Transcriptome and Phytochemical Analysis Reveals the Alteration of Plant Hormones, Characteristic Metabolites, and Related Gene Expression in Tea (Camellia sinensis L.) Leaves During Withering
Abstract
1. Introduction
2. Results and Discussion
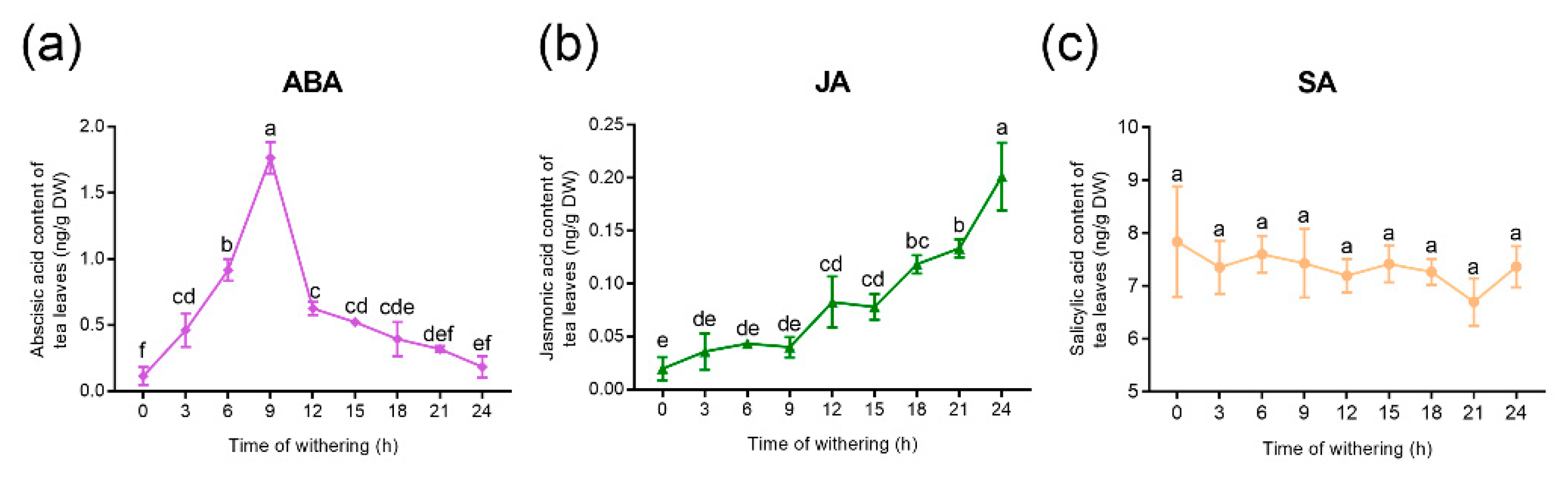
2.1. Changes in Hormone Content of Tea Leaves During the Withering Process
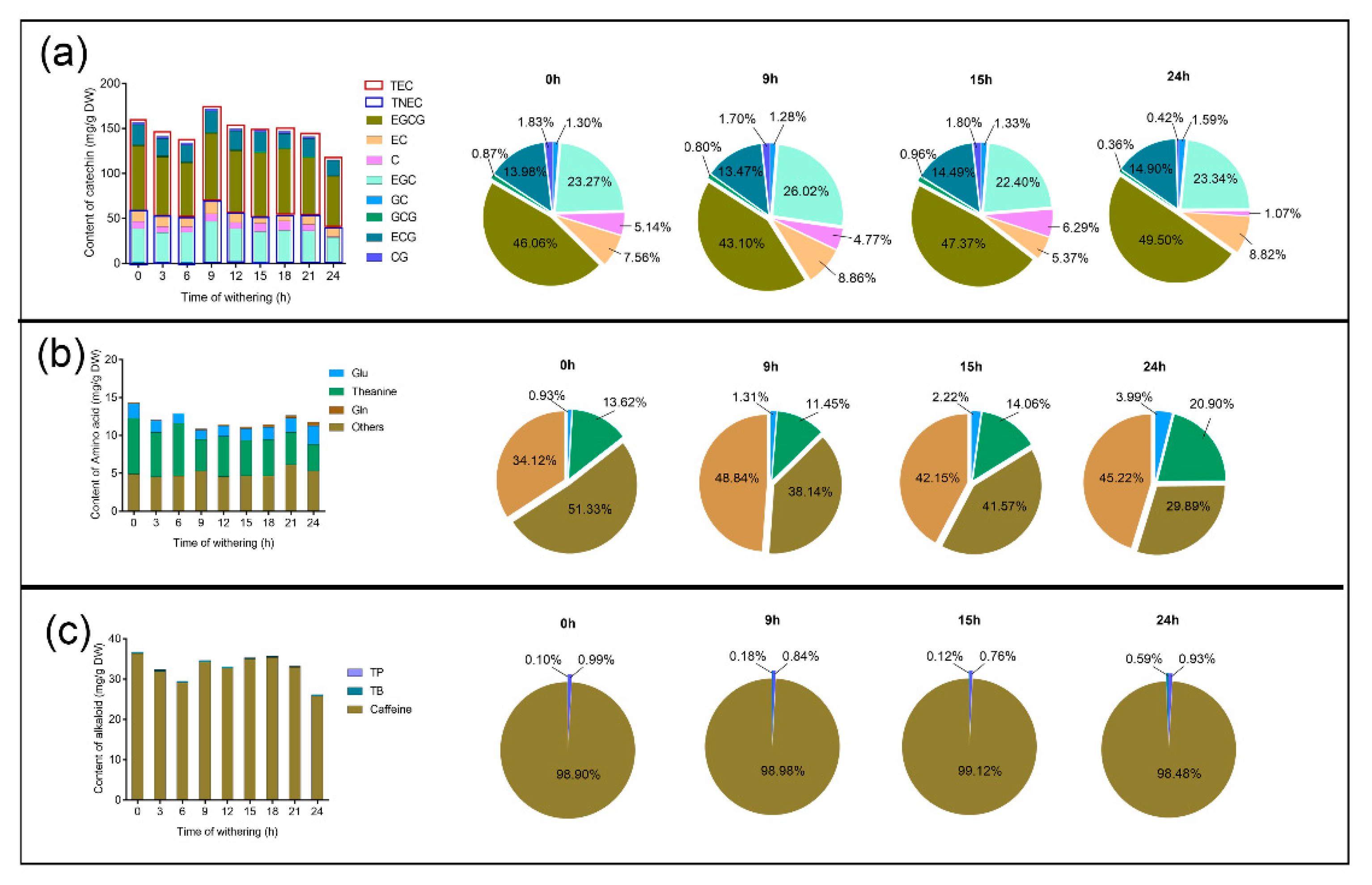
2.2. Determination of Catechins, Free Amino Acids, and Purine Alkaloids
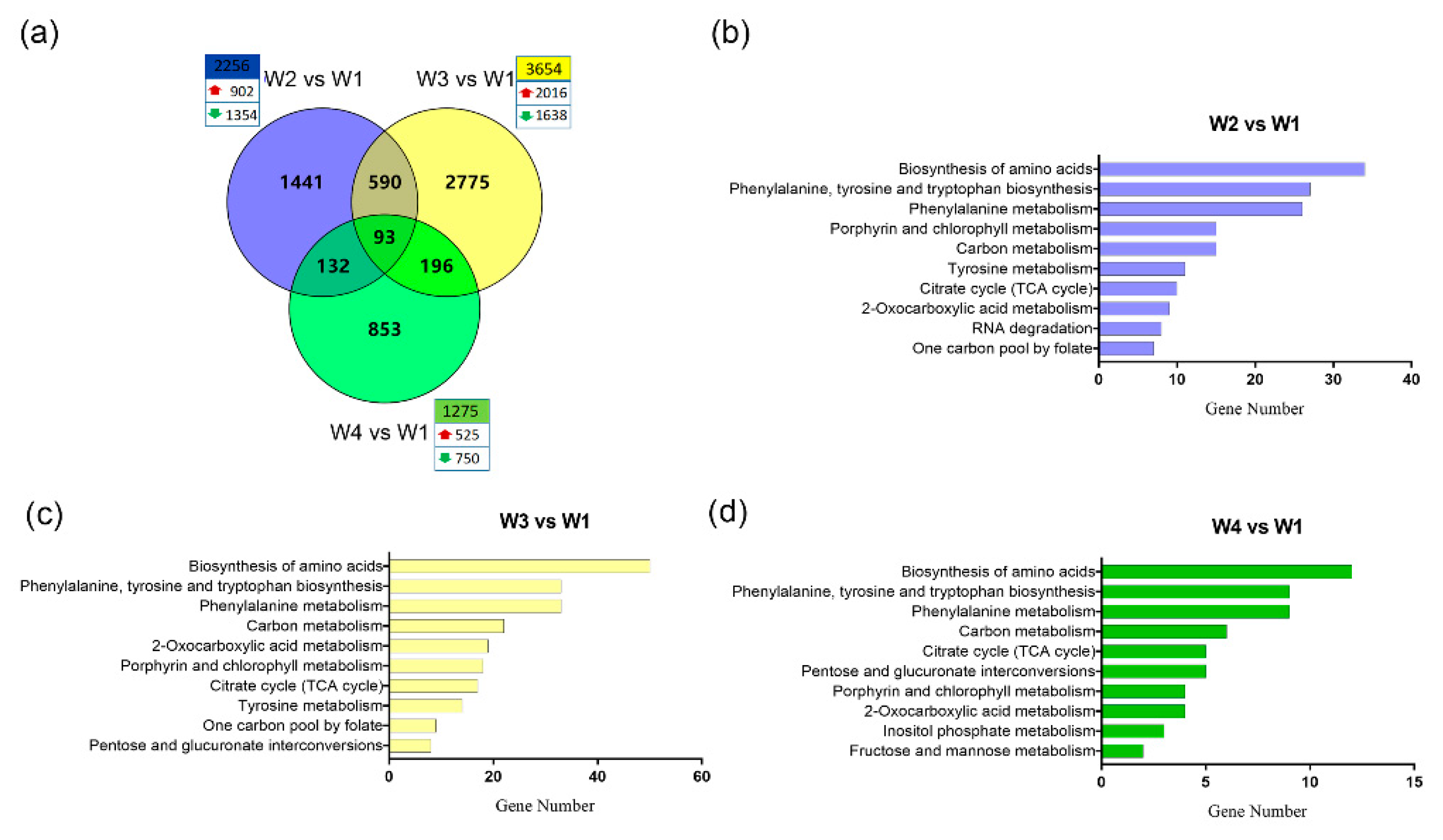
2.3. Global Transcriptome Analysis of Tea Leaves During the Withering Process
2.4. Construction of a Gene Co-Expression Network and Identification of Content-Related Modules
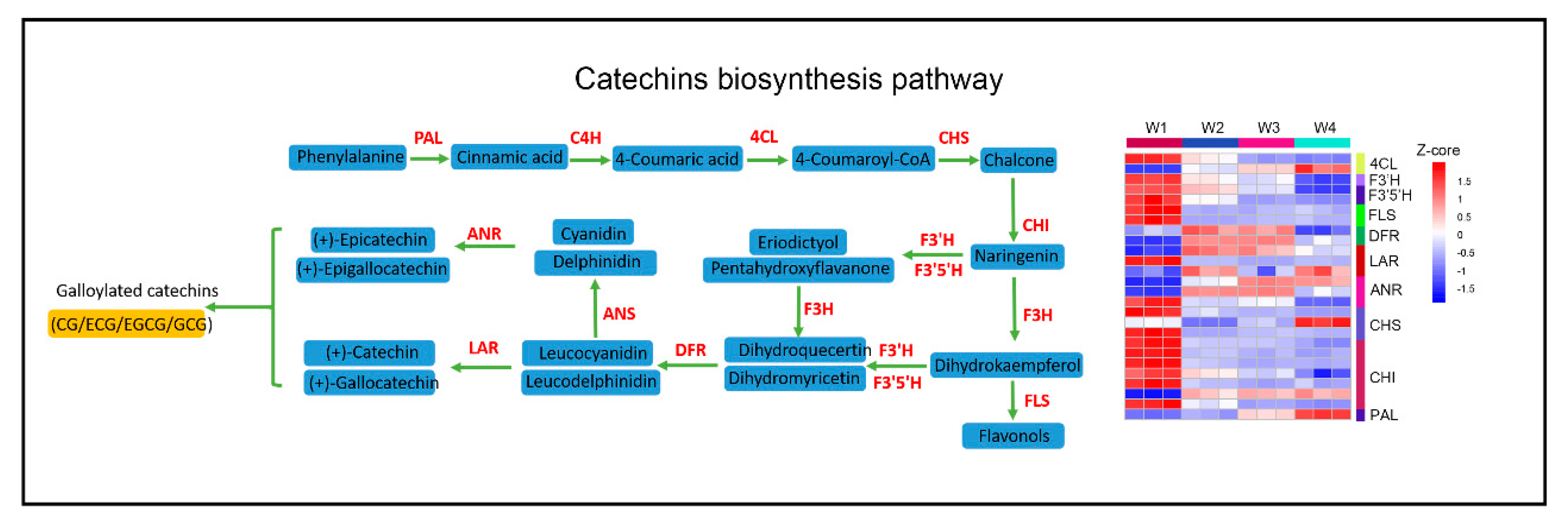
2.5. Effect of Withering on Catechins, Theanine, and Caffeine Metabolism
3. Materials and Methods
3.1. Plant Materials
3.2. Measuring Endogenous Salicylic Acid, Jasmonic Acid, and Abscisic Acid
3.3. Determination of Catechins and Purine Alkaloids
3.4. Determination of Free Amino Acid Content
3.5. RNA Extraction, Illumina Sequencing, and Transcriptome Data Processing
3.6. KEGG Pathway Enrichment Analyses
3.7. Co-Expression Networks
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yang, C.S.; Hong, J. Prevention of Chronic Diseases by Tea: Possible Mechanisms and Human Relevance. Annu. Rev. Nutr. 2013, 33, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Baldermann, S.; Watanabe, N. Recent studies of the volatile compounds in tea. Food Res. Int. 2013, 2, 585–599. [Google Scholar] [CrossRef]
