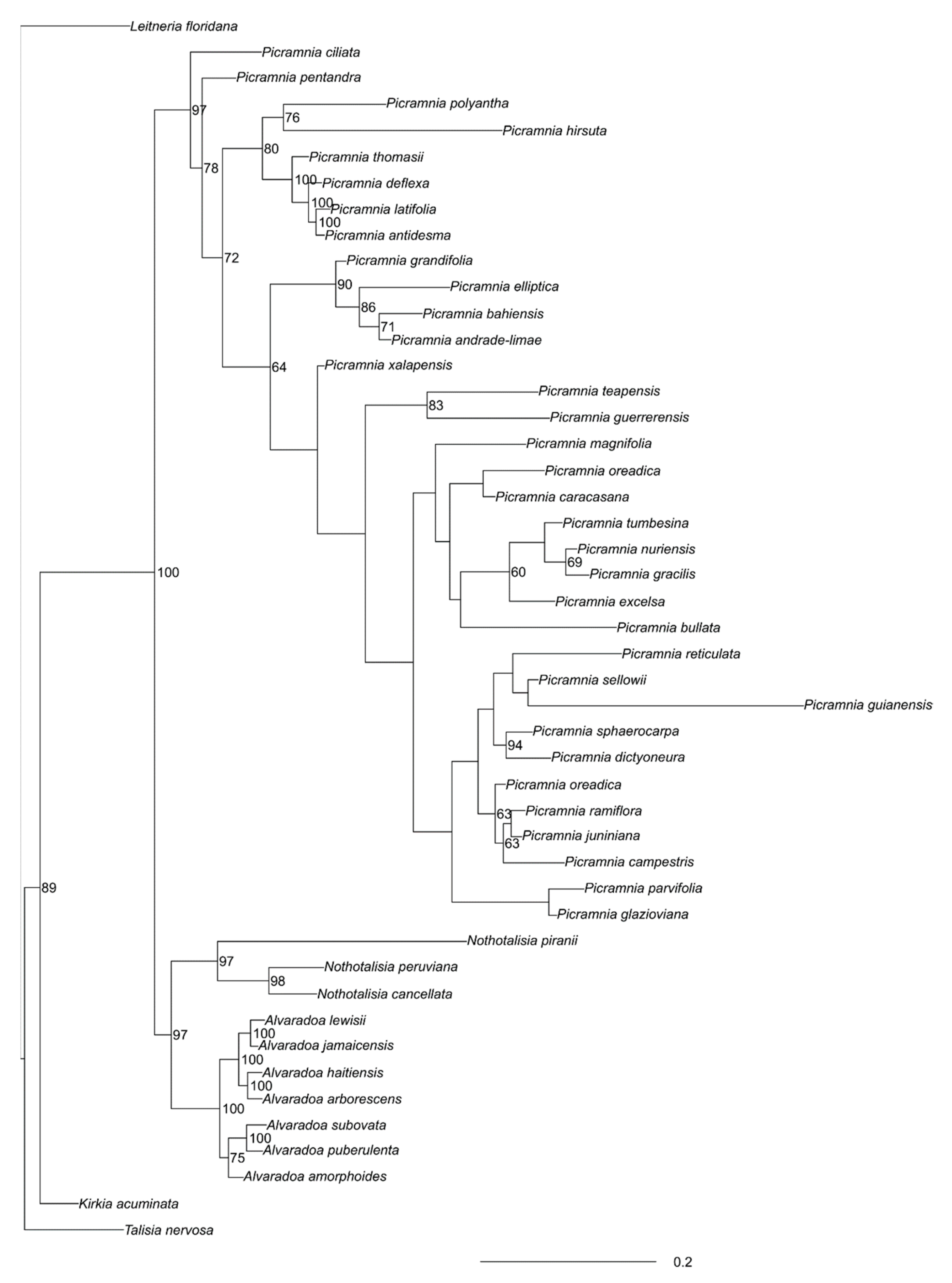
3.1. Picramniaceae in General
3.1.1. Alvaradoa
3.1.2. Nothotalisia
3.1.3. Picramnia
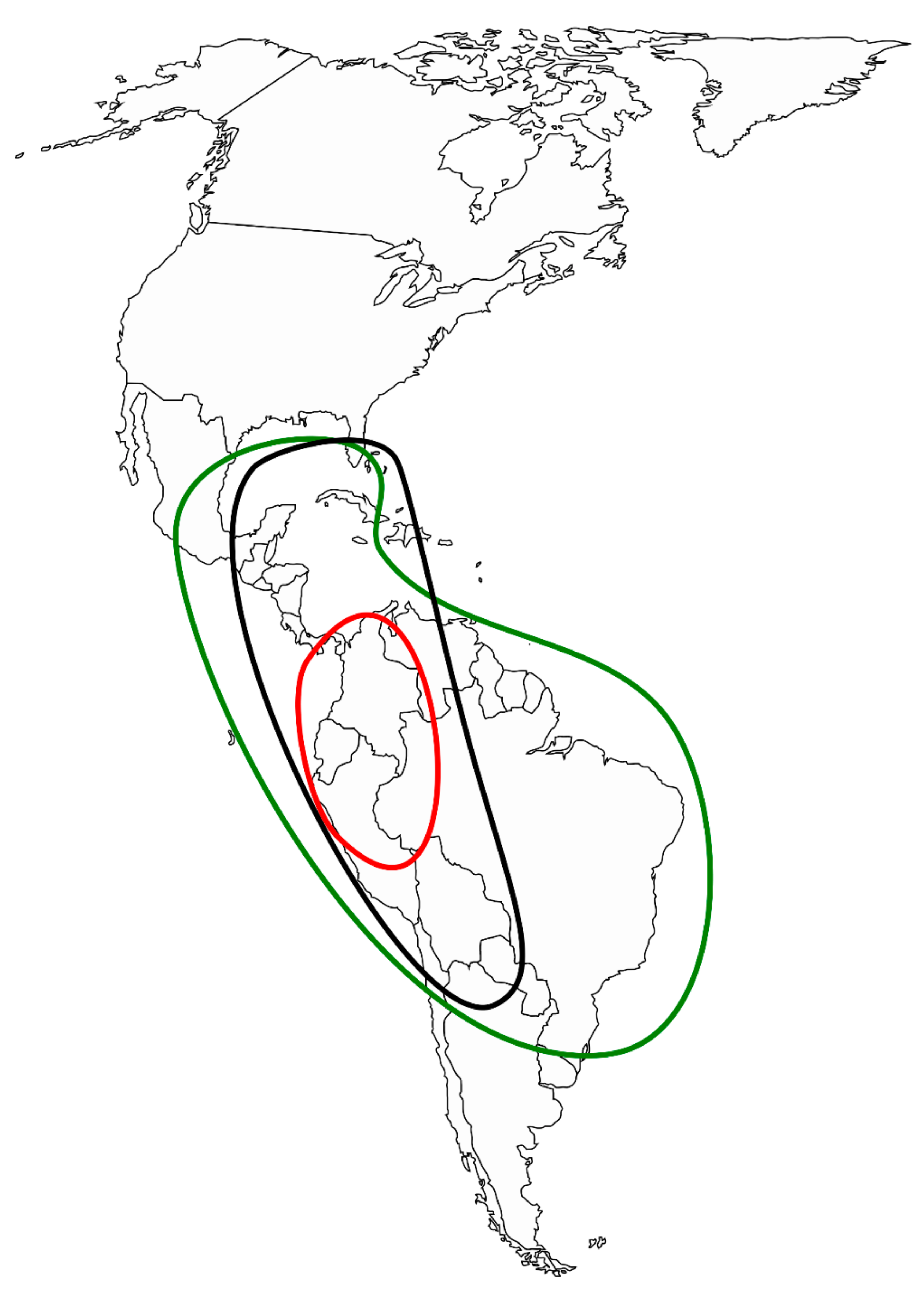
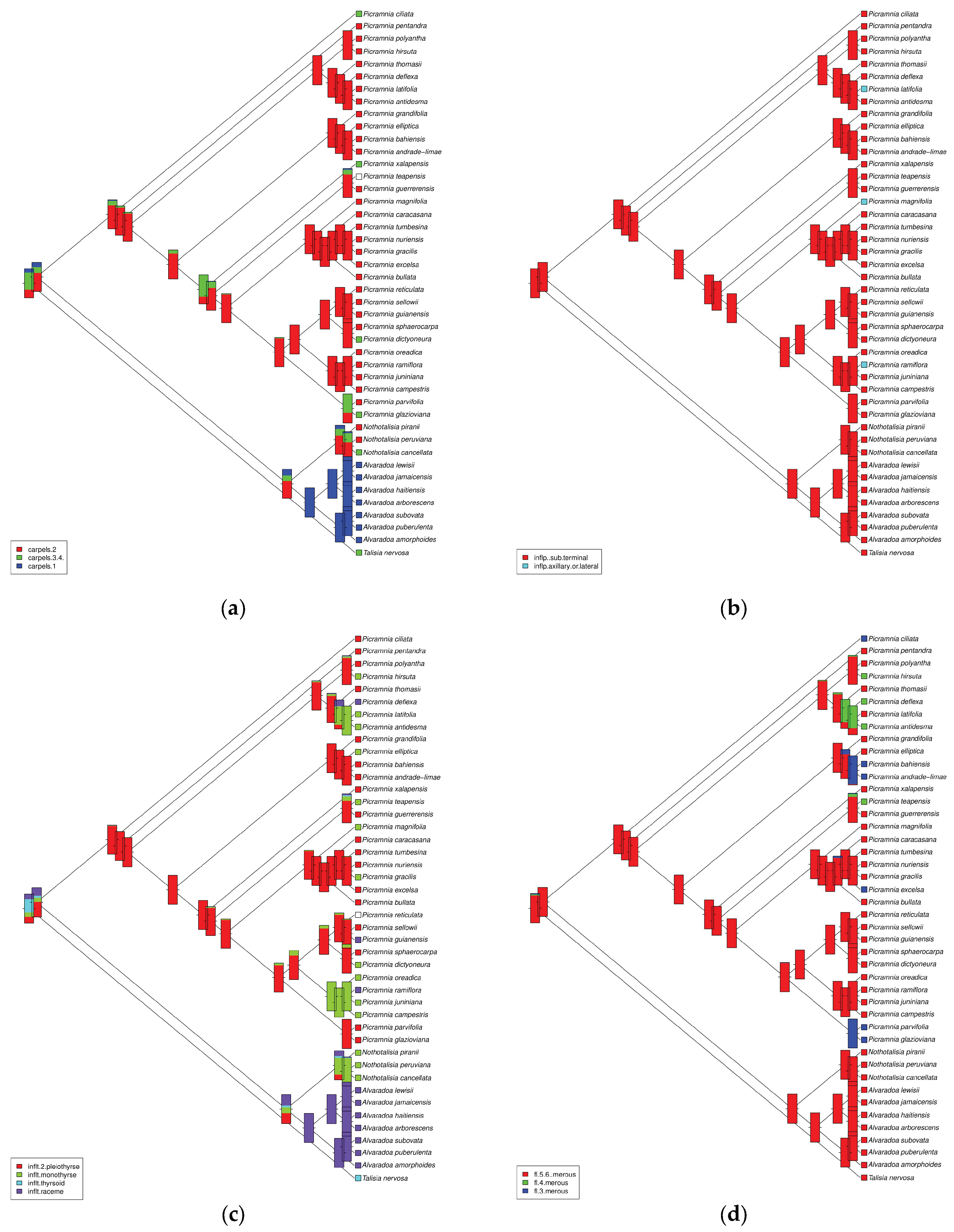
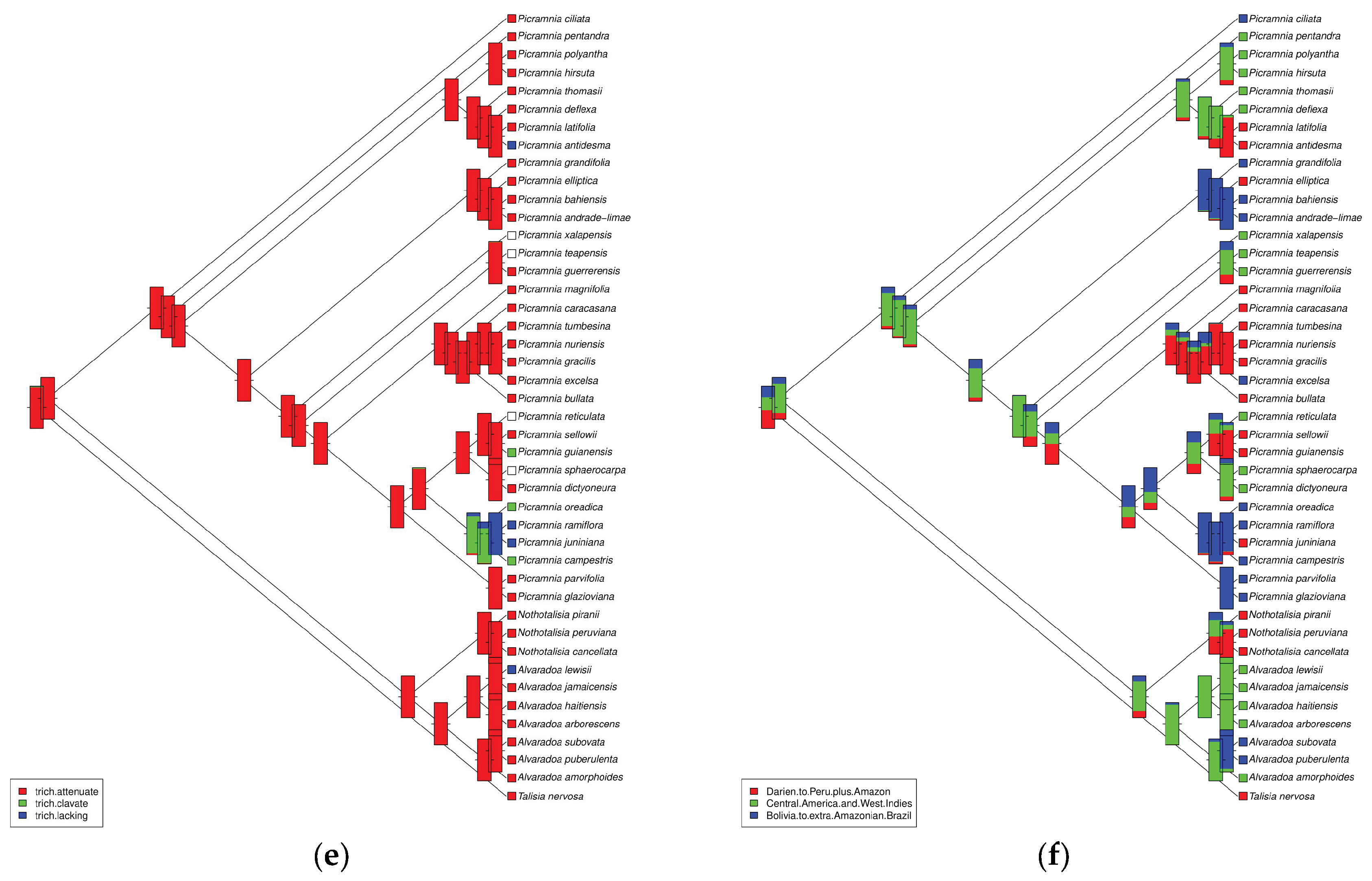
3.2. Morphological and Geographical Patterns
4. Discussion
4.1. Picramniaceae in General
4.2. Alvaradoa and Nothotalisia
4.3. Picramnia
Final Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fernando, E.S.; Gadek, P.A.; Quinn, C.J. Simaroubaceae, an artificial construct: Evidence from rbcL sequence variation. Am. J. Bot. 1995, 82, 92–103. [Google Scholar] [CrossRef]
- Fernando, E.S.; Quinn, C.J. Picramniaceae, a New Family, and a Recircumscription of Simaroubaceae. Taxon 1995, 44, 177–181. [Google Scholar] [CrossRef]
