Comparative Analysis of In Vitro Responses and Regeneration between Diverse Bioenergy Sorghum Genotypes
Abstract
1. Introduction
2. Results
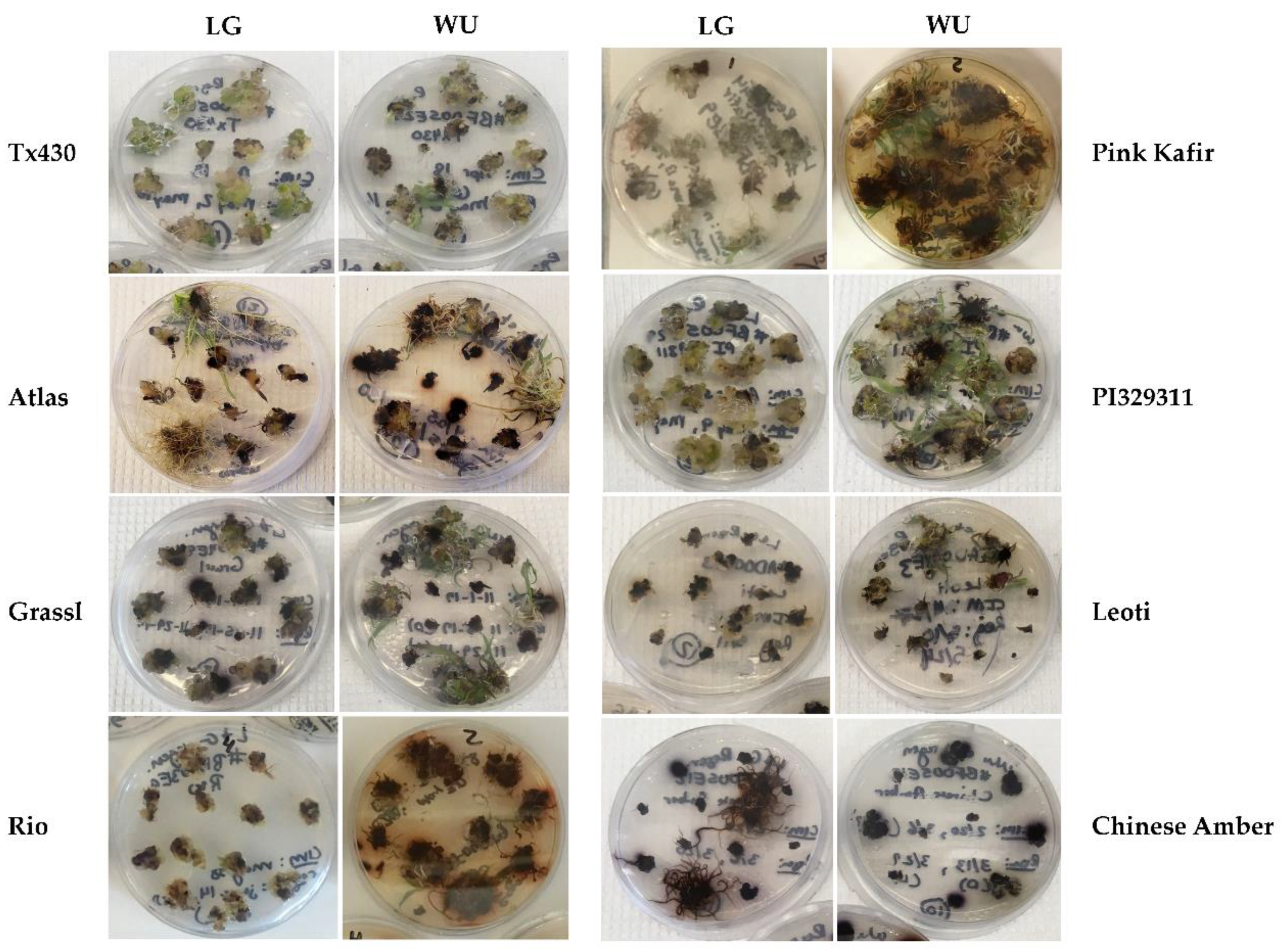
2.1. General Responses
2.2. CIM Browning Responses
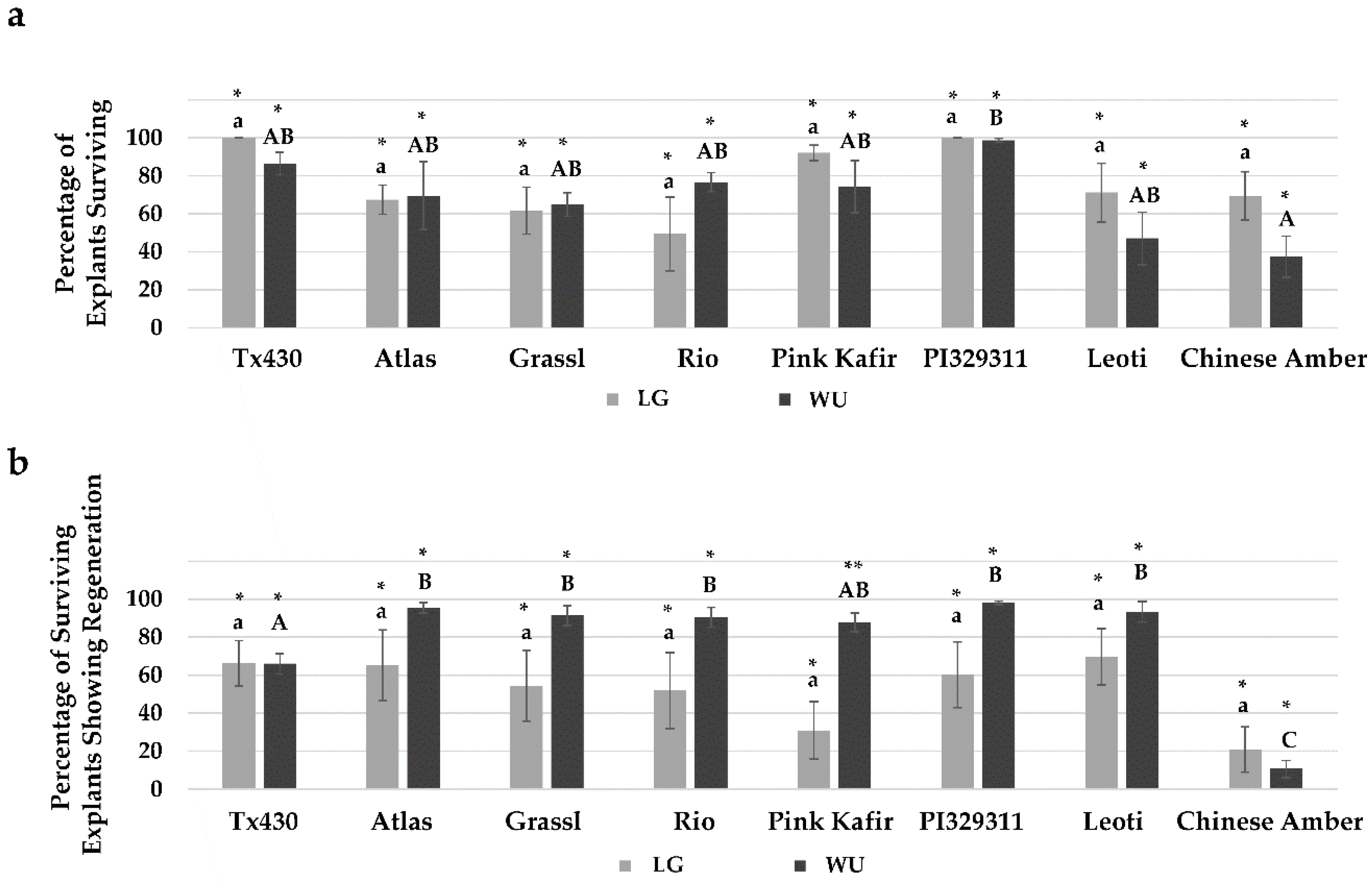
