The Role of Symbiotic Microorganisms, Nutrient Uptake and Rhizosphere Bacterial Community in Response of Pea (Pisum sativum L.) Genotypes to Elevated Al Concentrations in Soil
Abstract
1. Introduction
2. Results
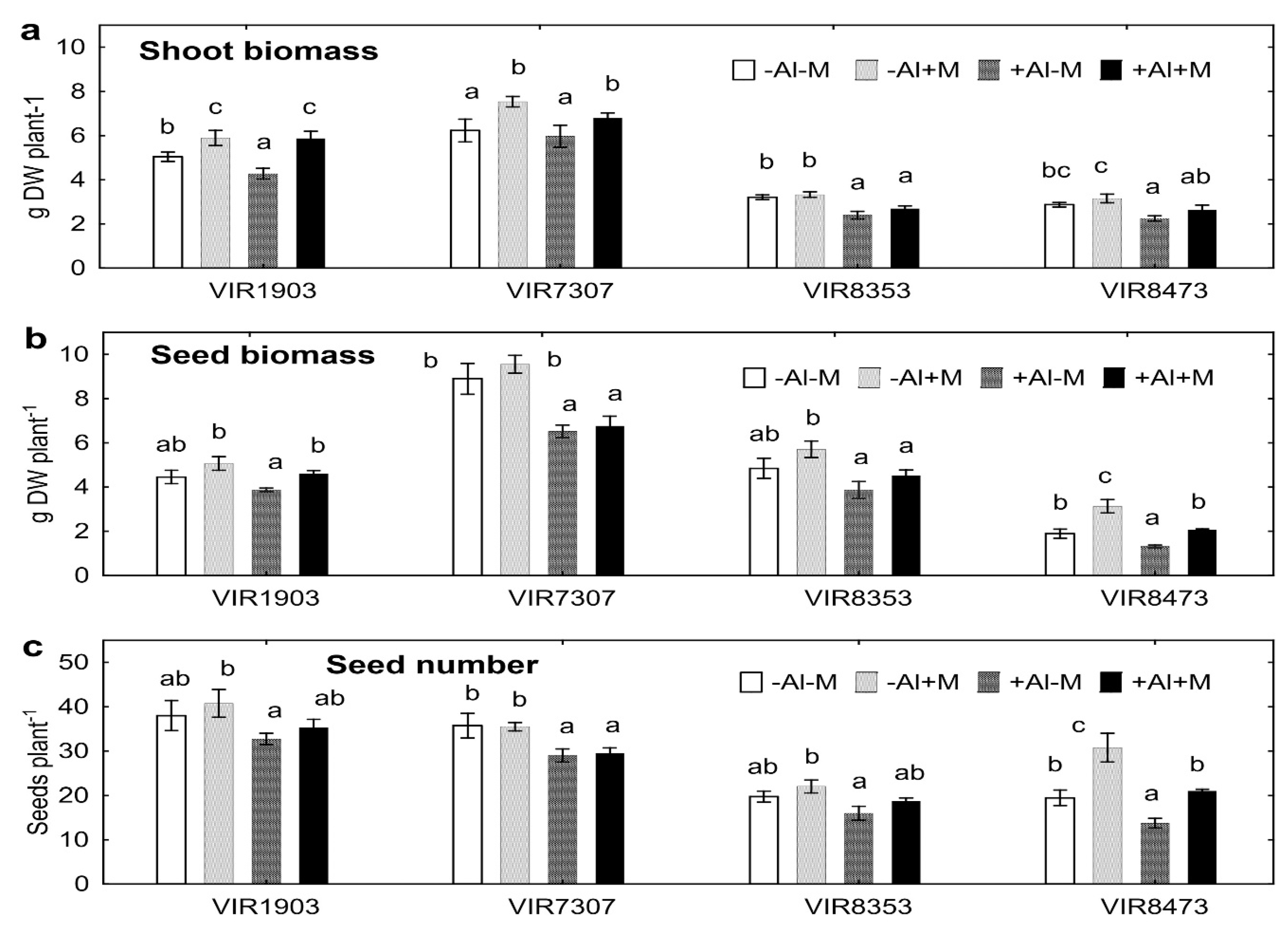
2.1. Plant Biomass
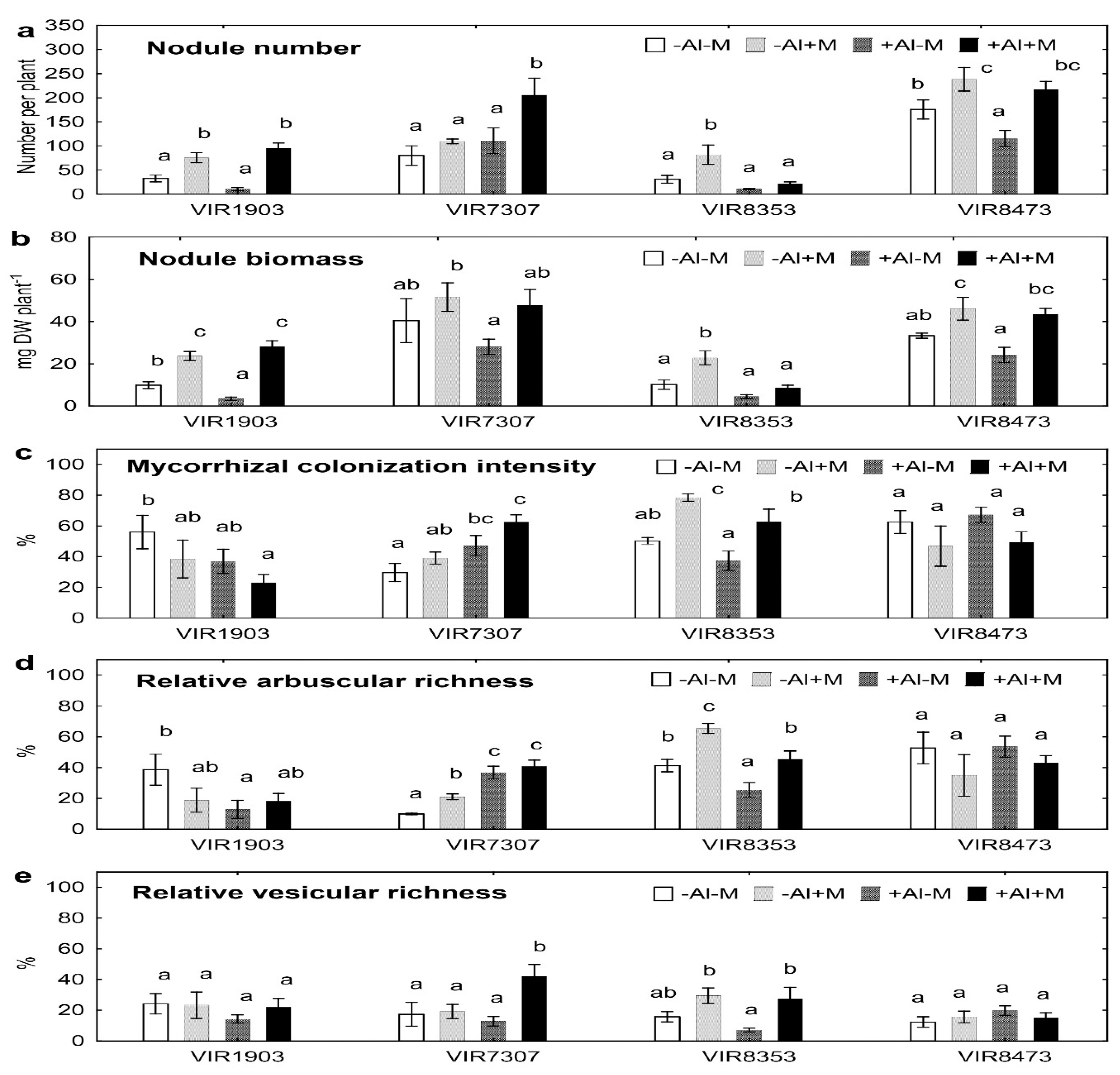
2.2. Symbiotic Structures
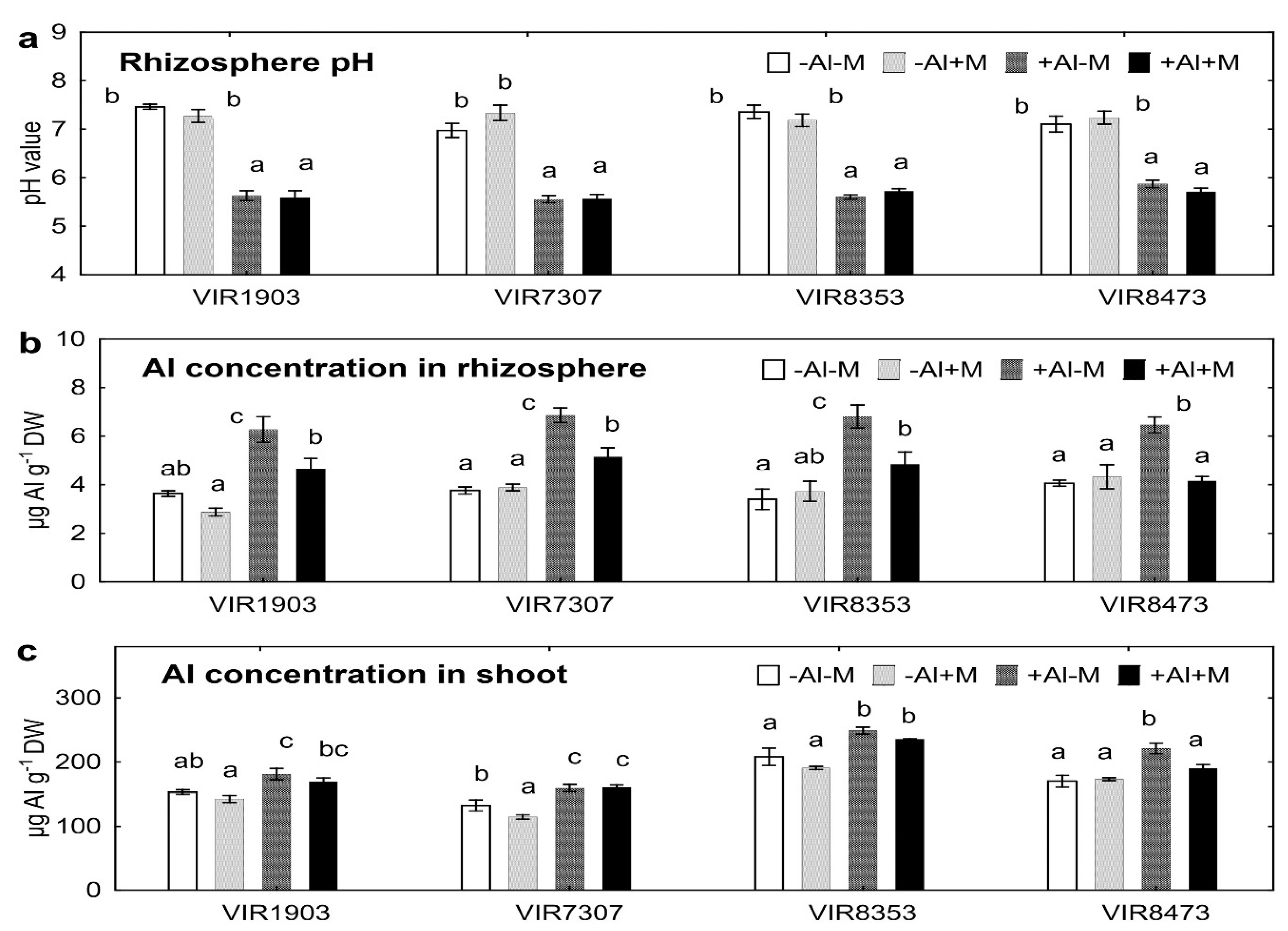
2.3. Rhizosphere pH and Al Concentrations
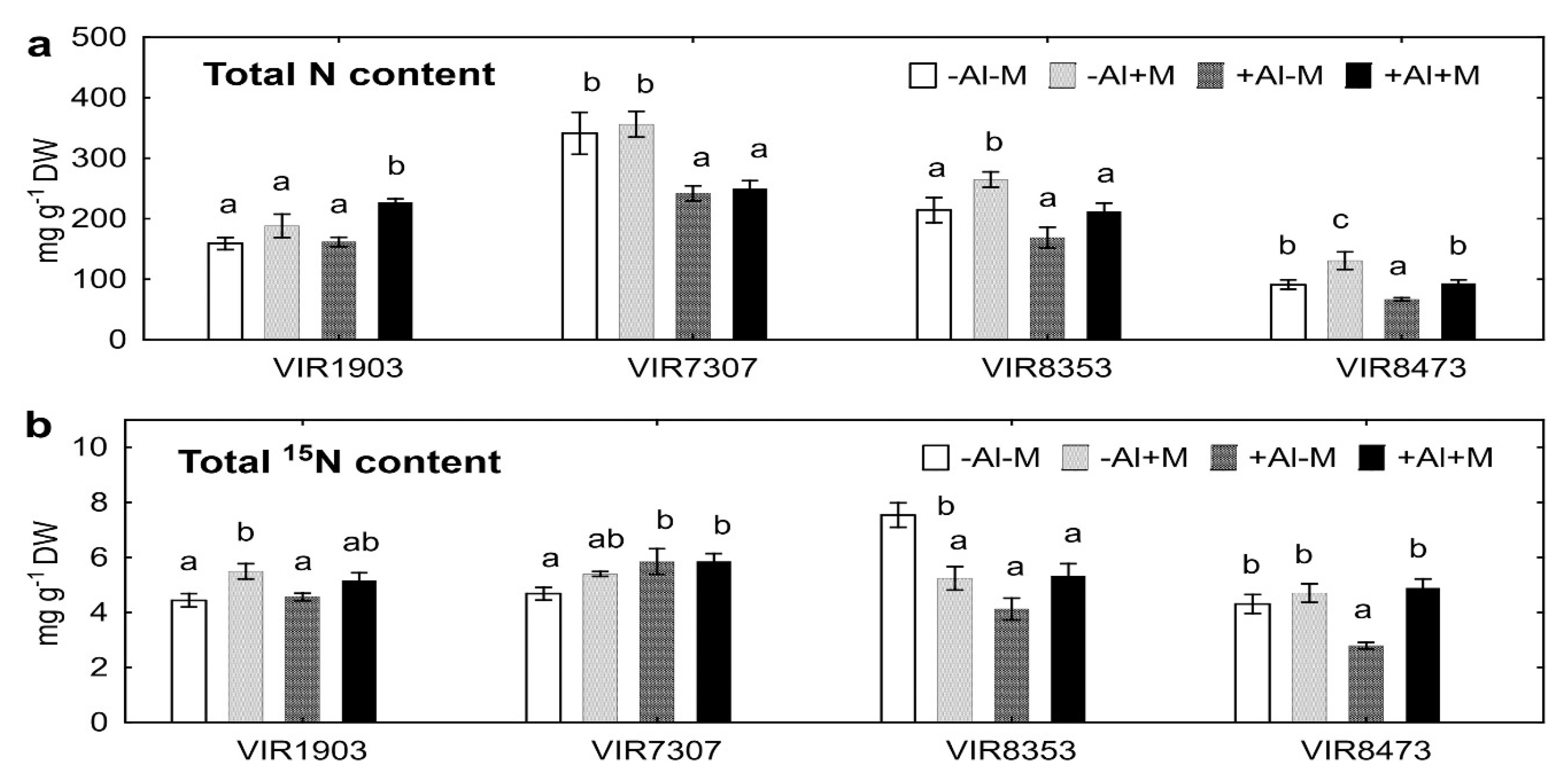
2.4. Nitrogen Uptake
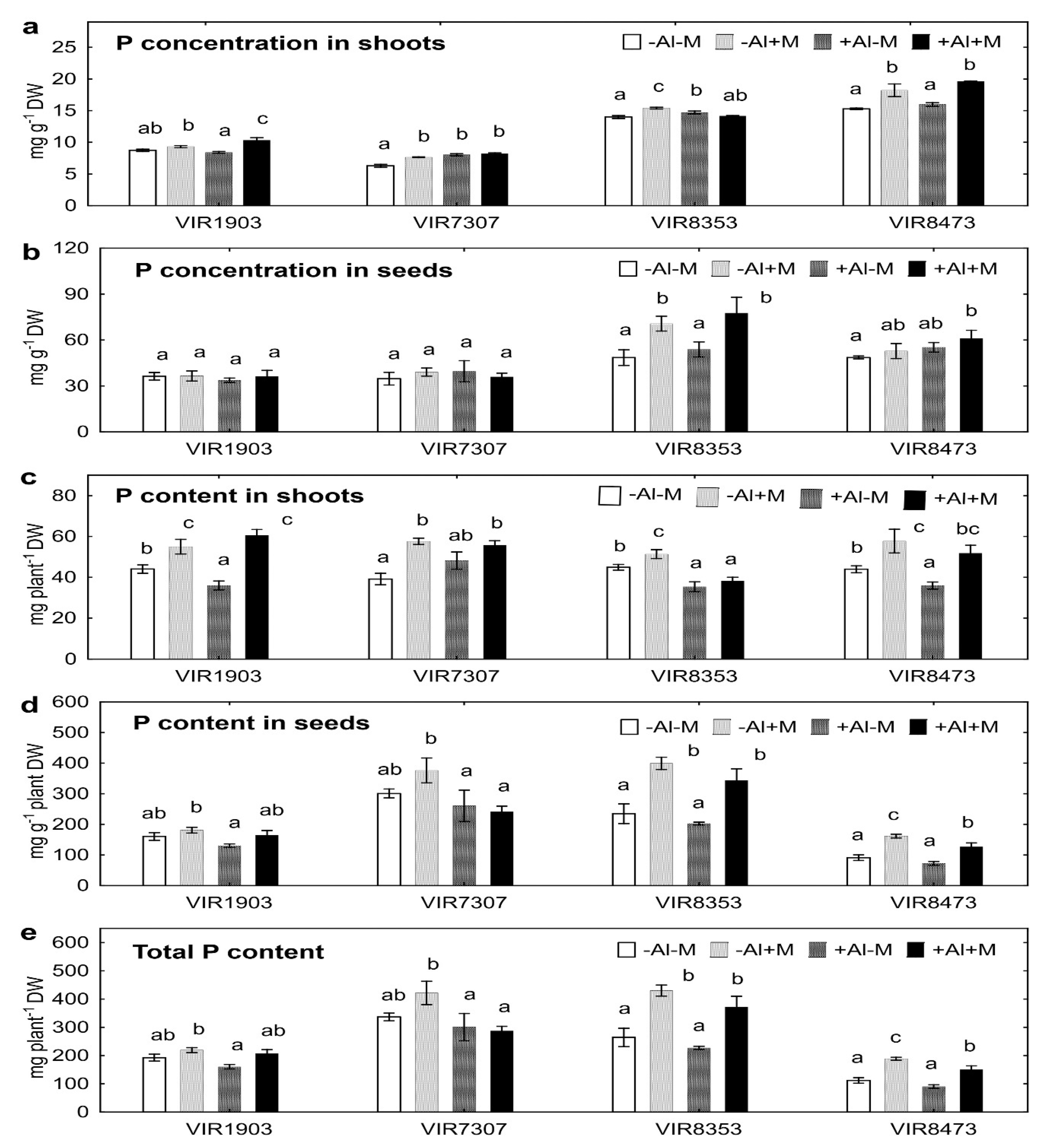
2.5. Phosphorus Uptake
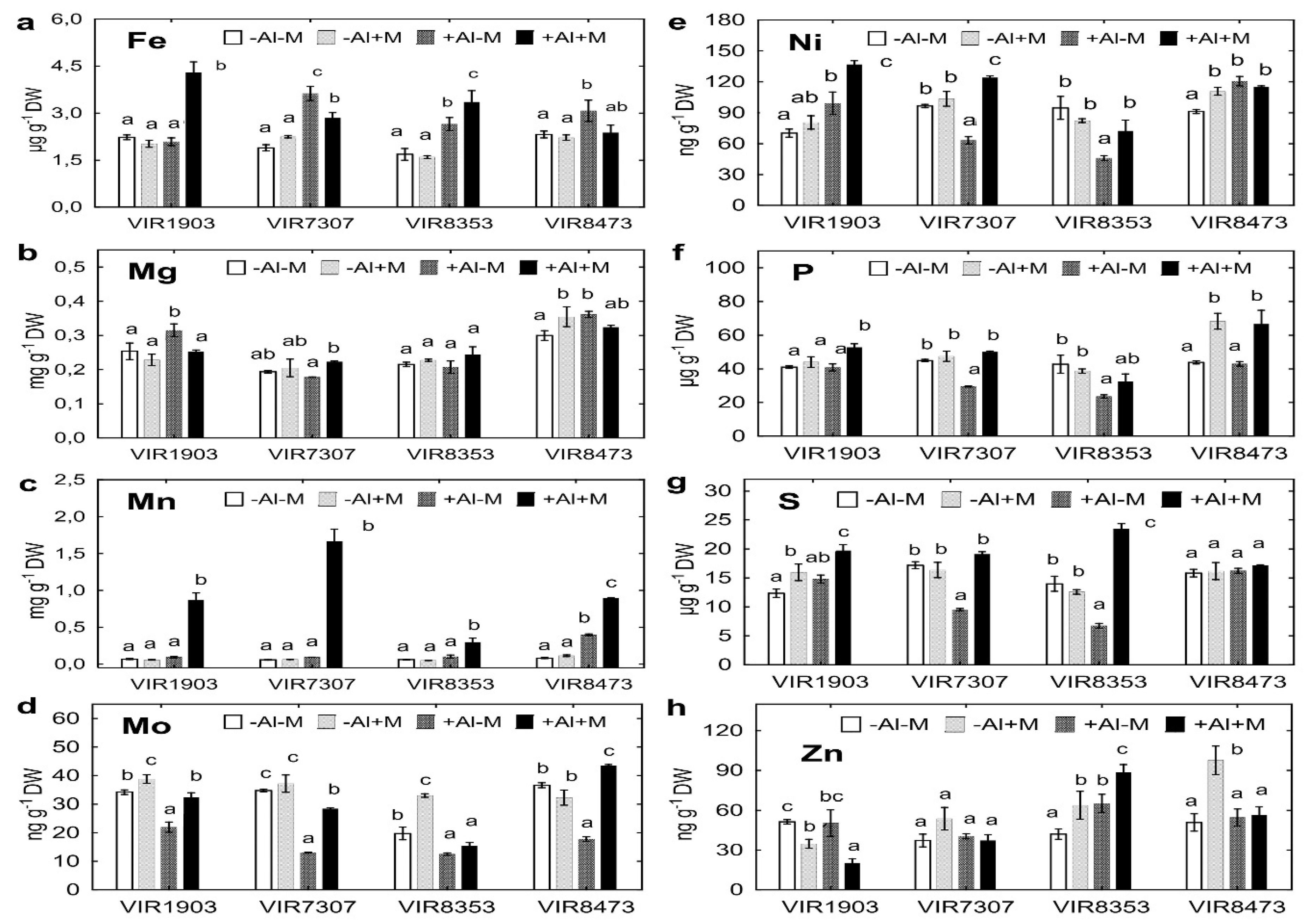
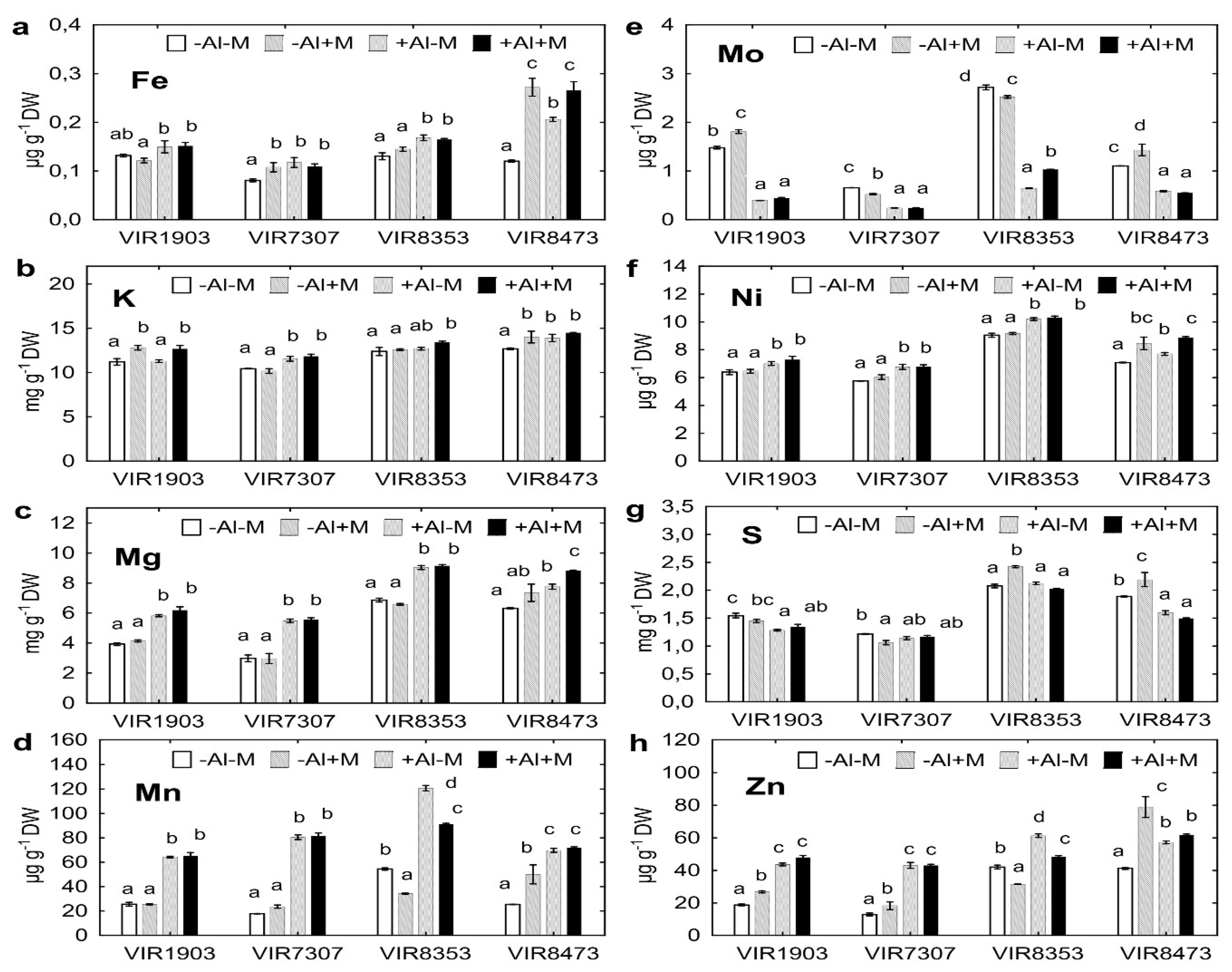
2.6. Concentration of Other Nutrients in the Rhizosphere
2.7. Uptake of Other Nutrients by Plants
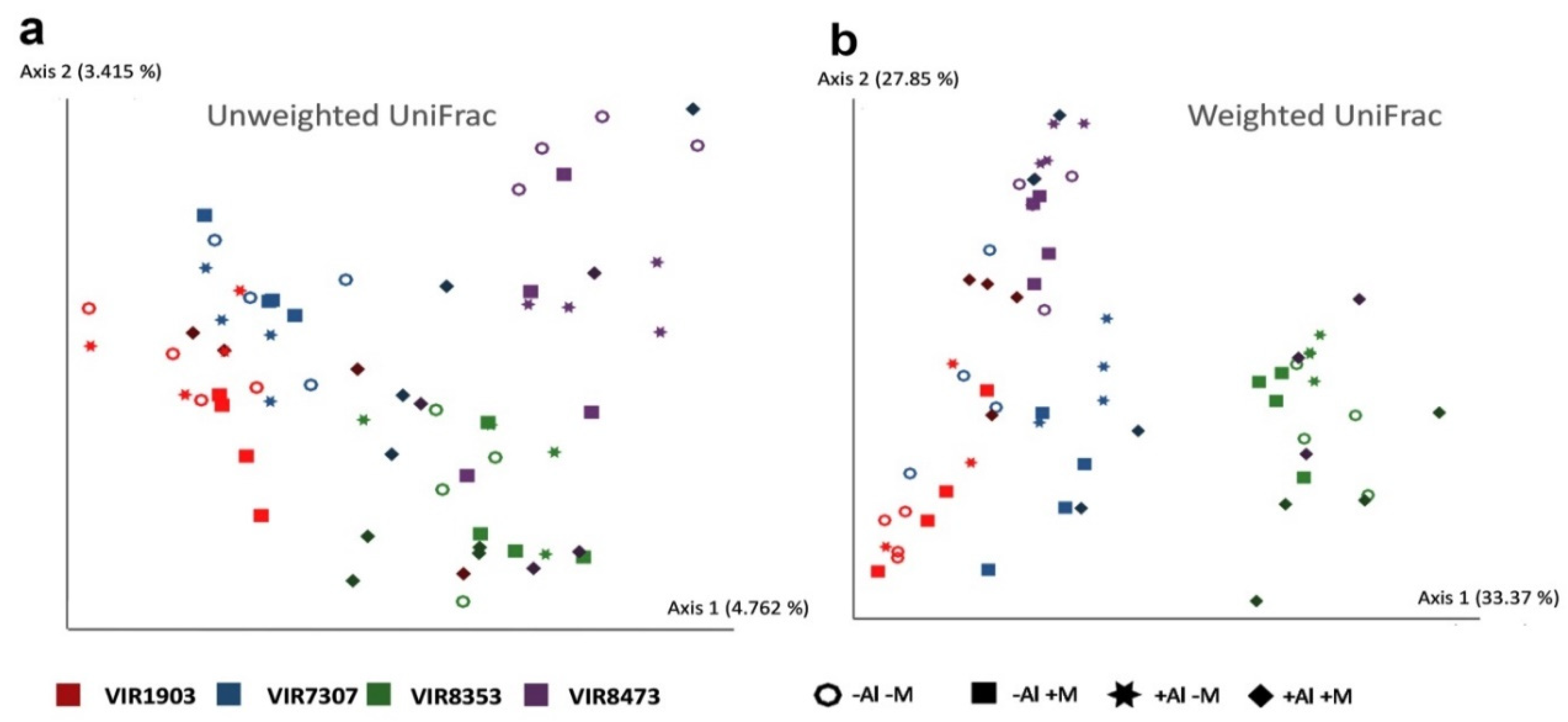
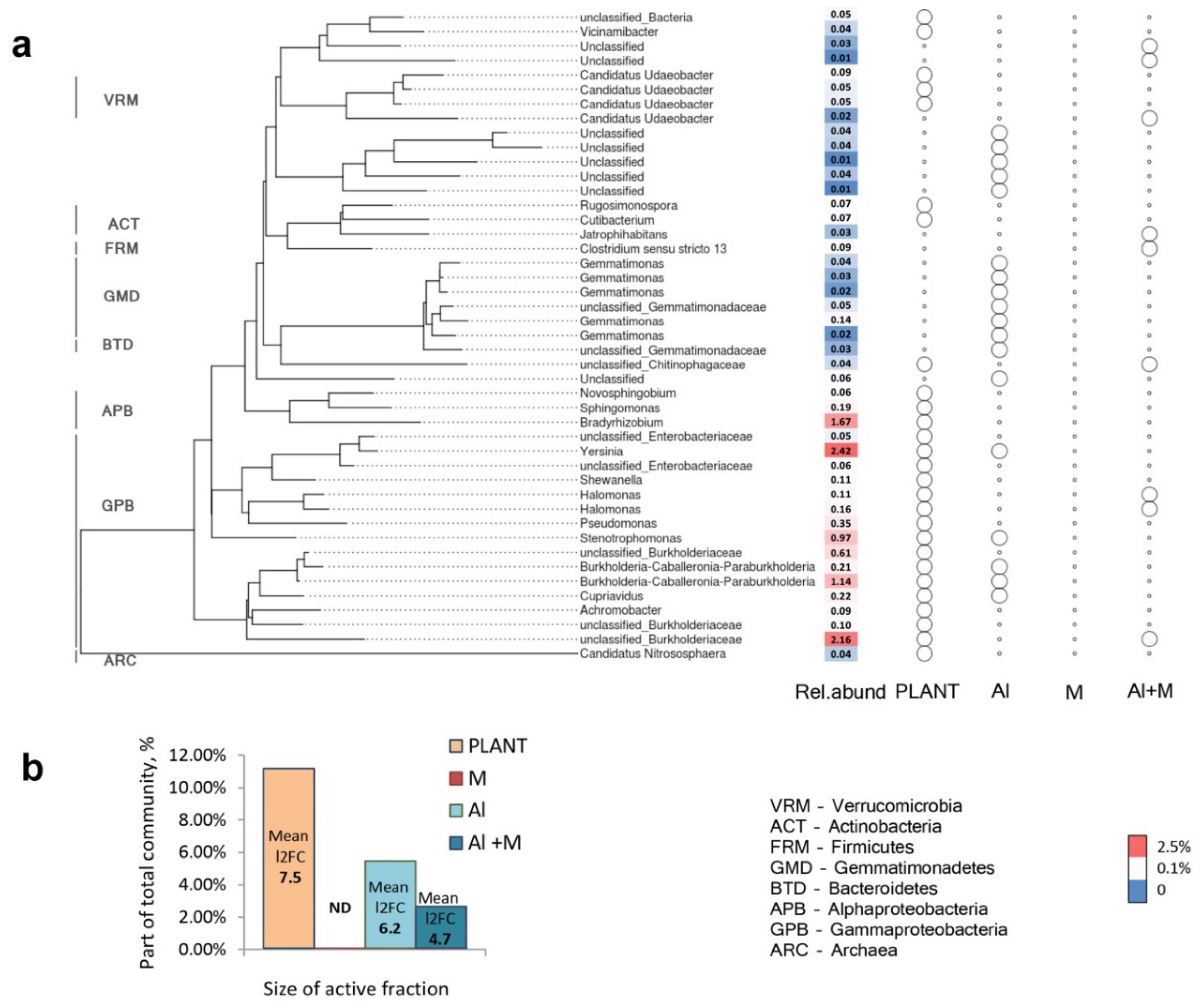
2.8. Rhizosphere Bacterial Communities
3. Discussion
3.1. Plant Biomass
3.2. Symbiotic Structures
3.3. Rhizosphere pH and Al Concentrations
3.4. Nitrogen Uptake
3.5. Phosphorus Uptake
3.6. Uptake of Other Nutrients by Plants
3.7. Rhizosphere Bacterial Communities
4. Materials and Methods
4.1. Plants and Microorganisms
4.2. Plant Growth Conditions
4.3. Symbiotic Parameters
4.4. DNA Extraction from Soil
4.5. Sequencing Data Processing
4.6. Aluminium and Nutrient Contents
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kochian, L.V. Cellular mechanisms of aluminium toxicity and resistance in plants. Annu. Rev. Plant Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Kochian, L.V.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Gaurav, S.S.; Kumar, A. Molecular basis of aluminium toxicity in plants: A review. Am. J. Plant. Sci. 2013, 4, 21–37. [Google Scholar] [CrossRef]
- Taylor, G.J. Current views of the aluminum stress response: The physiological basis of tolerance. Plant Physiol. Biochem. 1991, 10, 57–93. [Google Scholar]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Kochian, L.V.; Hoekenga, O.A.; Pineros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Motoda, H. Aluminum toxicity recovery processes in root apexes. Possible association with oxidative stress. Plant Sci. 2012, 185, 1–8. [Google Scholar] [CrossRef]
- Delhaize, E.; Ma, J.F.; Ryan, P.R. Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci. 2012, 17, 341–348. [Google Scholar] [CrossRef]
- Liu, J.; Piñeros, M.A.; Kochian, L.V. The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 2014, 56, 221–230. [Google Scholar] [CrossRef]
- Klimashevsky, E.L.; Markova, Y.A.; Malysheva, A.S. Genotypic specify in localization of Al3+ in cells of pea roots. Dokl. Akad. Nauk. 1972, 203, 711–713. [Google Scholar]
- Lazof, D.B.; Holland, M.J. Evaluation of the aluminium-induced root growth inhibition in isolation from low pH effects in Glycine max, Pisum sativum and Phaseolus vulgaris. Funct. Plant Biol. 1999, 26, 147–157. [Google Scholar] [CrossRef]
- Motoda, H.; Kano, Y.; Hiragami, F.; Kawamura, K.; Matsumoto, H. Morphological changes in the apex of pea roots during and after recovery from aluminium treatment. Plant Soil. 2010, 333, 49–58. [Google Scholar] [CrossRef]