- Thomas, W.W. Picramniaceae. In Flowering Plants of the Neotropics; Smith, N., Mori, S.A., Henderson, A., Stevenson, D.W., Heald, S.V., Eds.; Princeton University Press: Princeton, NJ, USA, 2004. [Google Scholar]
- Logacheva, M.; Shipunov, A. Phylogenomic analysis of Picramnia, Alvaradoa, and Leitneria supports the independent Picramniales. J. Syst. Evol. 2017, 55, 171–176. [Google Scholar] [CrossRef]
- Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Jacobs, H. Simaroubaceae. In Die Natürlichen Pflanzenfamilien; Engler, H.G.A., Prantl, K., Harms, H., Eds.; Verlag von Wilhelm Engelmann: Leipzig, Germany, 1931. [Google Scholar]
- Swartz, O. Picramnia. Nova Genera & Species Plantarum, seu, Prodromus Descriptionum Vegetabilium: Maximam Partem Incognitorum quæ sub Itinere in Indiam Occidentale Mannis. Available online: https://www.biodiversitylibrary.org/bibliography/433#/summary (accessed on 20 February 2020).
- Liebmann, F.M. Novorum plantarum mexicanarum generum decas. Videnskabelige Meddelelserfra den Naturhistoriske Foreningi Kjøbenhavn. Available online: https://www.biodiversitylibrary.org/bibliography/7547#/summary (accessed on 20 February 2020).
- Zanoni, T.A.; García, G.R.G. Casabitoa perfae (Euphorbiaceae)—A new synonym of Picramnia dictyoneura (Simaroubaceae). Brittonia 1994, 46, 81–82. [Google Scholar] [CrossRef]
- Thomas, W.W. The American genera of Simaroubaceae and their distributions. Acta Bot. Bras. 1990, 4, 11–18. [Google Scholar] [CrossRef]