2.3. RM Responses
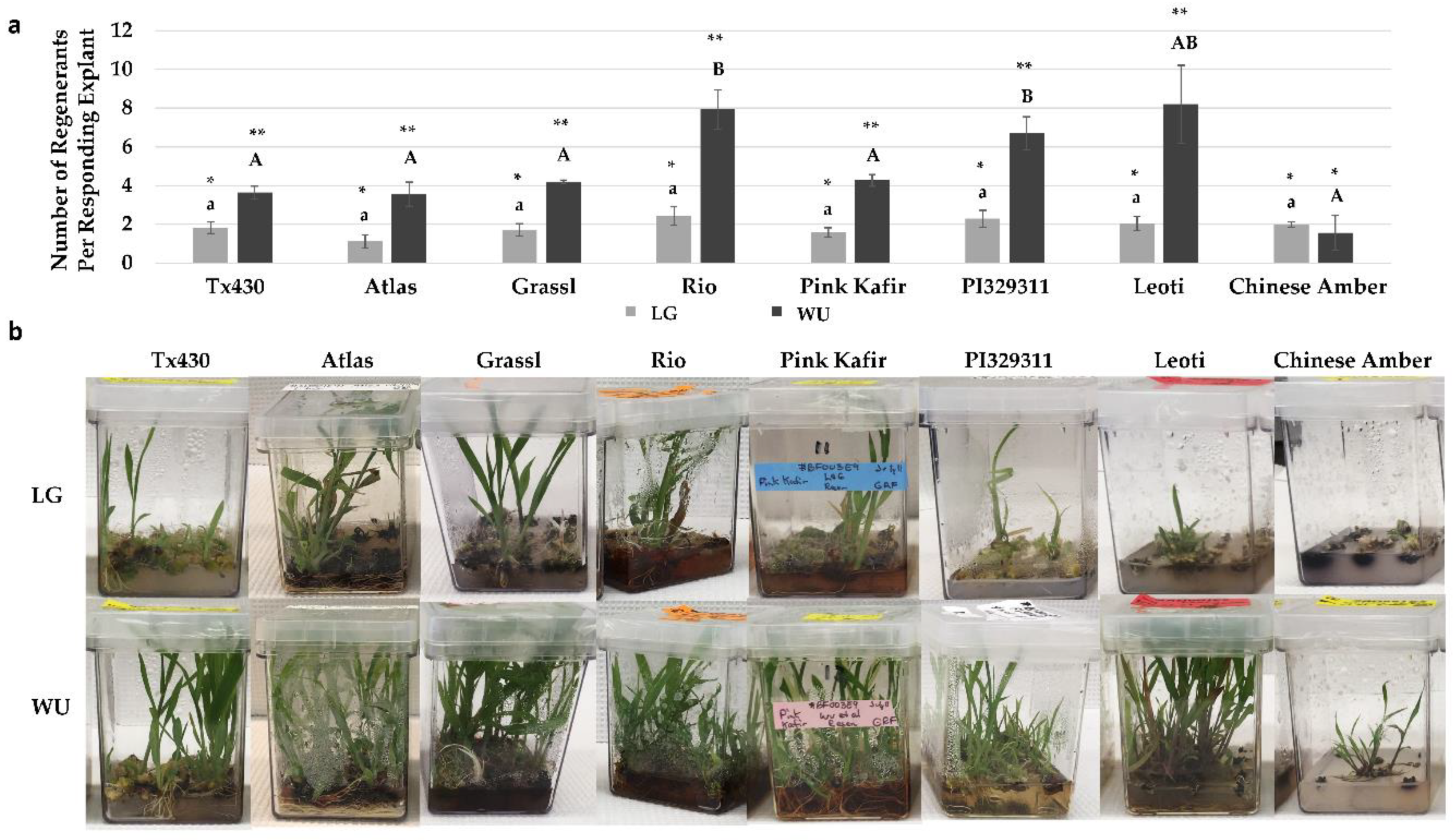
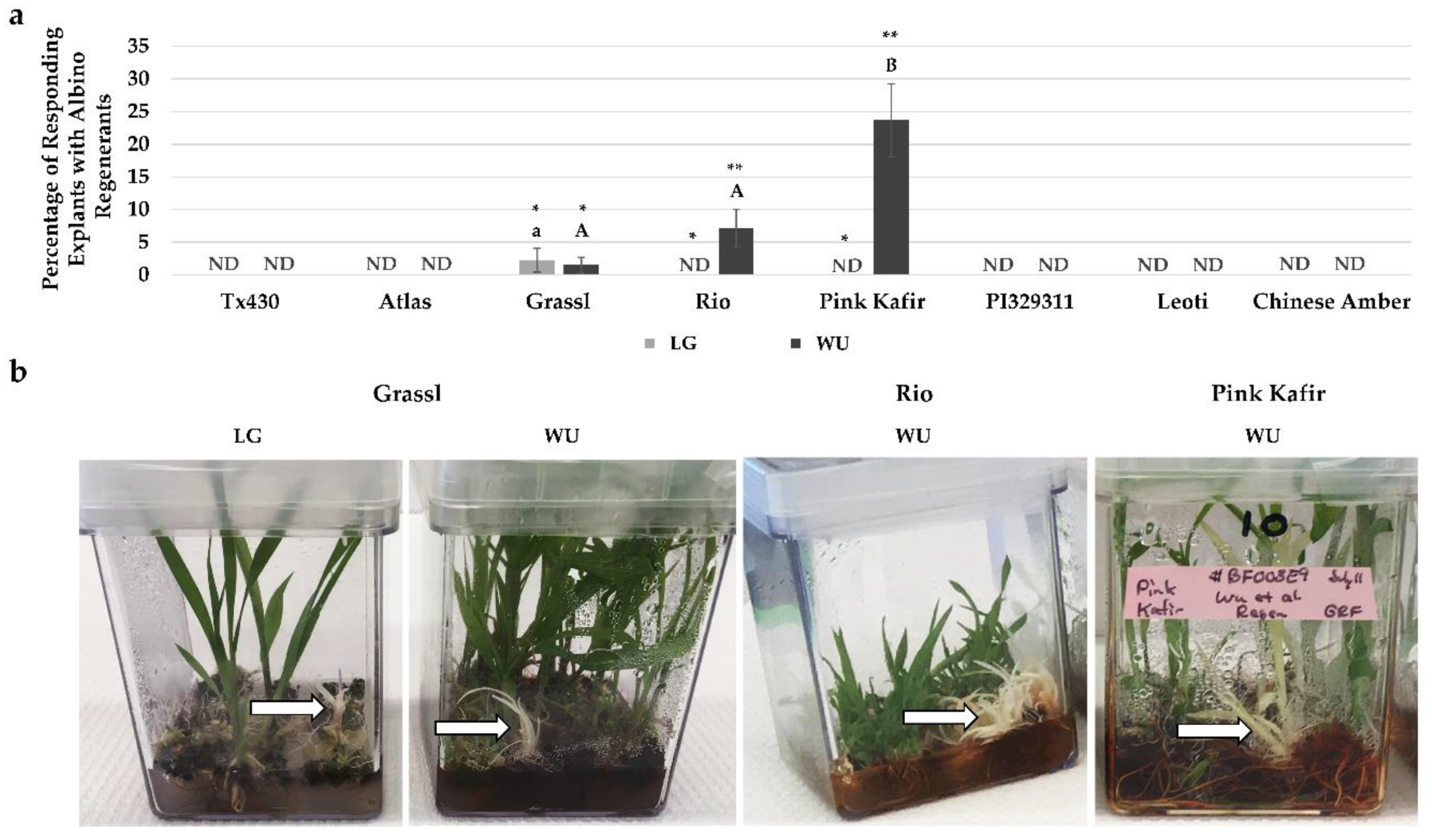
2.4. Performance Following ERM Culture
3. Discussion
3.1. In Vitro Performance
3.2. Medium Browning
3.3. Albino Regeneration
3.4. Summary
4. Materials and Methods
4.1. Plant Materials
4.2. Tissue Culture Media
4.3. Explant Culture
4.4. Tissue Culture Data Collection
4.5. Image and Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. FAOSTAT Database; FAO: Rome, Italy, 2017; Available online: http://faostat3.fao.org/home/E (accessed on 28 August 2019).