- Singh, N.B.; Yadav, K.; Amist, N. Phytotoxic effects of aluminum on growth and metabolism of Pisum sativum L. IJBCS 2011, 2, 10–21. [Google Scholar]
- Yu, M.; Shen, R.; Liu, J.; Chen, R.; Xu, M.; Yang, Y.; Xiao, H.; Wang, H.D.; Wang, H.Z.; Wang, C. The role of root border cells in aluminum resistance of pea (Pisum sativum) grown in mist culture. J. Plant Nutr. Soil. Sci. 2009, 172, 528–534. [Google Scholar] [CrossRef]
- Yu, M.; Shen, R.F.; Xiao, H.D.; Xu, M.M.; Wang, H.Z. Boron alleviates aluminium toxicity in pea (Pisum sativum L.). Plant Soil. 2009, 314, 87–98. [Google Scholar] [CrossRef]
- Kichigina, N.E.; Pukhalsky, Y.V.; Shaposhnikov, A.I.; Azarova, T.S.; Makarova, N.M.; Loskutov, S.I.; Safronova, V.I.; Tikhonovich, I.A.; Vishniyakova, M.A.; Semenova, E.V.; et al. Aluminum exclusion from root zone and maintenance of nutrient uptake are principal mechanisms of al tolerance in Pisum sativum L. Physiol. Mol. Biol. Plants 2017, 23, 851–863. [Google Scholar] [CrossRef]
- Akhter, A.; Wagatsuma, T.; Khan, M.S.H.; Tawaraya, K. Comparative studies on aluminum tolerance screening techniques for sorghum, soybean and maize in simple solution culture. Am. J. Plant Physiol. 2009, 4, 1–8. [Google Scholar] [CrossRef]
- Arunakumara, K.K.I.U.; Walpola, B.C.; Yoon, M. Aluminum toxicity and tolerance mechanism in cereals and legumes—A review. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 1–9. [Google Scholar] [CrossRef]
- Ezaki, B.; Jayaram, K.; Higashi, A.; Takahashi, K. A combination of five mechanisms confers a high tolerance for aluminum to a wild species of Poaceae, Andropogon virginicus L. Environ. Exp. Bot. 2013, 93, 35–44. [Google Scholar] [CrossRef]
- Mendoza-Soto, A.B.; Naya, L.; Leija, A.; Hernández, G. Responses of symbiotic nitrogen-fixing common bean to aluminum toxicity and delineation of nodule responsive microRNAs. Front. Plant Sci. 2015, 6, 587. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Naamala, J.; Dakora, F.D. Nature and mechanisms of aluminium toxicity, tolerance and amelioration in symbiotic legumes and rhizobia. Biol. Fertil. Soils. 2018, 54, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Paulino, V.T.; Olivares, J.; Bedmar, E.J. Nodulation and nitrogenase activity of pea nodules as affected by low pH and aluminium. Plant Soil. 1987, 101, 299–302. [Google Scholar] [CrossRef]
- Sujkowska-Rybkowska, M. Reactive oxygen species production and antioxidative defense in pea (Pisum sativum L.) root nodules after short-term aluminum treatment. Acta Physiol. Plant. 2012, 34, 1387–1400. [Google Scholar] [CrossRef]
- Sujkowska-Rybkowska, M.; Borucki, W. Pectins esterification in the apoplast of aluminum-treated pea root nodules. J. Plant Physiol. 2015, 184, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Thorton, F.C.; Davey, C.B. Acid tolerance of Rhizobium trifolii in culture media. Soil Sci. Soc. Am. J. 1983, 47, 496–501. [Google Scholar] [CrossRef]
- Munns, D.; Keyser, H.; Fogle, V.; Hohenberg, J.; Righetti, T.; Lauter, D.; Zaroug, M.; Clarkin, K.; Whitacre, K. Tolerance of soil acidity in symbioses of mung bean with rhizobia. Agron. J. 1979, 71, 256–260. [Google Scholar] [CrossRef]
