Diversity Patterns of Bermuda Grass along Latitudinal Gradients at Different Temperatures in Southeastern China
Abstract
1. Introduction
2. Results
2.1. Genetic Variation of 16 Populations of Cynodon along Latitudinal at Different Temperatures
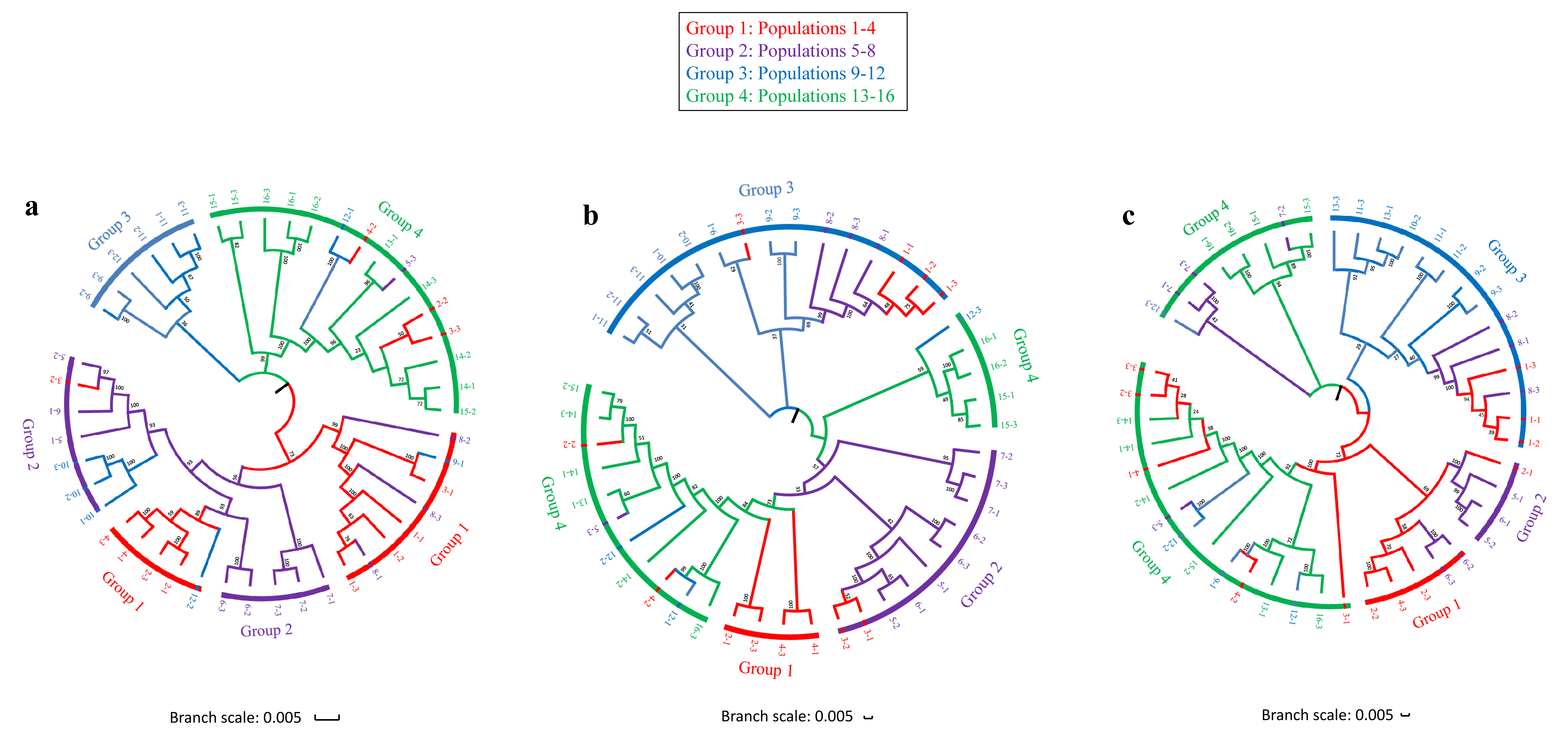
2.2. Phylogenetic Relationship along Latitudinal Gradients at Different Temperatures
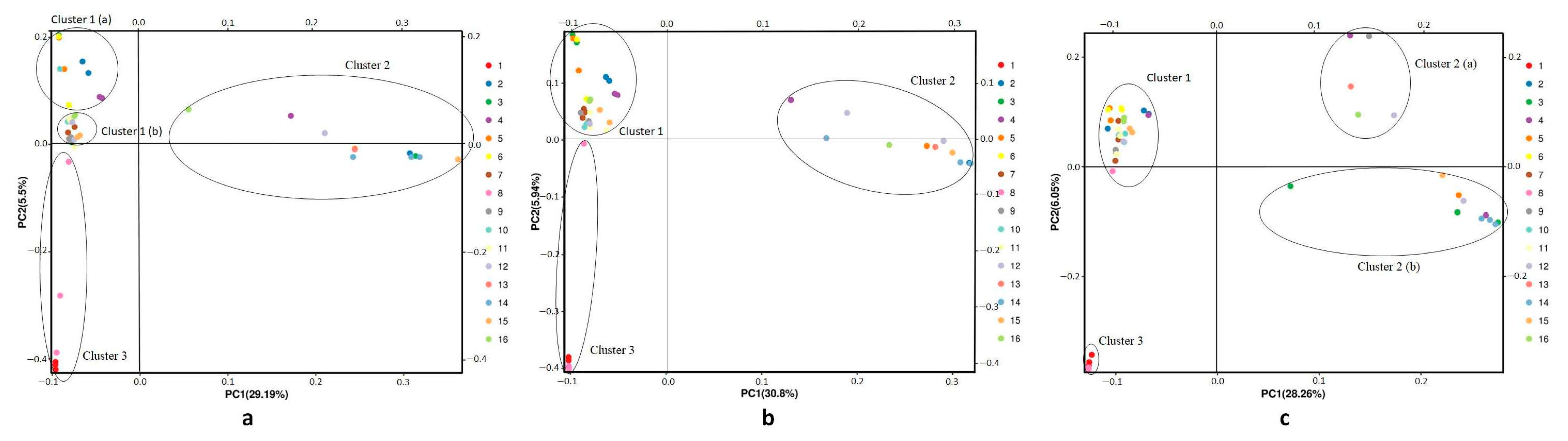
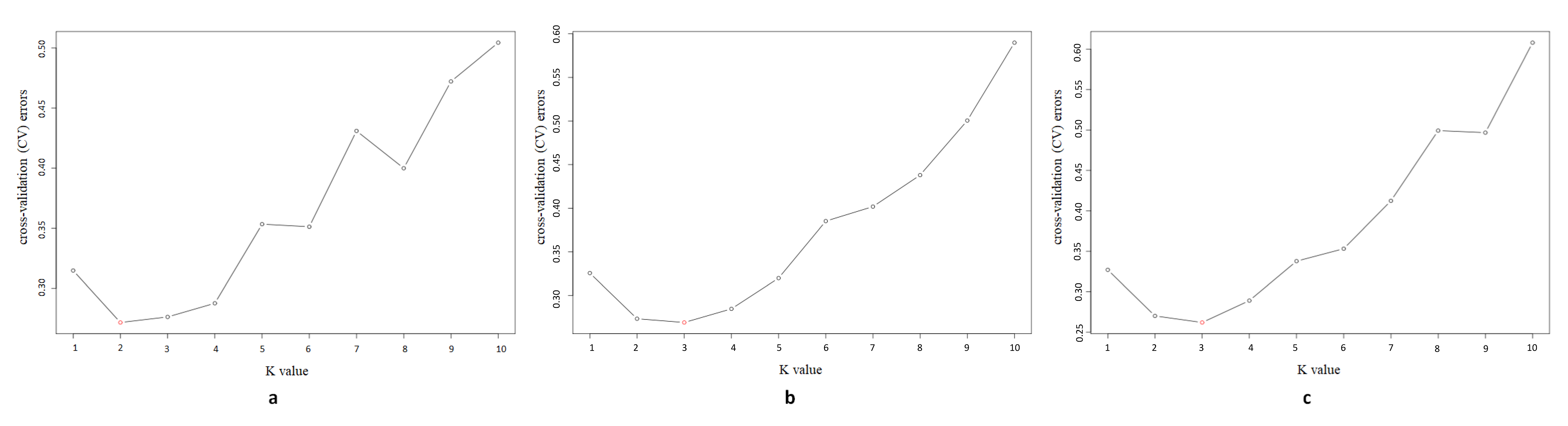
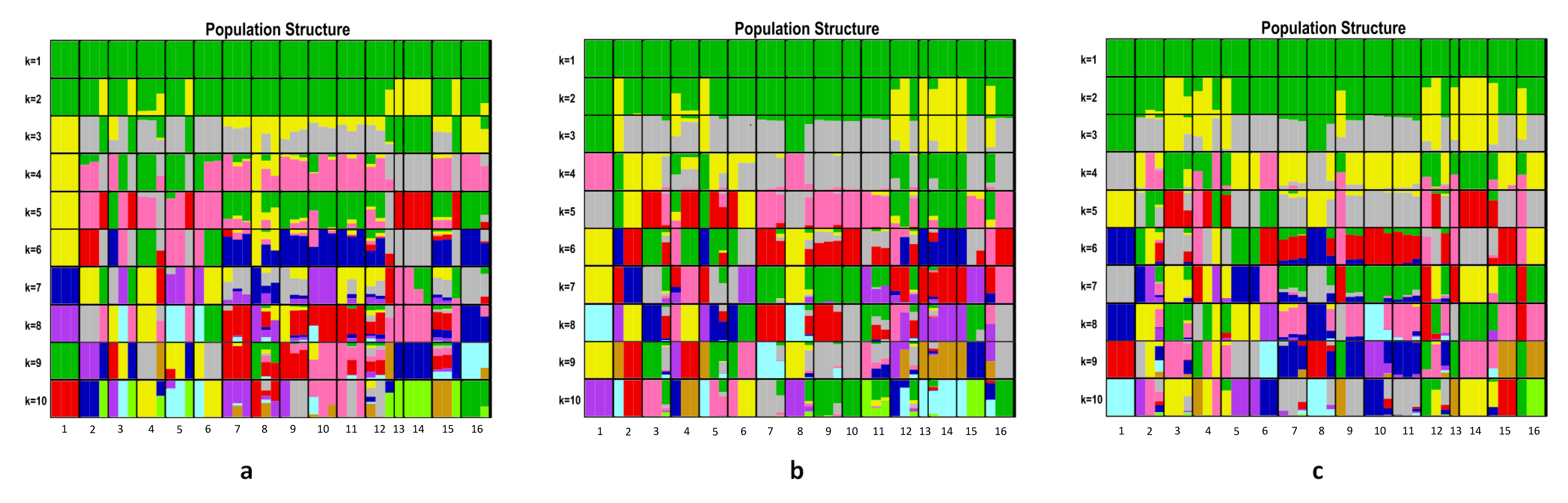
2.3. Population Genetic Structure
3. Discussion
3.1. Population Genetic Differentiation and Structure of C. dactylon
3.2. Analysis of C. dactylon Landscape Genetics
3.3. Adaptation of Genetic Variation along Latitudinal Gradients to Different Temperatures
4. Material and Methods
4.1. Plant Material and Experimental Design
4.2. RNA Extraction and Sequencing
4.3. Detection of SNP Population and Data Analysis
4.4. Population Structure and Phylogenetic Tree
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Avis, J.D.; Taliaferro, C.M.; Holbert, D. Genotype × interaction study of bermuda grass yields in Oklahoma. Proc. Okla. Acad. Sci. 1980, 60, 69–74. [Google Scholar]