- Kebede, H.; Subudhi, P.K.; Rosenow, D.T.; Nguyen, H.T. Quantitative trait loci influencing drought tolerance in grain sorghum (Sorghum bicolor L. Moench). Theor. Appl. Genet. 2001, 103, 266–276. [Google Scholar] [CrossRef]
- Mabelebele, M.; Gous, R.M.; O’Neil, H.M.; Iji, P.A. Whole sorghum inclusion and feed form on performance and nutrient digestibility of broiler chickens. J. Appl. Anim. Nutr. 2018, 6, e5. [Google Scholar] [CrossRef]
- Garzón, A.G.; Drago, S.R. Aptitude of sorghum (Sorghum bicolor (L) Moench) hybrids for brewery or bio-functional malted beverages. J. Food Biochem. 2018, 42, e12692. [Google Scholar] [CrossRef]
- Khazaeian, A.; Ashori, A.; Dizaj, M.Y. Suitability of sorghum stalk fibers for production of particleboard. Carbohydr. Polym. 2015, 120, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Asikin, Y.; Wada, K.; Imai, Y.; Kawamoto, Y.; Mizu, M.; Mutsuura, M.; Takahashi, M. Compositions, taste characteristics, volatile profiles, and antioxidant activities of sweet sorghum (Sorghum bicolor L.) and sugarcane (Saccharum officinarum L.) syrups. Food Meas. Charact. 2018, 12, 884–891. [Google Scholar] [CrossRef]
- Vanamala, J.K.; Massey, A.R.; Pinnamaneni, S.R.; Reddivari, L.; Reardon, K.F. Grain and sweet sorghum (Sorghum bicolor L. Moench) serves as a novel source of bioactive compounds for human health. Crit. Rev. Food Sci. Nutr. 2018, 58, 2867–2881. [Google Scholar] [CrossRef]
- Rooney, W.L.; Blumenthal, J.; Bean, B.; Mullet, J.E. Designing sorghum as a dedicated bioenergy feedstock. Biofuel Bioprod. Biorefining 2007, 1, 147–157. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef]
- Chopra, R.; Burow, G.; Hayes, C.; Emendack, Y.; Xin, Z.; Burke, J. Transcriptome profiling and validation of gene based single nucleotide polymorphisms (SNPs) in sorghum genotypes with contrasting responses to cold stress. BMC Genom. 2015, 16, 1040. [Google Scholar] [CrossRef]
- Ali, R.; Hash, C.T.; Damris, O.; Elhussein, A.; Mohamed, A.H. Introgression of striga resistance into popular Sudanese sorghum varieties using marker assisted selection. World J. Biotechnol. 2016, 1, 48–55. [Google Scholar] [CrossRef]
- Boyles, R.E.; Cooper, E.A.; Myers, M.T.; Brenton, Z.; Rauh, B.L.; Morris, G.P.; Kresovich, S. Genome-wide association studies of grain yield components in diverse sorghum germplasm. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Brenton, Z.W.; Cooper, E.A.; Myers, M.T.; Boyles, R.E.; Shakoor, N.; Zielinski, K.J.; Rauh, B.L.; Bridges, W.C.; Morris, G.P.; Kresovich, S. A genomic resource for the development, improvement, and exploitation of sorghum for bioenergy. Genetics 2016, 204, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Boyles, R.E.; Pfieffer, B.K.; Cooper, E.A.; Zielinski, K.J.; Myers, M.T.; Rooney, W.L.; Kresovich, S. Quantitative trait loci mapping of agronomic and yield traits in two grain sorghum biparental families. Crop Sci. 2017, 57, 2443–2456. [Google Scholar] [CrossRef]
- Mofokeng, M.A.; Shimelis, H.; Laing, M.; Shargie, N. Sorghum [‘Sorghum bicolor’ (L.) Moench] breeding for resistance to leaf and stalk anthracnose, Colletotrichum sublineolum, and improved yield: Progress and prospects. Aust. J. Crop Sci. 2017, 11, 1078–1085. [Google Scholar] [CrossRef]
- Disasa, T.; Feyissa, T.; Admassu, B.; Fetene, M.; Mendu, V. Mapping of QTLs associated with brix and biomass-related traits in Sorghum using SSR Markers. Sugar Tech 2018, 20, 275–285. [Google Scholar] [CrossRef]
- Boyles, R.E.; Brenton, Z.W.; Kresovich, S. Genetic and genomic resources of sorghum to connect genotype with phenotype in contrasting environments. Plant J. 2019, 97, 19–39. [Google Scholar] [CrossRef]
- Mace, E.; Innes, D.; Hunt, C.; Wang, X.; Tao, Y.; Baxter, J.; Hassall, M.; Hathorn, A.; Jordan, D. The Sorghum QTL Atlas: A powerful tool for trait dissection, comparative genomics and crop improvement. Theor. Appl. Genet. 2019, 132, 751–766. [Google Scholar] [CrossRef]
- Bhaskaran, S.; Smith, R.H.; Paliwal, S.; Schertz, K.F. Somaclonal variation from Sorghum bicolor (L.) Moench cell culture. Plant Cell Tissue Organ Cult. 1987, 9, 189–196. [Google Scholar] [CrossRef]
- Kumaravadivel, N.S.; Rangasamy, S.S. Plant regeneration from sorghum anther cultures and field evaluation of progeny. Plant Cell Rep. 1994, 13, 286–290. [Google Scholar] [CrossRef]