- Kingsley, M.T.; Bohlool, B.B. Extracellular polysaccharide is not responsible for aluminum tolerance of Rhizobium leguminosarum bv. phaseoli CIAT899. Appl. Environ. Microbiol. 1992, 58, 1095–1101. [Google Scholar] [CrossRef]
- Arora, N.; Khare, E.; Singh, S.; Maheshwari, D. Effect of Al and heavy metals on enzymes of nitrogen metabolism of fast and slow growing rhizobia under explanta conditions. World J. Microbiol. Biotechnol. 2010, 26, 811–816. [Google Scholar] [CrossRef]
- Avelar Ferreira, P.A.; Bomfeti, C.A.; Lima Soares, B.; de Souza Moreira, F.M. Efficient nitrogen-fixing Rhizobium strains isolated from amazonian soils are highly tolerant to acidity and aluminium. World J. Microbiol. Biotechnol. 2012, 28, 1947–1959. [Google Scholar] [CrossRef]
- Roy, N.; Chakrabartty, P.K. Effect of aluminum on the production of siderophore by Rhizobium sp. (Cicer arietinum). Curr. Microbiol. 2000, 41, 5–10. [Google Scholar] [CrossRef]
- Clarkson, D.T. Factors affecting mineral nutrient acquisition by plants. Annu. Rev. Plant Physiol. 1985, 36, 77–115. [Google Scholar] [CrossRef]
- Seguel, A.; Cumming, J.R.; Klugh-Stewart, K.; Cornejo, P.; Borie, F. The role of arbuscular mycorrhizas in decreasing aluminium phytotoxicity in acidic soils: A review. Mycorrhiza 2013, 23, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Koslowsky, S.D.; Boerner, R.E.J. Interactive effects of aluminium, phosphorus and mycorrhizae on growth and nutrient uptake of Panicum virgatum L. (Poaceae). Environ. Pollut. 1989, 61, 107–125. [Google Scholar] [CrossRef]
- Medeiros, C.A.B.; Clark, R.B.; Ellis, J.R. Effects of excess aluminium on mineral uptake in mycorrhizal sorghum. J. Plant Nutr. 1994, 17, 1399–1416. [Google Scholar] [CrossRef]
- Mendoza, J.; Borie, F. Effect of Glomus etunicatum inoculation on aluminium, phosphorus, calcium, and magnesium uptake of two barley genotypes with different aluminium tolerance. Comm. Soil Sci. Plant Anal. 1998, 29, 681–695. [Google Scholar] [CrossRef]
- Lux, H.B.; Cumming, J.R. Mycorrhizae confer aluminium resistance to tulip-poplar seedlings. Can. J. For. Res. 2001, 31, 694–702. [Google Scholar] [CrossRef]
- Wood, M. A Mechanism of Aluminum Toxicity to Soil Bacteria and Possible Ecological Implications. Plant-Soil Interactions at Low pH: Principles and Management; Springer: Dordrecht, The Netherland, 1995; pp. 173–179. [Google Scholar]
- Piña, R.G.; Cervantes, C. Microbial interactions with aluminium. Biometals 1996, 9, 311–316. [Google Scholar] [CrossRef]
- Thakur, R.; Sharma, K.C.; Gulati, A.; Sud, R.K.; Gulati, A. Stress-tolerant Viridibacillus arenosi strain IHB B 7171 from tea rhizosphere as a potential broad-spectrum microbial inoculant. Indian J. Microbiol. 2017, 57, 195–200. [Google Scholar] [CrossRef]
- Arora, P.; Singh, G.; Tiwari, A. Effect of Microbial inoculation in combating the aluminium toxicity effect on growth of Zea mays. Cell. Mol. Biol. (Noisy-le-Grand) 2017, 63, 79–82. [Google Scholar] [CrossRef]