- Salehi, M.; Salehi, H.; Niazi, A.; Ghobadi, C. Convergence of goals: Phylogenetical, morphological, and physiological characterization of tolerance to drought stress in tall fescue (Festuca arundinacea Schreb.). Mol. Biotechnol. 2013, 56, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Luo, N.; Liu, J.; Yu, X.; Jiang, Y. Natural variation of drought response in Brachypodium distachyon. Physiol. Plant. 2011, 141, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Fournier-Level, A.; Korte, A.; Cooper, M.D.; Nordborg, M.; Schmitt, J.; Wilczek, A.M. A map of local adaptation in arabidopsis thaliana. Science 2011, 334, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Ciais, P.; Reichstein, M.; Viovy, N.; Granier, A.; Ogée, J.; Allard, V.; Aubinet, M.; Buchmann, N.; Bernhofer, C.; Carrara, A.; et al. Europe-wide reduction in primary productivity caused by the heat and drought in 2003. Nature 2005, 437, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ye, T.; Chan, Z. Exogenous application of hydrogen sulfide donor sodium hydrosulfide enhanced multiple abiotic stress tolerance in bermuda grass (Cynodon dactylon (L). Pers.). Plant Physiol. Biochem. 2013, 71, 226–234. [Google Scholar] [CrossRef]
- Cahill, A.E.; Aiello-Lammens, M.E.; Fisher-Reid, M.C.; Hua, X.; Fisher-Reid, M.C.; Karanewsky, C.J.; Hae Yeong Ryu, C.J.; Sbeglia, G.C.; Spagnolo, F.; Waldron, J.B.; et al. How does climate change cause extinction? Proc. R. Soc. B Biol. Sci. 2013, 280, 20121890. [Google Scholar] [CrossRef]
- Merriam, C.H. Laws of temperature and control of the geographic distribution of terrestrial animals and plants. Natl. Geogr. Mag. 1894, 6, 229–238. [Google Scholar]
- Beard, J.B. Turfgrass: Science and Culture; Prentice-Hall, Inc.: Englewood Cliffs, NJ, USA, 1973; 672p. [Google Scholar]
- Anderson, J.; Taliaferro, C.; Martin, D. Freeze tolerance of bermuda grasses. Crop Sci. 2002, 42, 975–977. [Google Scholar]