- Reddy, P.S.; Jogeswar, G.; Rasineni, G.K.; Maheswari, M.; Reddy, A.R.; Varshney, R.K.; Kishor, P.K. Proline over-accumulation alleviates salt stress and protects photosynthetic and antioxidant enzyme activities in transgenic sorghum [Sorghum bicolor (L.) Moench]. Plant Physiol. Biochem. 2015, 94, 104–113. [Google Scholar] [CrossRef]
- Char, S.N.; Wei, J.; Mu, Q.; Li, X.; Zhang, Z.J.; Yu, J.; Yang, B. An Agrobacterium-delivered CRISPR/Cas9 system for targeted mutagenesis in sorghum. Plant Biotechnol. J. 2019, 18, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Bedell, J.A.; Budiman, M.A.; Nunberg, A.; Citek, R.W.; Robbins, D.; Jones, J.; Flick, E.; Rohlfing, T.; Fries, J.; Bradford, K.; et al. Sorghum genome sequencing by methylation filtration. PLoS Biol. 2005, 3, e13. [Google Scholar] [CrossRef] [PubMed]
- Cooper, E.A.; Brenton, Z.W.; Flinn, B.S.; Jenkins, J.; Shu, S.; Flowers, D.; Luo, F.; Wang, Y.; Xia, P.; Barry, K.; et al. A new reference genome for Sorghum bicolor reveals high levels of sequence similarity between sweet and grain genotypes: Implications for the genetics of sugar metabolism. BMC Genom. 2019, 20, 420. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Jia, S.; Yobi, A.; Ge, Z.; Sato, S.J.; Zhang, C.; Angelovici, R.; Clemente, T.E.; Holding, D.R. Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 2018, 177, 1425–1438. [Google Scholar] [CrossRef] [PubMed]
- Botella, J.R. Now for the hard ones: Is there a limit on CRISPR genome editing in crops? J. Exp. Bot. 2019, 70, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Kaeppler, H.F.; Pedersen, J.F. Evaluation of 41 elite and exotic inbred Sorghum genotypes for high quality callus production. Plant Cell Tissue Organ Cult. 1997, 48, 71–75. [Google Scholar] [CrossRef]
- Liu, G.; Gilding, E.K.; Godwin, I.D. A robust tissue culture system for sorghum [Sorghum bicolor (L.) Moench]. S. Afr. J. Bot. 2015, 98, 157–160. [Google Scholar] [CrossRef]
- Omer, R.A.; Asami, P.; Birungi, J. Callus induction and plant regeneration from immature embryos of sweet sorghum (Sorghum bicolor Moench). Biotechnology 2018, 17, 12–18. [Google Scholar] [CrossRef][Green Version]
- Elkonin, L.A.; Lopushanskaya, R.F.; Pakhomova, N.V. Initiation and maintenance of friable, embryogenic callus of sorghum (Sorghum bicolor (L.) Moench) by amino acids. Maydica 1995, 40, 153–157. [Google Scholar]
- Dreger, M.; Mól, R.; Deja, A.; Raj, E.; Mańkowska, G.; Wielgus, K. Improved plant regeneration in callus cultures of Sorghum bicolor (L.) Moench. In Vitro Cell. Dev. Biol. Plant 2019, 55, 190–198. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [PubMed]
- Arikita, F.N.; Azevedo, M.S.; Scotton, D.C.; de Siqueira Pinto, M.; Figueira, A.; Peres, L.E. Novel natural genetic variation controlling the competence to form adventitious roots and shoots from the tomato wild relative Solanum pennellii. Plant Sci. 2013, 199, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, N.; Dahleen, L.S.; Bregitzer, P. Candidate genes within tissue culture regeneration QTL revisited with a linkage map based on transcript-derived markers. Crop Sci. 2010, 50, 1697–1707. [Google Scholar] [CrossRef]
- Begheyn, R.F.; Yates, S.A.; Sykes, T.; Studer, B. Genetic Loci Governing Androgenic Capacity in Perennial Ryegrass (Lolium perenne L.). G3 Genes Genomes Genet. 2018, 8, 1897–1908. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liu, M.; Yan, Y.; Qing, C.; Zhang, X.; Zhang, Y.; Long, Y.; Wang, L.; Pan, L.; Zou, C.; et al. Genetic dissection of maize embryonic callus regenerative capacity using multi-locus genome-wide association studies. Front. Plant Sci. 2018, 9, 561. [Google Scholar] [CrossRef]
- Loyola-Vargas, V.M.; Ochoa-Alejo, N. An introduction to plant tissue culture: Advances and perspectives. In Plant Cell Culture Protocols; Humana Press: New York, NY, USA, 2018; pp. 3–13. [Google Scholar] [CrossRef]
- Cai, T.; Daly, B.; Butler, L. Callus induction and plant regeneration from shoot portions of mature embryos of high tannin sorghums. Plant Cell Tissue Organ Cult. 1987, 9, 245–252. [Google Scholar] [CrossRef]
- Sato, S.; Clemente, T.; Dweikat, I. Identification of an elite sorghum genotype with high in vitro performance capacity. In Vitro Cell. Dev. Biol. Plant 2004, 40, 57–60. [Google Scholar] [CrossRef]