- Schulzki, G.; Nusslein, B.; Sievers, H. Transition rates of selected metals determined in various types of teas (Camellia sinensis L. Kuntze) and herbal/fruit infusions. Food Chem. 2017, 215, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Campanella, L.; Bonanni, A.; Tomassetti, M. Determination of the antioxidant capacity of samples of different types of tea, or of beverages based on tea or other herbal products, using a superoxide dismutase biosensor. J. Pharm. Biomed. Anal. 2003, 32, 725–736. [Google Scholar] [CrossRef]
- Yu, X.; Hu, S.; He, C.; Zhou, J.; Qu, F.; Ai, Z.; Chen, Y.; Ni, D. Chlorophyll Metabolism in Postharvest Tea (Camellia sinensis L.) Leaves: Variations in Color Values, Chlorophyll Derivatives, and Gene Expression Levels under Different Withering Treatments. J. Agric. Food Chem. 2019, 67, 10624–10636. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.J.; Ma, H.Y.; Zhuang, J. iTRAQ-based proteomics monitors the withering dynamics in postharvest leaves of tea plant (Camellia sinensis). Mol. Genet. Genom. MGG 2018, 293, 45–59. [Google Scholar] [CrossRef]
- Allen, G.J.; Kuchitsu, K.; Chu, S.P.; Murata, Y.; Schroeder, J.I. Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 1999, 11, 1785–1798. [Google Scholar] [CrossRef]
- Owiti, J.; Grossmann, J.; Gehrig, P.; Dessimoz, C.; Laloi, C.; Hansen, M.B.; Gruissem, W.; Vanderschuren, H. iTRAQ-based analysis of changes in the cassava root proteome reveals pathways associated with post-harvest physiological deterioration. Plant J. Cell Mol. Biol. 2011, 67, 145–156. [Google Scholar] [CrossRef]