- Thomas, W.W. Nothotalisia, a new genus of Picramniaceae from tropical America. Brittonia 2011, 63, 51–61. [Google Scholar] [CrossRef]
- Ruiz Lopez, H.; Pavón, J. Florae Peruvianae et Chilensis; Gabrielis de Sancha: Madrid, Spain, 1802. [Google Scholar]
- Christenhusz, M.J.; Fay, M.F.; Chase, M.W. Plants of the World: An Illustrated Encyclopedia of Vascular Plants; University of Chicago Press: Chicago, IL, USA, 2017. [Google Scholar]
- Pirani, J.R. As especies de Picramnia Sw. (Simaroubaceae) do Brasil: Uma sinopse. Boletim Botanica Univ. Sao Paulo 1990, 12, 115–180. [Google Scholar] [CrossRef]
- Pirani, J.R. Diversidade taxonômica e padrões de distribuição geográfica em Picramnia (Simaroubaceae) no Brasil. Acta Bot. Bras. 1990, 4, 19–44. [Google Scholar] [CrossRef][Green Version]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Kuzmina, M.; Ivanova, N. Primer Sets for Plants and Fungi. Available online: http://ccdb.ca/site/wp-content/uploads/2016/09/CCDB_PrimerSets-Plants.pdf (accessed on 19 June 2019).
- Samarakoon, T.; Wang, S.Y.; Alford, M.H. Enhancing PCR amplification of DNA from recalcitrant plant specimens using a trehalose-based additive. Appl. Plant Sci. 2013, 1, 1200236. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: http://www.R-project.org/ (accessed on 24 January 2020).
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Shipunov, A. Shipunov: Miscellaneous Functions from Alexey Shipunov. R package version 1.2. Available online: https://CRAN.R-project.org/package=shipunov (accessed on 13 November 2019).