- Clará Valencia, R.; Rooney, W.L. Genetic Control of Sorghum Grain Color. INTSORMIL 2009, Presentation 10. Available online: https://digitalcommons.unl.edu/intsormilpresent/10/ (accessed on 8 February 2019).
- Chakaborti, P.; Ghosh, A. Variation in callus induction and root-shoot bud formation depend on seed coat of sesame genotypes. Res. J. Bot. 2010, 5, 14–19. [Google Scholar] [CrossRef]
- Ranch, J.P.; Oglesby, L.; Zielinski, A.C.; Horsch, R.B. Plant regeneration from embryo-derived tissue cultures of soybeans. In Vitro Cell. Dev. Biol. 1985, 21, 653–658. [Google Scholar] [CrossRef]
- Carvalho, C.H.; Zehr, U.B.; Gunaratna, N.; Anderson, J.; Kononowicz, H.H.; Hodges, T.K.; Axtell, J.D. Agrobacterium-mediated transformation of sorghum: Factors that affect transformation efficiency. Genet. Mol. Biol. 2004, 27, 259–269. [Google Scholar] [CrossRef]
- Howe, A.; Sato, S.; Dweikat, I.; Fromm, M.; Clemente, T. Rapid and reproducible Agrobacterium-mediated transformation of sorghum. Plant Cell Rep. 2006, 25, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.; Thu, T.T.; Claeys, M.; Angenon, G. Agrobacterium-mediated transformation of sorghum (Sorghum bicolor (L.) Moench) using an improved in vitro regeneration system. Plant Cell Tissue Organ Cult. 2007, 91, 155–164. [Google Scholar] [CrossRef]
- Gurel, S.; Gurel, E.; Miller, T.I.; Lemaux, P.G. Agrobacterium-mediated transformation of Sorghum bicolor using immature embryos. Methods Mol. Biol. 2012, 847, 109–122. [Google Scholar] [CrossRef]
- Chen, X.; Li, O.; Shi, L.; Wu, X.; Xia, B.; Pei, Z. To establish the regeneration system of sweet sorghum immature embryos. Adv. Appl. Biotechnol. 2015, 333, 83–91. [Google Scholar] [CrossRef]
- Do, P.T.; Lee, H.; Mookkan, M.; Folk, W.R.; Zhang, Z.J. Rapid and efficient Agrobacterium-mediated transformation of sorghum (Sorghum bicolor) employing standard binary vectors and bar gene as a selectable marker. Plant Cell Rep. 2016, 35, 2065–2076. [Google Scholar] [CrossRef] [PubMed]
- Belide, S.; Vanhercke, T.; Petrie, J.R.; Singh, S.P. Robust genetic transformation of sorghum (Sorghum bicolor L.) using differentiating embryogenic callus induced from immature embryos. Plant Methods 2017, 13, 109. [Google Scholar] [CrossRef]
- Espinoza-Sánchez, E.A.; Sánchez-Peña, Y.A.; Torres-Castillo, J.A.; García-Zambrano, E.A.; Ramírez, J.T.; Zavala-García, F.; Sinagawa-García, S.R. Somatic embryogenesis induction from immature embryos of Sorghum bicolor L. (Moench). Phyton-Int. J. Exp. Bot. 2018, 87, 105–112. [Google Scholar]
- Wernicke, W.; Brettell, R. Somatic embryogenesis from Sorghum bicolor leaves. Nature 1980, 287, 138–139. [Google Scholar] [CrossRef]
- Liu, G.; Godwin, I.D. Highly efficient sorghum transformation. Plant Cell Rep. 2012, 31, 999–1007. [Google Scholar] [CrossRef]
- Wu, E.; Lenderts, B.; Glassman, K.; Berezowska-Kaniewska, M.; Christensen, H.; Asmus, T.; Zhen, S.; Chu, U.; Cho, M.J.; Zhao, Z.Y. Optimized Agrobacterium-mediated sorghum transformation protocol and molecular data of transgenic sorghum plants. In Vitro Cell. Dev. Biol. Plant 2014, 50, 9–18. [Google Scholar] [CrossRef]
- Seetharama, N.; Sairam, R.V.; Rani, T.S. Regeneration of sorghum from shoot tip cultures and field performance of the progeny. Plant Cell Tissue Organ Cult. 2000, 61, 169–173. [Google Scholar] [CrossRef]
- Assem, S.K.; Zamzam, M.M.; Hussein, B.A. Evaluation of somatic embryogenesis and plant regeneration in tissue culture of ten sorghum (Sorghum bicolor L.) genotypes. Afr. J. Biotechnol. 2014, 13, 3672–3681. [Google Scholar] [CrossRef]
- Visarada, K.B.; Prasad, G.S.; Royer, M. Genetic transformation and evaluation of two sweet sorghum genotypes for resistance to spotted stemborer, Chilo partellus (Swinhoe). Plant Biotechnol. Rep. 2016, 10, 277–289. [Google Scholar] [CrossRef]
- Merkle, S.A.; Parrott, W.A.; Flinn, B.S. Morphogenic aspects of somatic embryogenesis. In In Vitro Embryogenesis in Plants; Thorpe, T.A., Ed.; Springer: Dordrecht, The Netherlands, 1995; pp. 155–203. [Google Scholar] [CrossRef]
- Ahmad, I.; Hussain, T.; Ashraf, I.; Nafees, M.; Maryam, R.M.; Iqbal, M. Lethal effects of secondary metabolites on plant tissue culture. Am. Eurasian J. Agric. Environ. Sci. 2013, 13, 539–547. [Google Scholar] [CrossRef]