- Page, T.; Griffiths, G.; Buchanan-Wollaston, V. Molecular and biochemical characterization of postharvest senescence in broccoli. Plant Physiol. 2001, 125, 718–727. [Google Scholar] [CrossRef]
- Reilly, K.; Bernal, D.; Cortes, D.F.; Gomez-Vasquez, R.; Tohme, J.; Beeching, J.R. Towards identifying the full set of genes expressed during cassava post-harvest physiological deterioration. Plant Mol. Biol. 2007, 64, 187–203. [Google Scholar] [CrossRef]
- Lee, S.; Seo, P.J.; Lee, H.J.; Park, C.M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. Cell Mol. Biol. 2012, 70, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, K.; Kaya, Z. Gene regulation network behind drought escape, avoidance and tolerance strategies in black poplar (Populus nigra L.). Plant Physiol. Biochem. PPB 2017, 115, 183–199. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guo, Y. Hormone Treatments in Studying Leaf Senescence. Methods Mol. Biol. 2018, 1744, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Wang, J.G.; Song, S.J.; Wang, Q.; Kang, H.; Zhang, Y.; Li, S. Precocious leaf senescence by functional loss of PROTEIN S-ACYL TRANSFERASE14 involves the NPR1-dependent salicylic acid signaling. Sci. Rep. 2016, 6, 20309. [Google Scholar] [CrossRef]
- Sarwat, M.; Naqvi, A.R.; Ahmad, P.; Ashraf, M.; Akram, N.A. Phytohormones and microRNAs as sensors and regulators of leaf senescence: Assigning macro roles to small molecules. Biotechnol. Adv. 2013, 31, 1153–1171. [Google Scholar] [CrossRef]
- Lin, C.H.; Shen, Y.A.; Hung, P.H.; Yu, Y.B.; Chen, Y.J. Epigallocathechin gallate, polyphenol present in green tea, inhibits stem-like characteristics and epithelial-mesenchymal transition in nasopharyngeal cancer cell lines. BMC Complement. Altern. Med. 2012, 12, 201. [Google Scholar] [CrossRef]