- Niu, H.; Leng, Y.; Ran, S.; Du, M.A.D.; Sun, J.; Chen, K.; Hong, S. Toxicity of soil labile aluminum fractions and aluminum species in soil water extracts on the rhizosphere bacterial community of tall fescue. Ecotoxicol. Environ. Saf. 2020, 187, 109828. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Ann. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, M.; Chang, S.X.; Qi, J.; Tang, C.; Wen, Z.; Hong, Z.; Yang, T.; Ma, Z.; Gao, Q.; et al. Changes of microbial functional capacities in the rhizosphere contribute to aluminum tolerance by genotype-specific soybeans in acid soils. Biol. Fert. Soils 2020, 56, 771–783. [Google Scholar] [CrossRef]
- Shi, Q.; Liu, Y.; Shi, A.; Cai, Z.; Nian, H.; Hartmann, M.; Lian, T. Rhizosphere soil fungal communities of aluminum-tolerant and –sensitive soybean genotypes respond differently to aluminum stress in an acid soil. Front. Microbiol. 2020, 11, 1177. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, L.M.; Kukalev, A.S.; Ushakov, K.V.; Tsyganov, V.E.; Naumkina, T.S.; Provorov, N.A.; Borisov, A.Y.; Tikhonovich, I.A. Polymorphism of garden pea (Pisum sativum L.) for the efficiency of symbiosis with endomycorrhizal fungus Glomus sp. under the conditions of rhizobia inoculation. Agric. Biol. 2000, 3, 94–102. (In Russian) [Google Scholar]
- Rivera-Becerril, F.; Calantzis, C.; Turnau, K.; Caussanel, J.P.; Belimov, A.A.; Gianinazzi, S.; Strasser, R.J.; Gianinazzi-Pearson, V. Cadmium accumulation and buffering of cadmium-induced stress by arbuscular mycorrhiza in three Pisum sativum L. genotypes. J. Exp. Bot. 2002, 53, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.C.; Graham, J.H.; Smith, F.A. Functioning of mycorrhizal associations along the mutualism-parasitism continuum. New Phytol. 1997, 135, 575–586. [Google Scholar] [CrossRef]
- Klironomos, J.N. Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 2003, 84, 2292–2301. [Google Scholar] [CrossRef]
- Janssen, S.; McDonald, D.; Gonzalez, A.; Navas-Molina, J.A.; Jiang, L.; Xu, Z.Z.; Winker, K.; Kado, D.M.; Orwoll, E.; Manary, M.; et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. MSystems 2018, 3. [Google Scholar] [CrossRef]
- Li, H.; Smith, F.A.; Dickson, S.; Holloway, R.E.; Smith, S.E. Plant growth depressions in arbuscular mycorrhizal symbioses: Not just caused by carbon drain? New Phytol. 2008, 178, 852–862. [Google Scholar] [CrossRef]
- Gavito, M.E.; Bruhn, D.; Jakobsen, I. Phosphorus uptake by arbuscular mycorrhizal hyphae does not increase when the host plant grows under atmospheric CO2 enrichment. New Phytol. 2002, 154, 751–760. [Google Scholar] [CrossRef]
- Xavier, L.J.C.; Germida, J.L. Selective interactions between arbuscular mycorrhizal fungi and Rhizobium leguminosarum bv. viceae enhance pea yield and nutrition. Biol. Fertil. Soils 2003, 37, 261–267. [Google Scholar] [CrossRef]
- Shtark, O.Y.; Puzanskiy, R.K.; Avdeeva, G.S.; Yurkov, A.P.; Smolikova, G.N.; Yemelyanov, V.V.; Kliukova, M.S.; Shavarda, A.L.; Kirpichnikova, A.A.; Zhernakov, A.I.; et al. Metabolic alterations in pea leaves during arbuscular mycorrhiza development. Peer J. 2019, 7, e7495. [Google Scholar] [CrossRef]
- Gyaneshwar, P.; James, E.K.; Reddy, P.M.; Ladha, J.K. Herbaspirillum colonization increases growth and nitrogen accumulation in aluminium-tolerant rice varieties. New Phytol. 2002, 154, 131–145. [Google Scholar] [CrossRef]