- Jones, A.M.; Saxena, P.K. Inhibition of phenylpropanoid biosynthesis in Artemisia annua L.: A novel approach to reduce oxidative browning in plant tissue culture. PLoS ONE 2013, 8, e76802. [Google Scholar] [CrossRef] [PubMed]
- Szabados, L.; Savoure, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S. The fermentation analogy: A point of view for understanding the intriguing role of proline accumulation in stressed plants. Front. Plant Sci. 2016, 7, 1339. [Google Scholar] [CrossRef]
- Wen, F.S.; Sorensen, E.L.; Barnett, F.L.; Liang, G.H. Callus induction and plant regeneration from anther and inflorescence culture of sorghum. Euphytica 1991, 52, 177–181. [Google Scholar] [CrossRef]
- Can, N.D.; Nakamura, S.; Haryanto, T.A.; Yoshida, T. Effects of physiological status of parent plants and culture medium composition on the anther culture of sorghum. Plant Prod. Sci. 1998, 1, 211–215. [Google Scholar] [CrossRef]
- Park, S.G.; Ubaidillah, M.; Kim, K.M. Effect of maltose concentration on plant regeneration of anther culture with different genotypes in rice (Oryza sativa L.). Am. J. Plant Sci. 2013, 4, 2265–2270. [Google Scholar] [CrossRef]
- Krzewska, M.; Czyczyło-Mysza, I.; Dubas, E.; Gołębiowska-Pikania, G.; Golemiec, E.; Stojałowski, S.; Chrupek, M.; Żur, I. Quantitative trait loci associated with androgenic responsiveness in triticale (× Triticosecale Wittm.) anther culture. Plant Cell Rep. 2012, 31, 2099–2108. [Google Scholar] [CrossRef] [PubMed]
- Sriskandarajah, S.; Sameri, M.; Lerceteau-Köhler, E.; Westerbergh, A. Increased recovery of green doubled haploid plants from barley anther culture. Crop Sci. 2015, 55, 2806–2812. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, K.; Zhang, W.; Liu, H.Y.; Du, L.P.; Hu, H.R.; Ye, X.G. Comprehensive analysis of differently expressed genes and proteins in albino and green plantlets from a wheat anther culture. Biol. Plant. 2017, 61, 255–265. [Google Scholar] [CrossRef]
- Ma, H.; Gu, M.; Liang, G.H. Plant regeneration from cultured immature embryos of Sorghum bicolor (L.) Moench. Theor. Appl. Genet. 1987, 73, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.M.; Xu, Z.H. Regeneration of fertile plants from embryogenic suspension culture protoplasts of Sorghum vulgare. Plant Cell Rep. 1990, 9, 51–53. [Google Scholar] [CrossRef]
- Ayed, O.S.; De Buyser, J.; Picard, E.; Trifa, Y.; Amara, H.S. Effect of pre-treatment on isolated microspores culture ability in durum wheat (Triticum turgidum subsp. durum Desf.). J. Plant Breed. Crop Sci. 2010, 2, 30–38. [Google Scholar]
- Khatun, R.; Islam, S.S.; Bari, M.A. Studies on plant regeneration efficiency through in vitro micropropagation and anther culture of twenty-five rice cultivars in Bangladesh. J. Appl. Sci. Res. 2010, 6, 1705–1711. [Google Scholar]
- Dewir, Y.H.; Naidoo, Y.; da Silva, J.A. Thidiazuron-induced abnormalities in plant tissue cultures. Plant Cell Rep. 2018, 37, 1451–1470. [Google Scholar] [CrossRef]
- Makowska, K.; Oleszczuk, S.; Zimny, J. The effect of copper on plant regeneration in barley microspore culture. Czech. J. Genet. Plant Breed. 2017, 53, 17–22. [Google Scholar] [CrossRef]
- Day, A.T.; Ellis, T.H. Chloroplast DNA deletions associated with wheat plants regenerated from pollen: Possible basis for maternal inheritance of chloroplasts. Cell 1984, 39, 359–368. [Google Scholar] [CrossRef]
- Dunford, R.; Walden, R.M. Plastid genome structure and plastid-related transcript levels in albino barley plants derived from anther culture. Curr. Genet. 1991, 20, 339–347. [Google Scholar] [CrossRef]
- Duarte-Aké, F.; Castillo-Castro, E.; Pool, F.B.; Espadas, F.; Santamaría, J.M.; Robert, M.L.; De-la-Peña, C. Physiological differences and changes in global DNA methylation levels in Agave angustifolia Haw. albino variant somaclones during the micropropagation process. Plant Cell Rep. 2016, 35, 2489–2502. [Google Scholar] [CrossRef]
- Isah, T. Changes in the biochemical parameters of albino, hyperhydric and normal green leaves of Caladium bicolor cv. “Bleeding hearts” in vitro long-term cultures. J. Photochem. Photobiol. B 2019, 191, 88–98. [Google Scholar] [CrossRef]