- Liang, Y.; Ma, W.; Lu, J.; Wu, Y. Comparison of chemical compositions of Ilex latifolia Thumb and Camellia sinensis L. Food Chem. 2001, 75, 339–343. [Google Scholar] [CrossRef]
- Hollman, P.C.; Feskens, E.J.; Katan, M.B. Tea flavonols in cardiovascular disease and cancer epidemiology. Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med. 1999, 220, 198–202. [Google Scholar] [CrossRef]
- Hodgson, J.M.; Croft, K.D.; Woodman, R.J.; Puddey, I.B.; Fuchs, D.; Draijer, R.; Lukoshkova, E.; Head, G.A. Black tea lowers the rate of blood pressure variation: A randomized controlled trial. Am. J. Clin. Nutr. 2013, 97, 943–950. [Google Scholar] [CrossRef]
- Auvichayapat, P.; Prapochanung, M.; Tunkamnerdthai, O.; Sripanidkulchai, B.O.; Auvichayapat, N.; Thinkhamrop, B.; Kunhasura, S.; Wongpratoom, S.; Sinawat, S.; Hongprapas, P. Effectiveness of green tea on weight reduction in obese Thais: A randomized, controlled trial. Physiol. Behav. 2008, 93, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.F.; Yong-Quan, X.U.; Yuan, H.B.; Shu-Ping, Y.U.; Wei, K.K.; Chen, J.X.; Wang, F.; Rong-Mei, W.U. Dynamic Change of Main Biochemical Components of Premium Green Tea Fresh Leaves during Spreading. J. Tea Sci. 2009, 29, 102–110. [Google Scholar]
- Yin, J.F.; Min, H.; Yong-Quan, X.U.; Yuan, H.B.; Wei, K.K. Effects of Spreading Environment on Tea Polyphenols and Catechin Components in Tea fresh Leaves of High Quality Green Tea. J. Tea Sci. 2008, 28, 22–27. [Google Scholar]