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Nixon, K.C. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 1999, 15, 407–414. [Google Scholar] [CrossRef]
- Shipunov, A. Ripeline: R-based sequence analysis pipeline. Available online: https://github.com/ashipunov/Ripeline (accessed on 24 January 2020).
- Ashkenazy, H.; Sela, I.; Levy Karin, E.; Landan, G.; Pupko, T. Multiple sequence alignment averaging improves phylogeny reconstruction. Syst. Biol. 2018, 68, 117–130. [Google Scholar] [CrossRef]
- Zander, R.H. A Framework for Post-Phylogenetic Systematics; Zetetic Publications: St. Louis, MO, USA, 2013. [Google Scholar]
- Thomas, W.W. A conspectus of Mexican and Central American Picramnia (Simaroubaceae). Brittonia 1988, 40, 89–105. [Google Scholar] [CrossRef]
- Thomas, W.W. A new species of Picramnia (Simaroubaceae) from Amazonian Peru. Brittonia 1990, 42, 171–174. [Google Scholar] [CrossRef]
- Raven, P.H. Amphitropical relationships in the floras of North and South America. Q. Rev. Biol. 1963, 38, 151–177. [Google Scholar] [CrossRef]
- Shipunov, A. Open Repository. Available online: http://ashipunov.info/shipunov/open/ (accessed on 24 January 2020).
- Simpson, M.G.; Johnson, L.A.; Villaverde, T.; Guilliams, C.M. American amphitropical disjuncts: Perspectives from vascular plant analyses and prospects for future research. Am. J. Bot. 2017, 104, 1600–1650. [Google Scholar] [CrossRef]
- Engler, H.G.A. Simarubaceae. In Flora brasiliensis; Martius, C.F.P., Eichler, A.G., Eds.; Fleischer: Leipzig, Germany, 1874. [Google Scholar]
- Morrone, J.J. Biogeographical regionalisation of the Neotropical region. Zootaxa 2014, 3781, 1–110. [Google Scholar] [CrossRef] [PubMed]
| Species to Place | Most Likely Neigbor | Probability, % | Most Likely Neigbor | Probability, % | Most Likely Neigbor | Probability, % |
|---|---|---|---|---|---|---|
| P. coccinea | P. latifolia | 31 | P. caracasana | 18 | P. juniniana | 16 |
| P. ferrea | P. juniniana | 18 | P. oreadica | 15 | P. campestris | 14 |
| P. gardneri | P. latifolia | 32 | P. caracasana | 19 | P. juniniana | 14 |
| P. gardneri | P. caracasana | 34 | P. latifolia | 27 | P. sellowii | 27 |
| P. grandiflora | P. latifolia | 26 | P. juniniana | 15 | P. oreadica | 15 |
| P. matudai | P. gracilis | 17 | P. hirsuta | 15 | P. teapensis | 14 |
| P. spruceana | P. latifolia | 36 | P. caracasana | 27 | P. sellowii | 20 |
| P. spruceana | P. caracasana | 18 | P. sellowii | 17 | P. latifolia | 15 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shipunov, A.; Carr, S.; Furniss, S.; Pay, K.; Pirani, J.R. First Phylogeny of Bitterbush Family, Picramniaceae (Picramniales). Plants 2020, 9, 284. https://doi.org/10.3390/plants9020284
Shipunov A, Carr S, Furniss S, Pay K, Pirani JR. First Phylogeny of Bitterbush Family, Picramniaceae (Picramniales). Plants. 2020; 9(2):284. https://doi.org/10.3390/plants9020284
Chicago/Turabian StyleShipunov, Alexey, Shyla Carr, Spencer Furniss, Kyle Pay, and José Rubens Pirani. 2020. "First Phylogeny of Bitterbush Family, Picramniaceae (Picramniales)" Plants 9, no. 2: 284. https://doi.org/10.3390/plants9020284
APA StyleShipunov, A., Carr, S., Furniss, S., Pay, K., & Pirani, J. R. (2020). First Phylogeny of Bitterbush Family, Picramniaceae (Picramniales). Plants, 9(2), 284. https://doi.org/10.3390/plants9020284






