Biochemical and Molecular Characterization of PvNTD2, a Nucleotidase Highly Expressed in Nodules from Phaseolus vulgaris
Abstract
1. Introduction
2. Results
2.1. Identification, Cloning, and Overexpression of PvNTD2
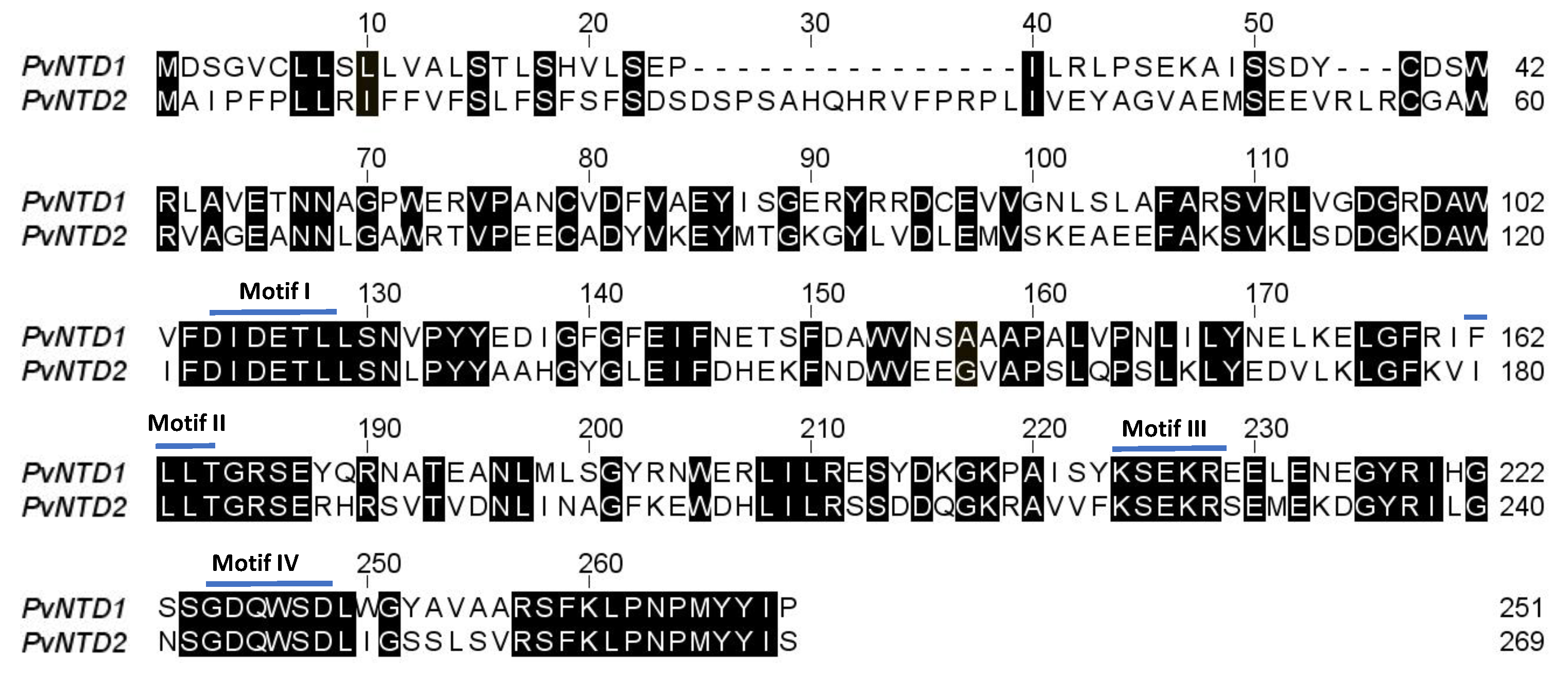
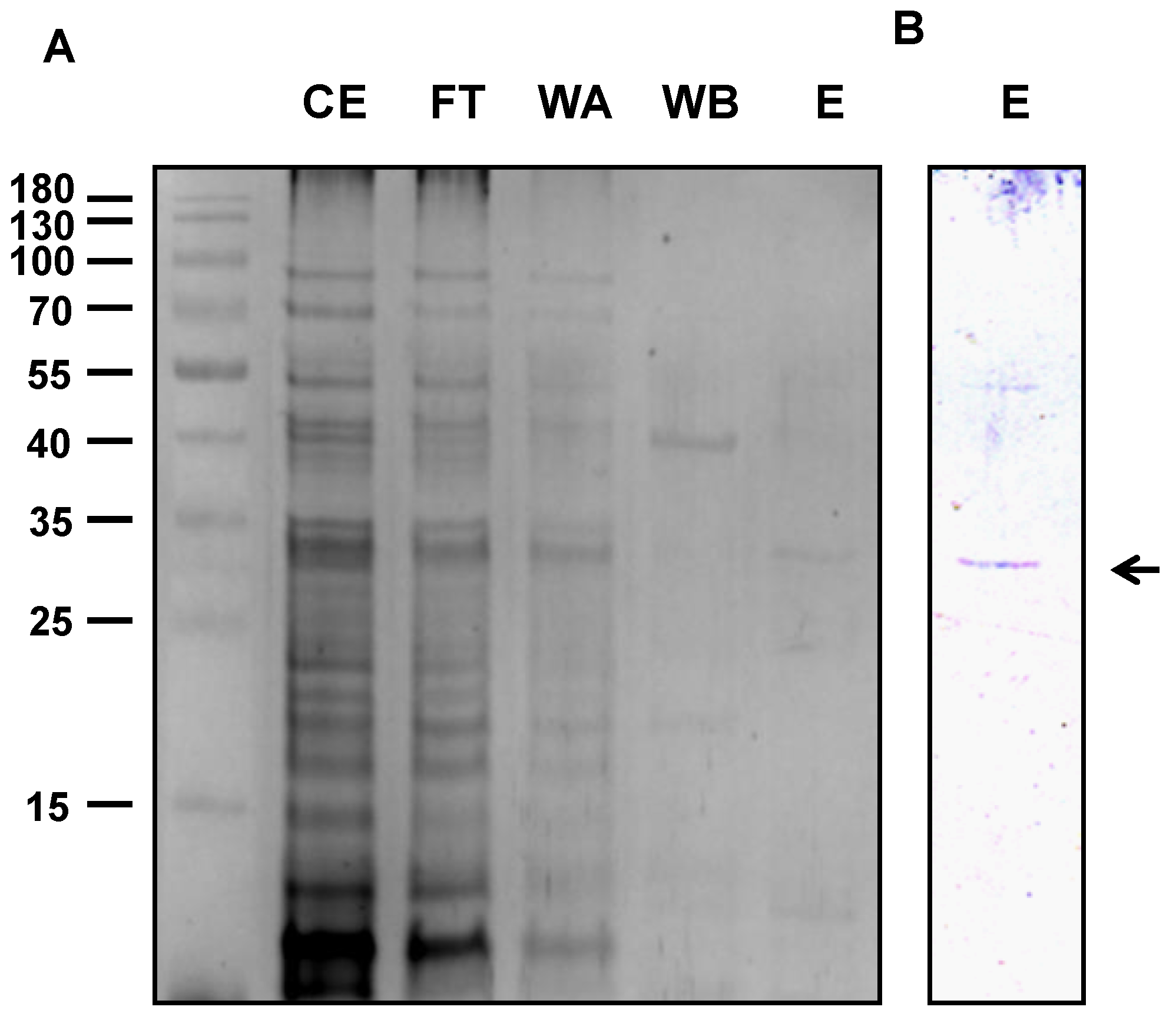
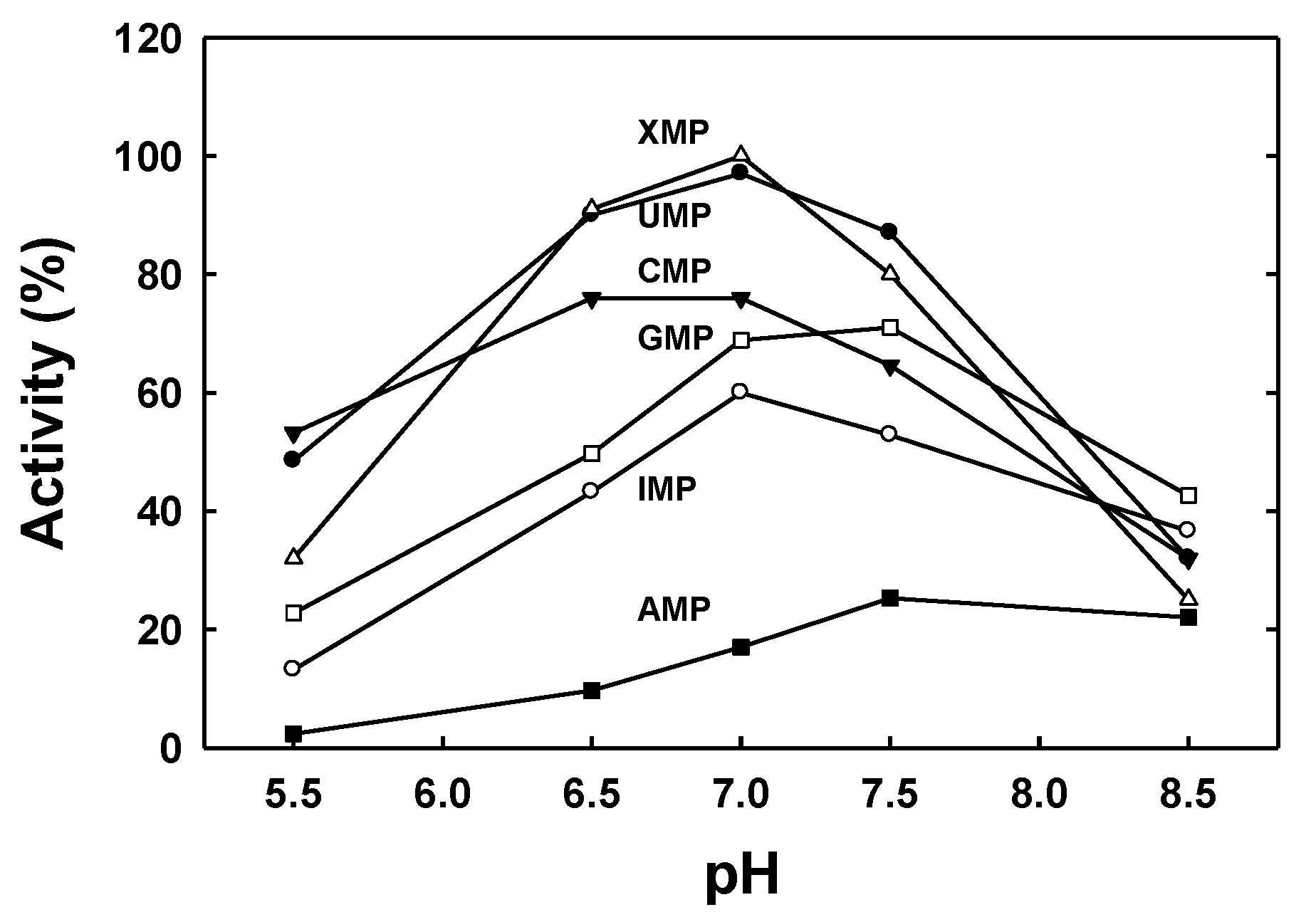
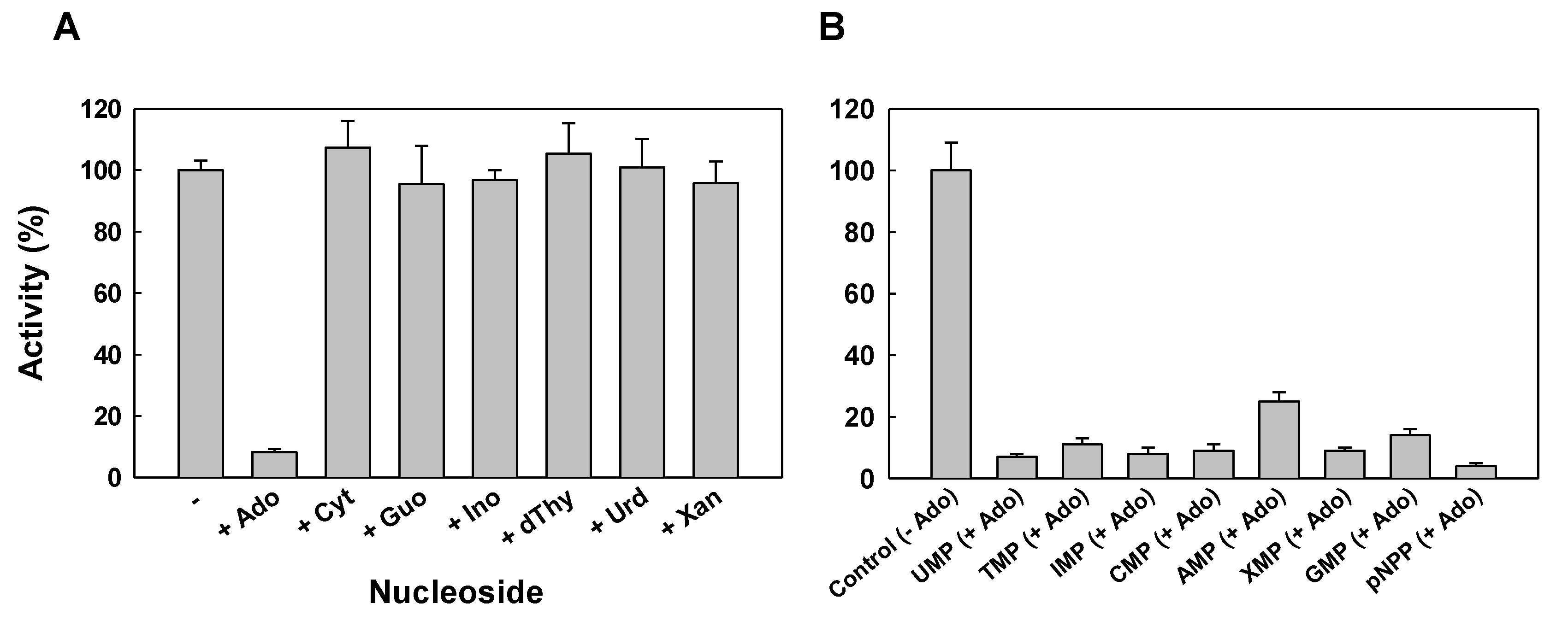
2.2. Protein Characterization
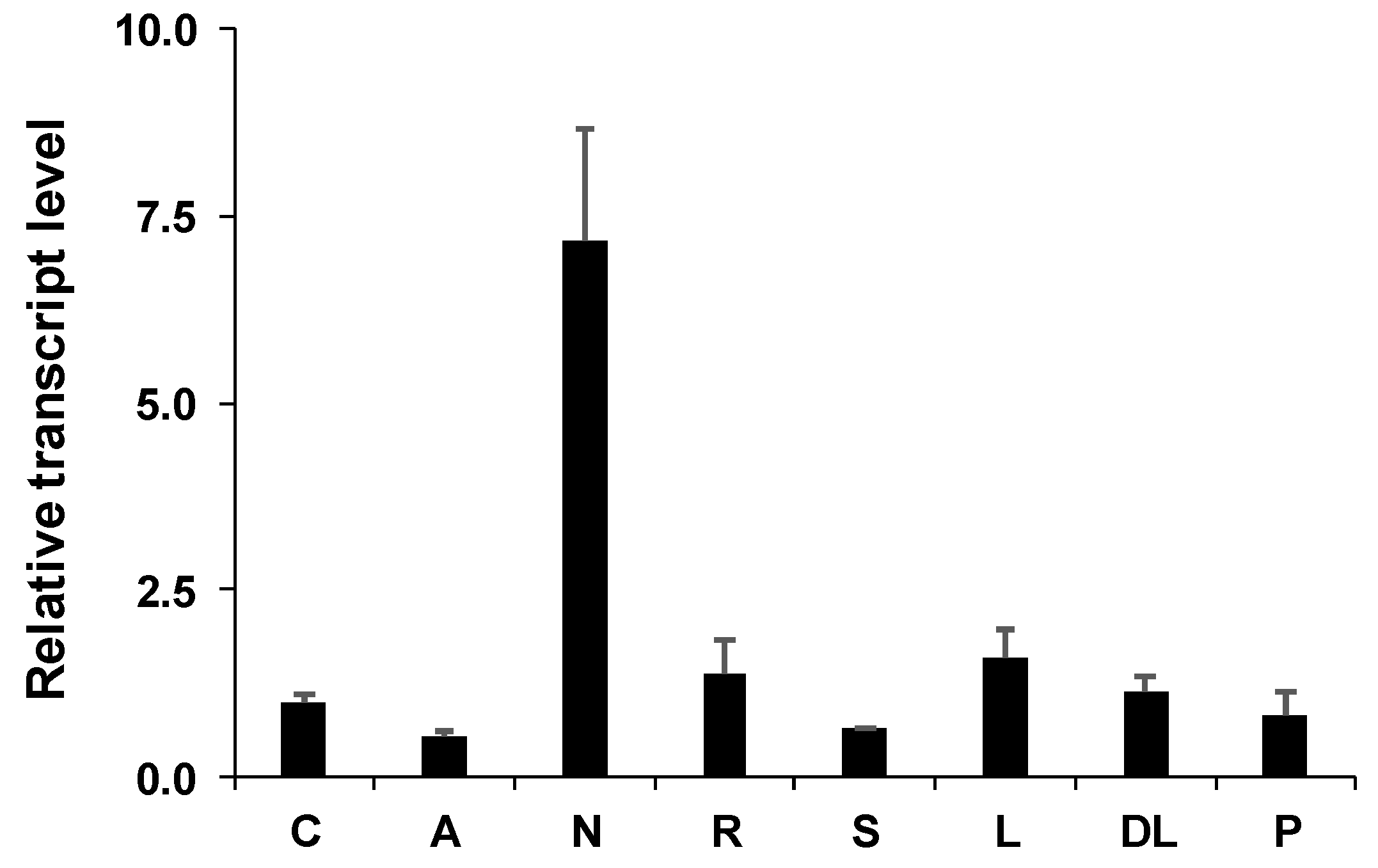
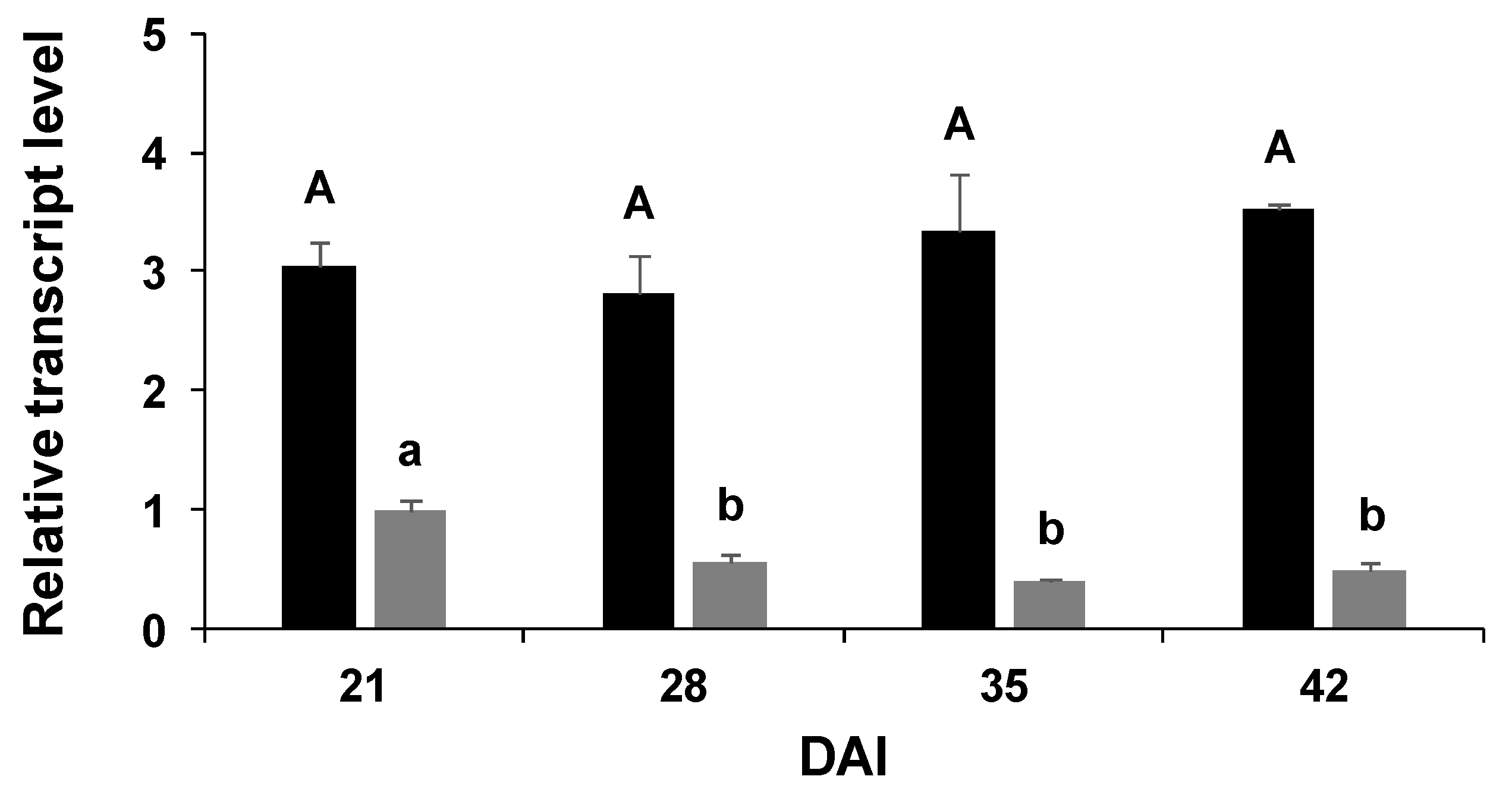
2.3. Gene Expression
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Condition
4.2. Cloning into the pET30b(+) Expression Vector
4.3. Expression and Purification of the Recombinant Protein
4.4. Gel Electrophoresis, Western Blot Analysis, and Protein Stain
4.5. Enzymatic Activity
4.6. RNA Isolation and cDNA Synthesis
4.7. Real-Time PCR