- Krzewska, M.; Czyczyło-Mysza, I.; Dubas, E.; Gołębiowska-Pikania, G.; Żur, I. Identification of QTLs associated with albino plant formation and some new facts concerning green versus albino ratio determinants in triticale (× Triticosecale Wittm.) anther culture. Euphytica 2015, 206, 263–278. [Google Scholar] [CrossRef]
- Bolibok, H.; Rakoczy-Trojanowska, M. Genetic mapping of QTLs for tissue-culture response in plants. Euphytica 2006, 149, 73–83. [Google Scholar] [CrossRef]
- Song, X.; Han, Y.; Teng, W.; Sun, G.; Li, W. Identification of QTL underlying somatic embryogenesis capacity of immature embryos in soybean (Glycine max (L.) Merr.). Plant Cell Rep. 2010, 29, 125–131. [Google Scholar] [CrossRef]
- Trujillo-Moya, C.; Gisbert, C.; Vilanova, S.; Nuez, F. Localization of QTLs for in vitro plant regeneration in tomato. BMC Plant Biol. 2011, 11, 140. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, T.; Yu, D.; Gai, J. Mapping QTLs for tissue culture response in soybean (Glycine max (L.) Merr.). Mol. Cells 2011, 32, 337–342. [Google Scholar] [CrossRef]
- Li, S.; Yan, S.; Wang, A.H.; Zou, G.; Huang, X.; Han, B.; Qian, Q.; Tao, Y. Identification of QTLs associated with tissue culture response through sequencing-based genotyping of RILs derived from 93-11 × Nipponbare in rice (Oryza sativa). Plant Cell Rep. 2013, 32, 103–116. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, H.; Li, W.; Wu, J.; Zhou, Z.; Zhou, F.; Chen, H.; Lin, Y. Genome-wide association study of callus induction variation to explore the callus formation mechanism of rice. J. Integr. Plant Biol. 2019, 61, 1134–1150. [Google Scholar] [CrossRef]
- Sheflin, A.M.; Kirkwood, J.S.; Wolfe, L.M.; Jahn, C.E.; Broeckling, C.D.; Schachtman, D.P.; Prenni, J.E. High-throughput quantitative analysis of phytohormones in sorghum leaf and root tissue by ultra-performance liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2019, 16, 4839–4848. [Google Scholar] [CrossRef]
- Watts-Williams, S.J.; Emmett, B.D.; Levesque-Tremblay, V.; MacLean, A.M.; Sun, X.; Satterlee, J.W.; Fei, Z.; Harrison, M.J. Diverse Sorghum bicolor accessions show marked variation in growth and transcriptional responses to arbuscular mycorrhizal fungi. Plant Cell Environ. 2019, 42, 1758–1774. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]

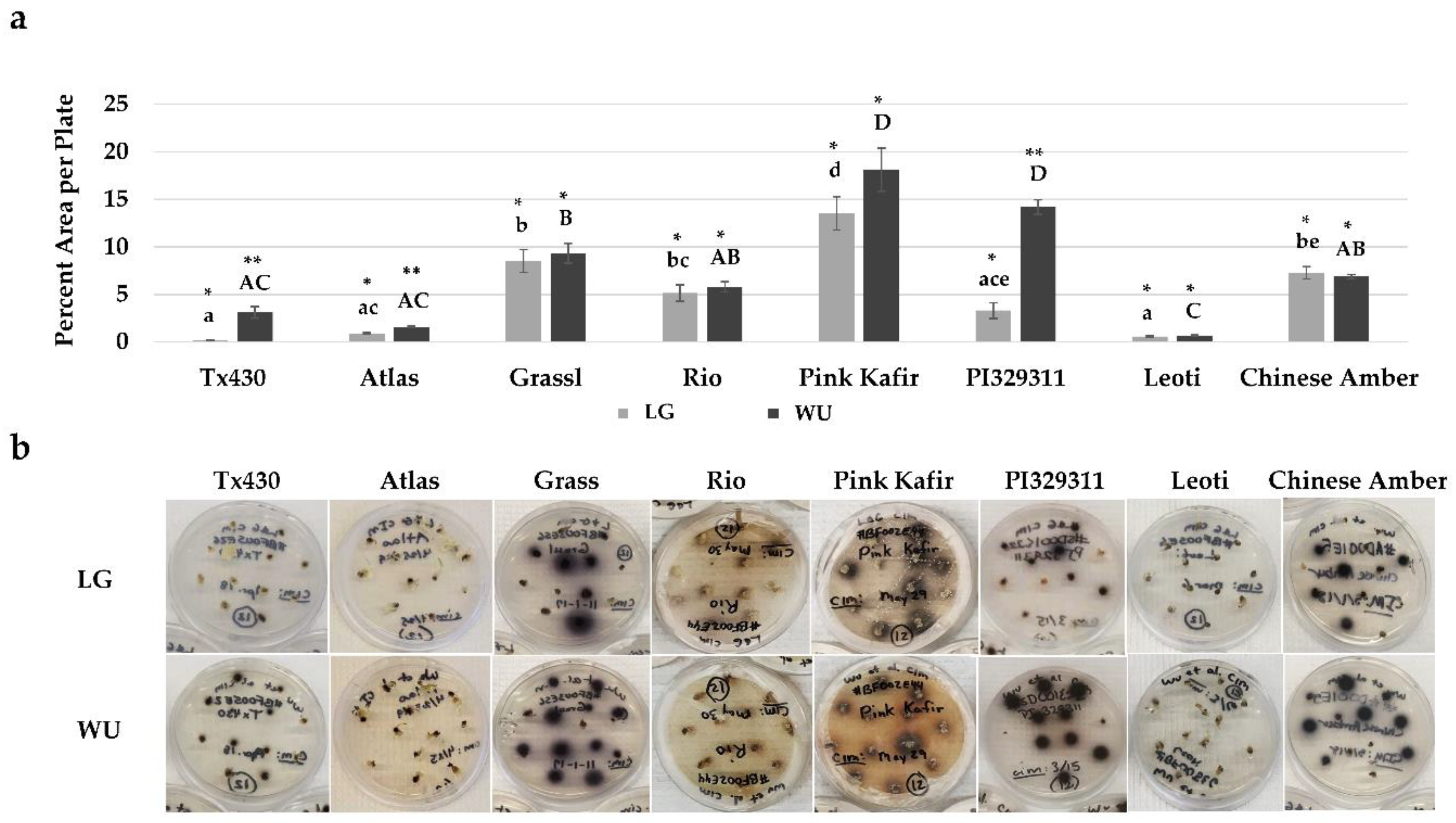
| Genotype | Protocol | Proliferation Index | Medium Browning |
|---|---|---|---|
| Tx430 | LG | ++++ | - |
| WU | +++ | + | |
| Atlas | LG | ++ | + |
| WU | +++ | ++ | |
| Grassl | LG | ++ | +++ |
| WU | +++ | +++ | |
| Rio | LG | ++ | + |
| WU | +++ | ++++ | |
| Pink Kafir | LG | +++ | + |
| WU | ++++ | ++++ | |
| PI329311 | LG | ++++ | + |
| WU | ++++ | ++ | |
| Leoti | LG | + | ++ |
| WU | + | ++ | |
| Chinese Amber | LG | + | +++ |
| WU | + | +++ |
| Genotype (Name and/or Identifier) | Sorghum Type | Seed Color/Tannin | Plant Color | Race | Origin Location |
|---|---|---|---|---|---|
| Tx430 | Grain | Yellow/no | Purple | Caudatum | USA |
| Atlas (PI641807) | Sweet | White/no | Purple | Kafir-Bicolor | USA |
| Grassl | Cellulosic | Brown/yes | Purple | Caudatum | Uganda |
| Rio | Sweet | White/yes | Purple | Caudatum | USA |
| Pink Kafir (PI655972) | Grain | Red-white/no | Purple | Kafir | USA |
| PI329311 | Cellulosic | Yellow/no | Purple | Durra | Ethiopia |
| Leoti (PI586454) | Sweet | Brown/yes | Purple | Kafir | Hungary |
| Chinese Amber (PI22913) | Sweet | Brown/yes | Purple | Bicolor | China |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flinn, B.; Dale, S.; Disharoon, A.; Kresovich, S. Comparative Analysis of In Vitro Responses and Regeneration between Diverse Bioenergy Sorghum Genotypes. Plants 2020, 9, 248. https://doi.org/10.3390/plants9020248
Flinn B, Dale S, Disharoon A, Kresovich S. Comparative Analysis of In Vitro Responses and Regeneration between Diverse Bioenergy Sorghum Genotypes. Plants. 2020; 9(2):248. https://doi.org/10.3390/plants9020248
Chicago/Turabian StyleFlinn, Barry, Savanah Dale, Andrew Disharoon, and Stephen Kresovich. 2020. "Comparative Analysis of In Vitro Responses and Regeneration between Diverse Bioenergy Sorghum Genotypes" Plants 9, no. 2: 248. https://doi.org/10.3390/plants9020248
APA StyleFlinn, B., Dale, S., Disharoon, A., & Kresovich, S. (2020). Comparative Analysis of In Vitro Responses and Regeneration between Diverse Bioenergy Sorghum Genotypes. Plants, 9(2), 248. https://doi.org/10.3390/plants9020248