- Vančura, V.; Přicryl, Z.; Kalachova, L.; Wurst, M. Some quantitative aspects on root exudation. Ecol. Bull. 1977, 25, 381–386. [Google Scholar]
- Bowen, G.D. Misconceptions, concepts and approaches in rhizosphere biology. In Contemporary Microbial Ecology; Ellwood, D.C., Hedger, J.N., Eds.; Academic Press: London, UK, 1980; pp. 283–304. [Google Scholar]
- Meharg, A.A.; Killham, K. Loss of exudates from the roots of perennial ryegrass inoculated with a range of micro-organisms. Plant Soil 1995, 170, 345–349. [Google Scholar] [CrossRef]
- Phillips, D.A.; Fox, C.T.; King, M.D.; Bhuvaneswari, T.V.; Teuber, L.R. Microbial products trigger amino acid exudation from plant roots. Plant Physiol. 2004, 136, 2887–2894. [Google Scholar] [CrossRef]
- Zhang, F.; Dashti, N.; Hynes, R.K.; Smith, D.L. Plant growth-promoting rhizobacteria and soybean (Glycine max (L.) Merr.) growth and physiology at suboptimal root zone temperatures. Ann. Bot. 1997, 79, 243–249. [Google Scholar] [CrossRef]
- Mia, M.A.B.; Shamsuddin, Z.H.; Wahab, Z.; Marziah, M. The effect of rhizobacterial inoculation on growth and nutrient accumulation of tissue-cultured banana plantlets under low N-fertilizer regime. Afr. J. Biotechnol. 2009, 8, 5855–5866. [Google Scholar] [CrossRef]
- Zheng, S.J.; Yang, J.L.; He, Y.F.; Yu, X.H.; Zhang, L.; You, J.F.; Shen, R.F.; Matsumoto, H. Immobilization of aluminum with phosphorus in roots is associated with high aluminum resistance in buckwheat. Plant Physiol. 2005, 138, 297–303. [Google Scholar] [CrossRef]
- Cumming, J.R.; Eckert, R.T.; Evans, L.S. Effect of aluminum on 32P uptake and translocation in red spruce seedlings. Can. J. For. Res. 1986, 16, 864–867. [Google Scholar] [CrossRef]
- Browne, P.; Rice, O.; Miller, S.H.; Burke, J.; Dowling, D.N.; Morrissey, J.P.; O’Gara, F. Superior inorganic phosphate solubilization is linked to phylogeny within the Pseudomonas fluorescens complex. Appl. Soil Ecol. 2009, 43, 131–138. [Google Scholar] [CrossRef]
- Li, G.E.; Wu, X.Q.; Ye, J.R.; Hou, L.; Zhou, A.D.; Zhao, L. Isolation and identification of phytate-degrading rhizobacteria with activity of improving growth of poplar and Masson pine. World J. Microbiol. Biotechnol. 2013, 29, 2181–2193. [Google Scholar] [CrossRef]
- Marschner, H. Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: London, UK, 1995. [Google Scholar] [CrossRef]
- Kabata-Pendias, A. Trace Elements in Soils and Plants; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar] [CrossRef]
- Burges, B.K.; Lowe, D.J. Mechanism of molybdenum nitrogenase. Chem. Rev. 1996, 96, 2983–3012. [Google Scholar] [CrossRef]
- Liao, H.; Wan, H.; Shaff, J.; Wang, X.; Yan, X.; Kochian, L.V. Phosphorus and aluminum interactions in soybean in relation to aluminum tolerance. Exudation of specific organic acids from different regions of the intact root system. Plant Physiol. 2006, 141, 674–684. [Google Scholar] [CrossRef]
- Liang, C.; Piñeros, M.A.; Tian, J.; Yao, Z.; Sun, L.; Liu, J.; Shaff, J.; Coluccio, A.; Kochian, L.V.; Liao, H. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef]