- Zhang, Y.G.; Chen, L.; Chen, Q.B.; Wang, X.P.; Wang, Z.K.; Ling-Sheng, W.U. Flavor Formation of White Tea during Natural Withering. Acta Tea Sin. 2016, 57, 80–84. [Google Scholar]
- Nobre, A.C.; Rao, A.; Owen, G.N. L-theanine, a natural constituent in tea, and its effect on mental state. Asia Pac. J. Clin. Nutr. 2008, 17 (Suppl. S1), 167–168. [Google Scholar]
- Guo, X.; Song, C.; Ho, C.T.; Wan, X. Contribution of l-theanine to the formation of 2,5-dimethylpyrazine, a key roasted peanutty flavor in Oolong tea during manufacturing processes. Food Chem. 2018, 263, 18–28. [Google Scholar] [CrossRef]
- Tsushida, T.; Takeo, T. An enzyme hydrolyzing L-theanine in tea leaves. J. Agric. Chem. Soc. Jpn. 1985, 49, 2913–2917. [Google Scholar] [CrossRef][Green Version]
- Tambasco-Studart, M.; Tews, I.; Amrhein, N.; Fitzpatrick, T.B. Functional analysis of PDX2 from Arabidopsis, a glutaminase involved in vitamin B6 biosynthesis. Plant Physiol. 2007, 144, 915–925. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef]
- Wu, J.; Hettenhausen, C.; Meldau, S.; Baldwin, I.T. Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 2007, 19, 1096–1122. [Google Scholar] [CrossRef]
- Tai, Y.; Wei, C.; Yang, H.; Zhang, L.; Chen, Q.; Deng, W.; Wei, S.; Zhang, J.; Fang, C.; Ho, C.; et al. Transcriptomic and phytochemical analysis of the biosynthesis of characteristic constituents in tea (Camellia sinensis) compared with oil tea (Camellia oleifera). BMC Plant Biol. 2015, 15, 190. