Molecular Characterization of the Dwarf53 Gene Homolog in Dasypyrum Villosum
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Iwata, N. Linkage studies in rice (Oryza sativa L.). Linkage groups for six genes newly described. Jpn. J. Breed. 1977, 27, 250–251. [Google Scholar]
- Wei, L.-R.; Xu, J.-C.; Li, X.-B.; Qian, Q.; Zhu, L.-H. Genetic Analysis and Mapping of the Dominant Dwarfing Gene D-53 in Rice. J. Integr. Plant Biol. 2006, 48, 447–452. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef]
- Ruyter-Spira, C.; Al-Babili, S.; Van der Krol, S.; Bouwmeester, H. The biology of strigolactones. Trends Plant Sci. 2013, 18, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Navabi, A.; Iqbal, M.; Strenzke, K.; Spaner, D. The relationship between lodging and plant height in a diverse wheat population. Can. J. Plant Sci. 2006, 86, 723–726. [Google Scholar] [CrossRef]
- Xie, Q.; Mayes, S.; Sparkes, D.L. Optimizing tiller production and survival for grain yield improvement in a bread wheat × spelt mapping population. Ann. Bot. 2016, 117, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Palta, J.; Fillery, I. Root characteristics of vigorous wheat improve early nitrogen uptake. Aust. J. Agric. Res. 2006, 57, 1097–1107. [Google Scholar] [CrossRef]
- Qin, L.; Zhao, J.; Li, T.; Hou, J.; Zhang, X.; Hao, C. TaGW2, a Good Reflection of Wheat Polyploidization and Evolution. Front. Plant Sci. 2017, 8, 318. [Google Scholar] [CrossRef]
- Li, S.; Wang, J.; Wang, K.; Chen, J.; Wang, K.; Du, L.; Ni, Z.; Lin, Z.; Ye, X. Development of PCR markers specific to Dasypyrum villosum genome based on transcriptome data and their application in breeding Triticum aestivum-D. villosum#4 alien chromosome lines. BMC Genomics 2019, 20, 289. [Google Scholar]
- Tajima, F.; Nei, M. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1984, 1, 269–285. [Google Scholar] [PubMed]
- URGI BLAST. Available online: https://urgi.versailles.inra.fr/blast/ (accessed on 22 December 2019).
- Grądzielewska, A. The genus Dasypyrum—part 2. Dasypyrum villosum—a wild species used in wheat improvement. Euphytica 2006, 152, 441–454. [Google Scholar]
- Sears, E.R. Addition of the Genome of Haynaldia villosa to Triticum aestivum. Am. J. Bot. 1953, 40, 168–174. [Google Scholar] [CrossRef]
- Bie, T.; Zhao, R.; Zhu, S.; Chen, S.; Cen, B.; Zhang, B.; Gao, D.; Jiang, Z.; Chen, T.; Wang, L.; et al. Development and characterization of marker MBH1 simultaneously tagging genes Pm21 and PmV conferring resistance to powdery mildew in wheat. Mol. Breed. 2015, 35, 189. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Q.; Wang, X.; Wang, H.; Lang, S.; Wang, Y.; Wang, S.; Chen, P.; Liu, D. Development and characterization of a Triticum aestivum-Haynaldia villosa translocation line T4VS⋅4DL conferring resistance to wheat spindle streak mosaic virus. Euphytica 2005, 145, 317–320. [Google Scholar] [CrossRef]
- Zhang, R.; Fan, Y.; Kong, L.; Wang, Z.; Wu, J.; Xing, L.; Cao, A.; Feng, Y. Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor. Appl. Genet. 2018, 131, 2613–2620. [Google Scholar] [CrossRef]
- Zhang, R.; Sun, B.; Chen, J.; Cao, A.; Xing, L.; Feng, Y.; Lan, C.; Chen, P. Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 2016, 129, 1975–1984. [Google Scholar] [CrossRef]
- Qi, L.L.; Pumphrey, M.O.; Friebe, B.; Zhang, P.; Qian, C.; Bowden, R.L.; Rouse, M.N.; Jin, Y.; Gill, B.S. A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor. Appl. Genet. 2011, 123, 159–167. [Google Scholar] [CrossRef]
- Zhang, R.; Mingyi, Z.; Xiue, W.; Peidu, C. Introduction of chromosome segment carrying the seed storage protein genes from chromosome 1V of Dasypyrum villosum showed positive effect on bread-making quality of common wheat. Theor. Appl. Genet. 2014, 127, 523–533. [Google Scholar]
- Zhang, R.; Hou, F.; Feng, Y.; Zhang, W.; Zhang, M.; Chen, P. Characterization of a Triticum aestivum-Dasypyrum villosum T2VS·2DL translocation line expressing a longer spike and more kernels traits. Theor. Appl. Genet. 2015, 128, 2415–2425. [Google Scholar] [CrossRef]
- Djanaguiraman, M.; Prasad, P.V.V.; Kumari, J.; Sehgal, S.K.; Friebe, B.; Djalovic, I.; Chen, Y.; Siddique, K.H.M.; Gill, B.S. Alien chromosome segment from Aegilops speltoides and Dasypyrum villosum increases drought tolerance in wheat via profuse and deep root system. BMC Plant Biol. 2019, 19, 242. [Google Scholar] [CrossRef] [PubMed]
- Doyle, P.J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; NATO ASI Series; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. ISBN 978-3-642-83964-1. [Google Scholar]
- BLAST–WHEAT URGI. Available online: https://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST (accessed on 19 December 2019).