- Alarcón-Poblete, E.; Inostroza-Blancheteau, C.; Alberdi, M.; Rengel, Z.; Reyes-Díaz, M. Molecular regulation of aluminum resistance and sulfur nutrition during root growth. Planta 2018, 247, 27–39. [Google Scholar] [CrossRef]
- Borie, F.; Rubio, R. Effects of arbuscular mycorrhizae and liming on growth and mineral acquisition of Al-tolerant barley cultivars. J. Plant Nutr. 1999, 22, 121–137. [Google Scholar] [CrossRef]
- Belimov, A.A.; Shaposhnikov, A.I.; Azarova, T.S.; Makarova, N.M.; Safronova, V.I.; Litvinskiy, V.A.; Nosikov, V.V.; Zavalin, A.A.; Tikhonovich, I.A. Microbial consortium of PGPR, rhizobia and arbuscular mycorrhizal fungus makes pea mutant SGECdt comparable with Indian mustard in cadmium tolerance and accumulation. Plants 2020, 9, 975. [Google Scholar] [CrossRef]
- Zverev, O.; Pershina, E.V.; Provorov, N.A.; Andronov, E.E.; Serikova, E.N. Metagenomic characteristic of rhizosphere effect on cereals in black and sod-podzolic soils. Agric. Biol. 2016, 51, 654–663. [Google Scholar] [CrossRef]
- Zverev, O.; Pershina, E.V.; Shapkin, V.M.; Kichko, A.K.; Mitrofanova, O.P.; Kobylyanskii, V.D.; Yuzikhin, O.S.; Belimov, A.A.; Andronov, E.E. Molecular analysis of the rhizosphere microbial communities from gramineous plants grown on contrasting soils. Microbiology 2020, 89, 231–241. [Google Scholar] [CrossRef]
- Chaudhari, D.; Rangappa, K.; Das, A.; Layek, J.; Basavaraj, S.; Kandpal, B.K.; Shouche, Y.; Rahi, P. Pea (Pisum sativum L.) plant shapes its rhizosphere microbiome for nutrient uptake and stress amelioration in acidic soils of the North-East region of India. Front. Microbiol. 2020, 11, 968. [Google Scholar] [CrossRef] [PubMed]
- Harkes, P.; van Steenbrugge, J.J.M.; van den Elsen, S.J.J.; Suleiman, A.K.A.; de Haan, J.J.; Holterman, M.H.M.; Helder, J. Shifts in the active rhizobiome paralleling low Meloidogyne chitwoodi densities in fields under prolonged organic soil management. Front. Plant Sci. 2020, 10, 1697. [Google Scholar] [CrossRef] [PubMed]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, N.; Zvobgo, G.; Zhang, G.P. A Review: The beneficial effects and possible mechanisms of aluminum on plant growth in acidic soil. J. Integr. Agric. 2019, 18, 1518–1528. [Google Scholar] [CrossRef]
- Changey, F.; Meglouli, H.; Fontaine, J.; Magnin-Robert, M.; Tisserant, B.; Lerch, T.Z.; Lounès-Hadj Sahraoui, A. Initial microbial status modulates mycorrhizal inoculation effect on rhizosphere microbial communities. Mycorrhiza 2019, 29, 475–487. [Google Scholar] [CrossRef]
- Castro-Sowinski, S.; Herschkovitz, Y.; Okon, Y.; Jurkevitch, E. Effects of inoculation with plant growth-promoting rhizobacteria on resident rhizosphere microorganisms. FEMS Microbiol. Lett. 2007, 276, 1–11. [Google Scholar] [CrossRef]
- Belimov, A.A.; Puhalsky, I.V.; Safronova, V.I.; Shaposhnikov, A.I.; Vishnyakova, M.A.; Semenova, E.V.; Zinovkina, N.Y.; Makarova, N.M.; Wenzel, W.; Tikhonovich, I.A. Role of plant genotype and soil conditions in symbiotic plant-microbe interactions for adaptation of plants to cadmium polluted soils. Water Air Soil Poll. 2015, 226, 264. [Google Scholar] [CrossRef]