4.8. Analytical Determination
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zrenner, R.; Stitt, M.; Sonnewald, U.; Boldt, R. Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 2006, 57, 805–836. [Google Scholar] [CrossRef] [PubMed]
- Quiles, F.A.; Raso, M.J.; Pineda, M.; Piedras, P. Ureide metabolism during seedling development in French bean (Phaseolus vulgaris). Physiol. Plant 2009, 135, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Lambert, R.; Cabello-Diaz, J.M.; Quiles, F.A.; Piedras, P. Identification of nucleases related to nutrient mobilization in senescing cotyledons from French bean. Acta Physiol. Plant 2016, 38, 266. [Google Scholar] [CrossRef]
- Lambert, R.; Quiles, F.A.; Galvez-Valdivieso, G.; Piedras, P. Nucleases activities during French bean leaf aging and dark-induced senescence. J. Plant Physiol. 2017, 218, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Brychkova, G.; Alikulov, Z.; Fluhr, R.; Sagi, M. A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J. 2008, 54, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Irani, S.; Todd, C.D. Ureide metabolism under abiotic stress in Arabidopsis thaliana. J. Plant Physiol. 2016, 199, 87–95. [Google Scholar] [CrossRef]
- Irani, S.; Todd, C.D. Exogenous allantoin increases Arabidopsis seedlings tolerance to NaCl stress and regulates expression of oxidative stress response genes. J. Plant Physiol. 2018, 221, 43–50. [Google Scholar] [CrossRef]
- Watanabe, S.; Matsumoto, M.; Hakomori, Y.; Takagi, H.; Shimada, H.; Sakamoto, A. The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ. 2014, 37, 1022–1036. [Google Scholar] [CrossRef]