- Primer designing tool. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 19 December 2019).
- SantaLucia, J. A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 1998, 95, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics–FastQC A Quality Control tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 19 December 2019).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- GeneStudio.com | Free molecular biology software. Available online: http://genestudio.com/ (accessed on 19 December 2019).
- GitHub–MikhailBazhenov/Transform-RTF-to-FASTA. Available online: https://github.com/MikhailBazhenov/Transform-RTF-to-FASTA (accessed on 19 December 2019).
- Nicholas, K.B.; Nikolas, H.B. Jr. GeneDoc: A tool for editing and annotating multiple sequence alignments. 1997. Available online: https://genedoc.software.informer.com/download/ (accessed on 19 December 2019).
- Zaharia, M.; Bolosky, W.J.; Curtis, K.; Fox, A.; Patterson, D.; Shenker, S.; Stoica, I.; Karp, R.M.; Sittler, T. Faster and More Accurate Sequence Alignment with SNAP. arXiv 2011, arXiv:1111.5572. Available online: https://arxiv.org/abs/1111.5572 (accessed on 19 December 2019).
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. Available online: https://arxiv.org/abs/1207.3907 (accessed on 19 December 2019).
- GitHub–samtools/bcftools. Available online: https://github.com/samtools/bcftools (accessed on 19 December 2019).
- PHYLIP Home Page. Available online: http://evolution.genetics.washington.edu/phylip.html (accessed on 19 December 2019).
- Stanke, M. The AUGUSTUS gene prediction tool. Available online: http://bioinf.uni-greifswald.de/augustus/ (accessed on 19 December 2019).
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
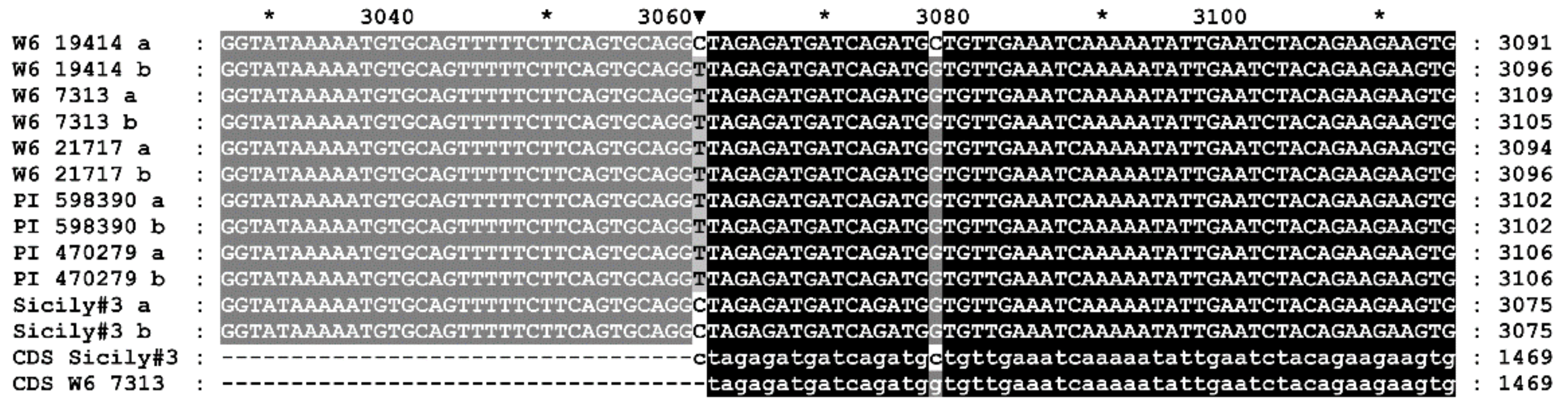
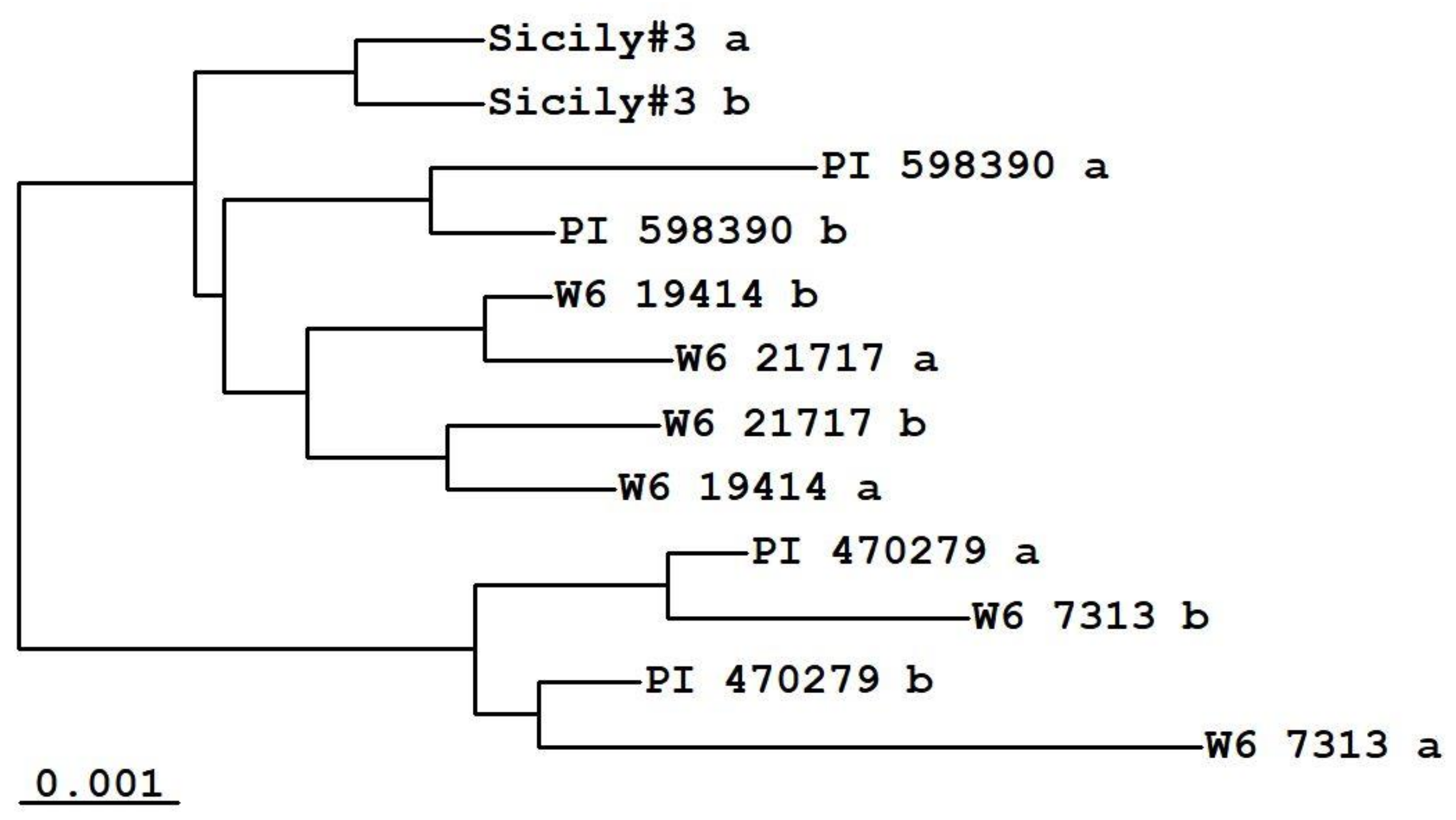
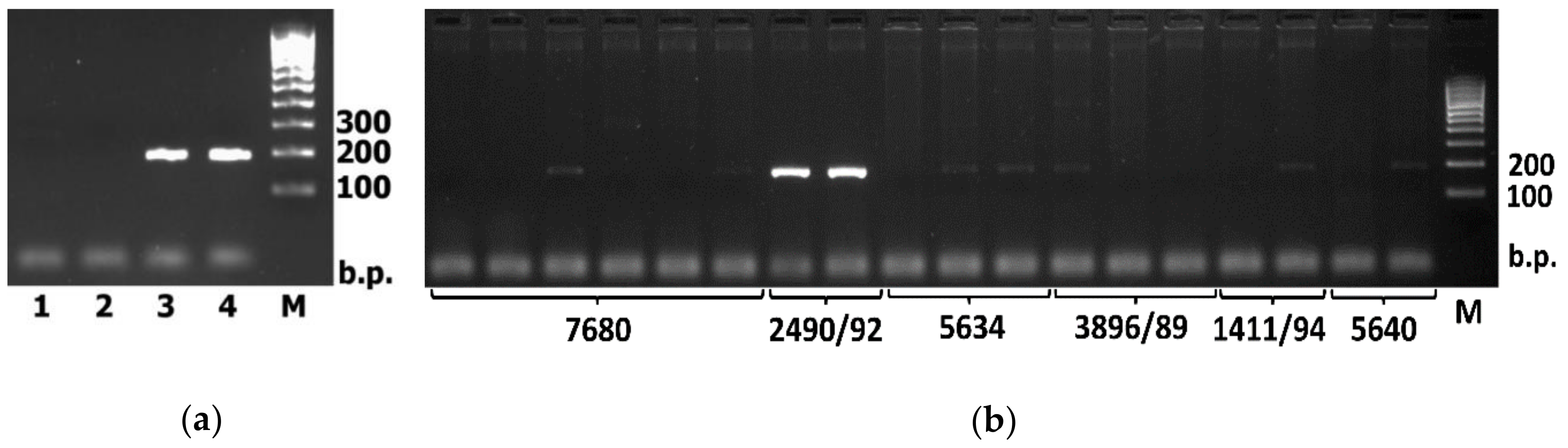
| Accession | Place of Collection | Source |
|---|---|---|
| W6 19414 | Bulgaria | W6 |
| W6 7313 | Greece | W6 |
| W6 21717 | Crimea | W6 |
| PI 598390 | former USSR | W6 |
| PI 470279 | Turkey | W6 |
| Sicily#3 | Italy, Sicily | AJL |
| Primer Pairs, Sequence Direction is 5’ -> 3’ | Tm | Expected Amplicon Size for Wheat Subgenomes | ||
|---|---|---|---|---|
| A | B | D | ||
| D53-F1: CGTGGTTTATAAGCAAGCAATCCA D53-R1: GGGGAACTTGGACAGGAAGG | 60 | 1239 | 1281 | 1263 |
| D53-F2: TACCTCACCTTCCTGTCCAAGTT D53-R2: TCTTACCCTTTTCATCAAGCTGT | 58 | 1619 | 1459 | 1248 |