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gong, L.H.; Chen, C.J.; Han, H.B.; Li, H.H. Column-chromatographic extraction and separation of polyphenols, caffeine and theanine from green tea. J. Crohns Colitis 2012, 131, 1539–1545. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, P.; Su, H.; Zhao, S.; Jin, R.; Cheng, H.; Xu, A.; Lai, W.; Yin, X.; Wang, Y. Transcriptome and Phytochemical Analysis Reveals the Alteration of Plant Hormones, Characteristic Metabolites, and Related Gene Expression in Tea (Camellia sinensis L.) Leaves During Withering. Plants 2020, 9, 204. https://doi.org/10.3390/plants9020204
Xu P, Su H, Zhao S, Jin R, Cheng H, Xu A, Lai W, Yin X, Wang Y. Transcriptome and Phytochemical Analysis Reveals the Alteration of Plant Hormones, Characteristic Metabolites, and Related Gene Expression in Tea (Camellia sinensis L.) Leaves During Withering. Plants. 2020; 9(2):204. https://doi.org/10.3390/plants9020204
Chicago/Turabian StyleXu, Ping, Hui Su, Shiqi Zhao, Rong Jin, Haiyan Cheng, Anan Xu, Wanyi Lai, Xueren Yin, and Yuefei Wang. 2020. "Transcriptome and Phytochemical Analysis Reveals the Alteration of Plant Hormones, Characteristic Metabolites, and Related Gene Expression in Tea (Camellia sinensis L.) Leaves During Withering" Plants 9, no. 2: 204. https://doi.org/10.3390/plants9020204
APA StyleXu, P., Su, H., Zhao, S., Jin, R., Cheng, H., Xu, A., Lai, W., Yin, X., & Wang, Y. (2020). Transcriptome and Phytochemical Analysis Reveals the Alteration of Plant Hormones, Characteristic Metabolites, and Related Gene Expression in Tea (Camellia sinensis L.) Leaves During Withering. Plants, 9(2), 204. https://doi.org/10.3390/plants9020204