- Quiles, F.A.; Galvez-Valdivieso, G.; Guerrero-Casado, J.; Pineda, M.; Piedras, P. Relationship between ureidic/amidic metabolism and antioxidant enzymatic activities in legume seedlings. Plant Physiol. Biochem. 2019, 138, 1–8. [Google Scholar] [CrossRef]
- Atkins, C.A.; Smith, P.M.C. Ureide synthesis in legume nodules. In Prokaryotic Nitrogen Fixation: A Model System for the Analysis of a Biological Process; Triplett, E.W., Ed.; Horizon Scientific Press: Norfolk, UK, 2000; pp. 559–587. [Google Scholar]
- Bogan, K.L.; Brenner, C. 5’-nucleotidases and their new roles in NAD(+) and phosphate metabolism. New J. Chem. 2010, 34, 845–853. [Google Scholar] [CrossRef]
- Cabello-Díaz, J.M.; Quiles, F.A.; Lambert, R.; Pineda, M.; Piedras, P. Identification of a novel phosphatase with high affinity for nucleotides monophosphate from common bean (Phaseolus vulgaris). Plant Physiol. Biochem. 2012, 53, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Cabello-Diaz, J.M.; Galvez-Valdivieso, G.; Caballo, C.; Lambert, R.; Quiles, F.A.; Pineda, M.; Piedras, P. Identification and characterization of a gene encoding for a nucleotidase from Phaseolus vulgaris. J. Plant Physiol. 2015, 185, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Duff, S.M.G.; Sarath, G.; Plaxton, W.C. The role of acid phosphatases in plant phosphorus metabolism. Physiol. Plant. 1994, 90, 791–800. [Google Scholar] [CrossRef]
- Penheiter, A.R.; Duff, S.M.G.; Sarath, G. Soybean root nodule acid phosphatase. Plant Physiol. 1997, 114, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Penheiter, A.R.; Klucas, R.V.; Sarath, G. Purification and characterization of a soybean root nodule phosphatase expressed in Pichia pastoris. Protein Expres. Purif. 1998, 14, 125–130. [Google Scholar] [CrossRef]