- Belimov, A.A.; Puhalsky, Y.V.; Shaposhnikov, A.I.; Azarova, T.S.; Makarova, N.M.; Safronova, V.I.; Tikhonovich, I.A.; Nosikov, V.V.; Litvinsky, V.A.; Zavalin, A.A. Plant-microbe symbiotic system based on cadmium tolerant pea mutant, mycorrhizal fungus, rhizobia and PGPR. In Proceedings of the 18-th International Conference “Heavy Metals in the Environment”, Ghent University, Ghent, Belgium, 12–15 September 2016; pp. 621–622. [Google Scholar]
- Shaposhnikov, A.I.; Vishnevskaya, N.A.; Shakhnazarova, V.Y.; Belimov, A.A.; Strunnikova, O.K. The role of barley root exudates as a food source in the relationship between Fusarium culmorum and Pseudomonas fluorescens. Mycol. Phytopathol. 2019, 53, 311–318. [Google Scholar] [CrossRef]
- Vincent, J.M. A manual for the practical study of root nodule bacteria. In IBP Handbook No 15; Blackwell Scientific Publ: Oxford, UK, 1970; 164p. [Google Scholar] [CrossRef]
- Turnau, K.; Anielska, T.; Ryszka, P.; Gawroński, S.; Ostachowicz, B.; Jurkiewicz, A. Establishment of arbuscular mycorrhizal plants originating from xerothermic grasslands on heavy metal rich industrial wastes—New solution for waste revegetation. Plant Soil 2008, 305, 267–280. [Google Scholar] [CrossRef]
- Trouvelot, A.; Kough, J.L.; Gianinazzi-Pearson, V. Mesure du taux de mycorhization VA d’un système radiculaire. Recherche de méthode d’estimation ayant une signification fonctionnelle. In Physiological and Genetical Aspects of Mycorrhizae, Proceedings of the 1st European Symposium on Mycorrhizae, Dijon, France, 1–5 July 1985; INR: Paris, France, 1986; pp. 217–221. [Google Scholar]
- Bates, S.T.; Berg–Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2010, 5, 908–917. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R. RStudio; PBC: Boston, MA, USA, 2020. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 1–14. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microb. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-3-319-24277-4. [Google Scholar]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]

| Treatments | Shoots | Seeds | ||||||
|---|---|---|---|---|---|---|---|---|
| N Concentration (mg g−1 DW) | 15N Fraction (%) | N Content (mg plant−1) | 15N Content (mg plant−1) | N Concentration (mg g−1 DW) | 15N Fraction (%) | N Content (mg plant−1) | 15N Content (mg plant−1) | |
| Pea genotype VIR1903 | ||||||||
| -Al −M | 7.1 ± 0.2 a | 3.50 ± 0.01 c | 36 ± 1 a | 1.25 ± 0.03 b | 27.9 ± 2.0 a | 2.60 ± 0.16 a | 123 ± 11 a | 3.19 ± 0.27 a |
| -Al +M | 7.6 ± 0.1 a | 4.44 ± 0.02 d | 45 ± 3 b | 1.98 ± 0.13 d | 28.1 ± 1.3 a | 2.52 ± 0.21 a | 143 ± 17 ab | 3.51 ± 0.20 a |
| +Al −M | 7.9 ± 0.1 a | 3.25 ± 0.01 b | 34 ± 2 a | 1.11 ± 0.07 a | 33.0 ± 1.6 b | 2.72 ± 0.18 a | 128 ± 7 ab | 3.46 ± 0.20 a |
| +Al +M | 10.0 ± 0.2 b | 2.79 ± 0.03 a | 59 ± 3 c | 1.64 ± 0.08 c | 36.8 ± 1.8 c | 2.10 ± 0.20 a | 168 ± 6 b | 3.53 ± 0.35 a |
| Pea genotype VIR7307 | ||||||||
| -Al −M | 6.4 ± 0.1 ab | 2.02 ± 0.01 a | 40 ± 3 a | 0.81 ± 0.06 a | 33.6 ± 1.4 b | 1.31 ± 0.09 a | 301 ± 33 b | 3.87 ± 0.19 a |
| -Al +M | 5.6 ± 0.2 a | 2.00 ± 0.01 a | 42 ± 1 ab | 0.84 ± 0.03 a | 32.8 ± 0.9 ab | 1.48 ± 0.12 a | 314 ± 21 b | 4.56 ± 0.09 a |