| D53-F3: ACTTACTGCATCTGGGTTGATAA D53-R3: ATAGCTTCACACCTTGATTGCAT | 58 | 810 | 1013 | 811 |
| D53-F4: CGGTGTCAACAGTGCAATGAT D53-R4: GCCTCCTGAAGCTGGTGAAT | 60 | 959 | 958 | 959 |
| D53-F5: ATTCACCAGCTTCAGGAGGC D53-R5: ACTACCGTGGACTAGCTACC | 60 | 1403 | 1397 | 1398 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bazhenov, M.; Chernook, A.; Kroupin, P.; Karlov, G.; Divashuk, M. Molecular Characterization of the Dwarf53 Gene Homolog in Dasypyrum Villosum. Plants 2020, 9, 186. https://doi.org/10.3390/plants9020186
Bazhenov M, Chernook A, Kroupin P, Karlov G, Divashuk M. Molecular Characterization of the Dwarf53 Gene Homolog in Dasypyrum Villosum. Plants. 2020; 9(2):186. https://doi.org/10.3390/plants9020186
Chicago/Turabian StyleBazhenov, Mikhail, Anastasiya Chernook, Pavel Kroupin, Gennady Karlov, and Mikhail Divashuk. 2020. "Molecular Characterization of the Dwarf53 Gene Homolog in Dasypyrum Villosum" Plants 9, no. 2: 186. https://doi.org/10.3390/plants9020186
APA StyleBazhenov, M., Chernook, A., Kroupin, P., Karlov, G., & Divashuk, M. (2020). Molecular Characterization of the Dwarf53 Gene Homolog in Dasypyrum Villosum. Plants, 9(2), 186. https://doi.org/10.3390/plants9020186