- Veljanovski, V.; Major, I.T.; Patton, J.J.; Bol, E.; Louvet, S.; Hawkins, B.J.; Constabel, C.P. Induction of acid phosphatase transcripts, protein and enzymatic activity by simulated herbivory of hybrid poplar. Phytochemistry 2010, 71, 619–626. [Google Scholar] [CrossRef]
- Liu, Y.L.; Ahn, J.E.; Datta, S.; Salzman, R.A.; Moon, J.; Huyghues-Despointes, B.; Pittendrigh, B.; Murdock, L.L.; Koiwa, H.; Zhu-Salzman, K. Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiol. 2005, 139, 1545–1556. [Google Scholar] [CrossRef]
- Gohla, A. Do metabolic HAD phosphatases moonlight as protein phosphatases? BBA-Mol. Cell Res. 2019, 1866, 153–166. [Google Scholar] [CrossRef]
- Seifried, A.; Schultz, J.; Gohla, A. Human HAD phosphatases: Structure, mechanism, and roles in health and disease. FEBS J. 2013, 280, 549–571. [Google Scholar] [CrossRef]
- Baldwin, J.C.; Karthikeyan, A.S.; Cao, A.; Raghothama, K.G. Biochemical and molecular analysis of LePS2;1: A phosphate starvation induced protein phosphatase gene from tomato. Planta 2008, 228, 273–280. [Google Scholar] [CrossRef]
- Liang, C.Y.; Chen, Z.J.; Yao, Z.F.; Tian, J.; Liao, H. Characterization of two putative protein phosphatase genes and their involvement in phosphorus efficiency in Phaseolus vulgaris. J. Integr. Plant Biol. 2012, 54, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.K.; Mehra, P.; Verma, L.; Bhadouria, J.; Giri, J. OsHAD1, a haloacid dehalogenase-like APase, enhances phosphate accumulation. Plant Physiol. 2017, 174, 2316–2332. [Google Scholar] [CrossRef] [PubMed]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 19 June 2019).