| +Al −M | 7.1 ± 0.2 b | 2.61 ± 0.01 b | 43 ± 4 ab | 1.12 ± 0.12 b | 30.5 ± 1.1 ab | 2.38 ± 0.23 b | 199 ± 13 a | 4.73 ± 0.46 a |
| +Al +M | 7.1 ± 0.1 b | 2.57 ± 0.01 b | 48 ± 2 b | 1.24 ± 0.06 b | 29.8 ± 0.6 a | 2.34 ± 0.24 b | 201 ± 15 a | 4.62 ± 0.28 a |
| Pea genotype VIR8353 | ||||||||
| -Al −M | 13.4 ± 0.1 a | 5.60 ± 0.01 d | 43 ± 1 b | 2.41 ± 0.07 b | 35.2 ± 0.8 a | 3.08 ± 0.34 b | 171 ± 20 a | 5.13 ± 0.43 c |
| -Al +M | 13.3 ± 0.1 a | 2.72 ± 0.01 a | 44 ± 2 b | 1.21 ± 0.05 a | 38.7 ± 0.9 a | 1.82 ± 0.09 a | 220 ± 11 b | 4.04 ± 0.37 b |
| +Al −M | 13.1 ± 0.3 a | 3.36 ± 0.01 c | 32 ± 3 a | 1.06 ± 0.09 a | 35.5 ± 1.2 a | 2.28 ± 0.25 a | 137 ± 15 a | 3.07 ± 0.35 a |
| +Al +M | 13.8 ± 0.2 a | 3.12 ± 0.01 b | 37 ± 2 ab | 1.16 ± 0.05 a | 38.7 ± 0.6 a | 2.39 ± 0.20 a | 175 ± 12 a | 4.17 ± 0.44 b |
| Pea genotype VIR8473 | ||||||||
| -Al −M | 12.4 ± 0.2 b | 4.17 ± 0.01 b | 36 ± 2 b | 1.49 ± 0.08 b | 29.4 ± 2.0 a | 5.10 ± 0.17 b | 55 ± 6 ab | 2.81 ± 0.29 b |
| -Al +M | 10.9 ± 0.7 ab | 3.42 ± 0.03 a | 34 ± 4 b | 1.10 ± 0.15 a | 30.4 ± 0.8 a | 3.98 ± 0.60 a | 96 ± 13 b | 3.61 ± 0.27 bc |
| +Al −M | 10.3 ± 0.2 a | 4.36 ± 0.01 c | 23 ± 1 a | 1.01 ± 0.04 a | 33.0 ± 0.9 a | 4.12 ± 0.34 a | 44 ± 3 a | 1.78 ± 0.14 a |
| +Al +M | 9.8 ± 0.9 a | 4.46 ± 0.01 d | 26 ± 2 a | 1.14 ± 0.10 a | 32.5 ± 2.0 a | 5.54 ± 0.16 b | 68 ± 4 ab | 3.74 ± 0.25 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belimov, A.A.; Shaposhnikov, A.I.; Syrova, D.S.; Kichko, A.A.; Guro, P.V.; Yuzikhin, O.S.; Azarova, T.S.; Sazanova, A.L.; Sekste, E.A.; Litvinskiy, V.A.; et al. The Role of Symbiotic Microorganisms, Nutrient Uptake and Rhizosphere Bacterial Community in Response of Pea (Pisum sativum L.) Genotypes to Elevated Al Concentrations in Soil. Plants 2020, 9, 1801. https://doi.org/10.3390/plants9121801
Belimov AA, Shaposhnikov AI, Syrova DS, Kichko AA, Guro PV, Yuzikhin OS, Azarova TS, Sazanova AL, Sekste EA, Litvinskiy VA, et al. The Role of Symbiotic Microorganisms, Nutrient Uptake and Rhizosphere Bacterial Community in Response of Pea (Pisum sativum L.) Genotypes to Elevated Al Concentrations in Soil. Plants. 2020; 9(12):1801. https://doi.org/10.3390/plants9121801
Chicago/Turabian StyleBelimov, Andrey A., Alexander I. Shaposhnikov, Darya S. Syrova, Arina A. Kichko, Polina V. Guro, Oleg S. Yuzikhin, Tatiana S. Azarova, Anna L. Sazanova, Edgar A. Sekste, Vladimir A. Litvinskiy, and et al. 2020. "The Role of Symbiotic Microorganisms, Nutrient Uptake and Rhizosphere Bacterial Community in Response of Pea (Pisum sativum L.) Genotypes to Elevated Al Concentrations in Soil" Plants 9, no. 12: 1801. https://doi.org/10.3390/plants9121801
APA StyleBelimov, A. A., Shaposhnikov, A. I., Syrova, D. S., Kichko, A. A., Guro, P. V., Yuzikhin, O. S., Azarova, T. S., Sazanova, A. L., Sekste, E. A., Litvinskiy, V. A., Nosikov, V. V., Zavalin, A. A., Andronov, E. E., & Safronova, V. I. (2020). The Role of Symbiotic Microorganisms, Nutrient Uptake and Rhizosphere Bacterial Community in Response of Pea (Pisum sativum L.) Genotypes to Elevated Al Concentrations in Soil. Plants, 9(12), 1801. https://doi.org/10.3390/plants9121801