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Sonderby, C.K.; Sonderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Allen, K.N.; Dunaway-Mariano, D.; Aravind, L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 2006, 361, 1003–1034. [Google Scholar] [CrossRef]
- Kuznetsova, E.; Proudfoot, M.; Gonzalez, C.F.; Brown, G.; Omelchenko, M.V.; Borozan, I.; Carmel, L.; Wolf, Y.I.; Mori, H.; Savchenko, A.V.; et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J. Biol. Chem. 2006, 281, 36149–36161. [Google Scholar] [CrossRef]
- Stasolla, C.; Katahira, R.; Thorpe, T.A.; Ashihara, H. Purine and pyrimidine nucleotide metabolism in higher plants. J. Plant Physiol. 2003, 160, 1271–1295. [Google Scholar] [CrossRef]
- Lambert, R.; Quiles, F.A.; Cabello-Diaz, J.M.; Piedras, P. Purification and identification of a nuclease activity in embryo axes from French bean. Plant Sci. 2014, 224, 137–143. [Google Scholar] [CrossRef]
- Coleto, I.; Trenas, A.T.; Erban, A.; Kopka, J.; Pineda, M.; Alamillo, J.M. Functional specialization of one copy of glutamine phosphoribosyl pyrophosphate amidotransferase in ureide production from symbiotically fixed nitrogen in Phaseolus vulgaris. Plant Cell Environ. 2016, 39, 1767–1779. [Google Scholar] [CrossRef]
- Garcia, N.A.T.; Olivera, M.; Iribarne, C.; Lluch, C. Partial purification and characterization of a non-specific acid phosphatase in leaves and root nodules of Phaseolus vulgaris. Plant Physiol. Biochem. 2004, 42, 585–591. [Google Scholar] [CrossRef]
- Sulieman, S.; Tran, L.S.P. Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci. 2015, 239, 36–43. [Google Scholar] [CrossRef]
- Galvez-Valdivieso, G.; Alamillo, J.M.; Fernandez, J.; Pineda, M. Molecular characterization of PVAS3: An asparagine synthetase gene from common bean prevailing in developing organs. J. Plant Physiol. 2013, 170, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]


| Substrate | Km (mM) | Vmax (U/mg) | Vmax/Km |
|---|---|---|---|
| AMP | 0.011 | 1.46 | 132 |
| UMP | 0.039 | 4.00 | 103 |
| TMP | 0.063 | 4.08 | 64 |
| XMP | 0.095 | 4.22 | 44 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galvez-Valdivieso, G.; Delgado-Garcia, E.; Diaz-Baena, M.; Montaño, O.; Quiles, F.A.; Pineda, M.; Piedras, P. Biochemical and Molecular Characterization of PvNTD2, a Nucleotidase Highly Expressed in Nodules from Phaseolus vulgaris. Plants 2020, 9, 171. https://doi.org/10.3390/plants9020171
Galvez-Valdivieso G, Delgado-Garcia E, Diaz-Baena M, Montaño O, Quiles FA, Pineda M, Piedras P. Biochemical and Molecular Characterization of PvNTD2, a Nucleotidase Highly Expressed in Nodules from Phaseolus vulgaris. Plants. 2020; 9(2):171. https://doi.org/10.3390/plants9020171
Chicago/Turabian StyleGalvez-Valdivieso, Gregorio, Elena Delgado-Garcia, Mercedes Diaz-Baena, Oscar Montaño, Francisco A. Quiles, Manuel Pineda, and Pedro Piedras. 2020. "Biochemical and Molecular Characterization of PvNTD2, a Nucleotidase Highly Expressed in Nodules from Phaseolus vulgaris" Plants 9, no. 2: 171. https://doi.org/10.3390/plants9020171
APA StyleGalvez-Valdivieso, G., Delgado-Garcia, E., Diaz-Baena, M., Montaño, O., Quiles, F. A., Pineda, M., & Piedras, P. (2020). Biochemical and Molecular Characterization of PvNTD2, a Nucleotidase Highly Expressed in Nodules from Phaseolus vulgaris. Plants, 9(2), 171. https://doi.org/10.3390/plants9020171