- Munshaw, G.C.; Ervin, E.H.; Beasley, J.S.; Shang, C.; Zhang, X.; Parrish, D.J. Effects of late-season ethephon applications on cold tolerance parameters of four bermuda grass cultivars. Crop Sci. 2011, 50, 1022–1029. [Google Scholar] [CrossRef]
- Akbari, M.; Salehi, H.; Niazi, A. Evaluation of Diversity Based on Morphological Variabilities and ISSR Molecular Markers in Iranian Cynodon dactylon (L.) Pers. Accessions to Select and Introduce Cold-Tolerant Genotypes. Mol. Biotechnol. 2018, 60, 259–270. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 2014, 56, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Cibrian-Jaramillo, A.; Kolokotronis, S.O.; Katari, M.S.; Stamatakis, A.; Ott, M.; Chiu, J.C.; Little, D.P.; Stevenson, D.W.; Richard McCombie, W.; et al. A functional phylogenomic view of the seed plants. PLoS Genet. 2011, 7, e1002411. [Google Scholar] [CrossRef] [PubMed]
- Dohm, J.C.; Minoche, A.E.; Holtgrawe, D.; Capella-Gutiérrez, S.; Zakrzewski, F.; Tafer, H.; Rupp, O.; Sörensen, T.R.; Stracke, R.; Reinhardt, R.; et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 2014, 505, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Sveinsson, S.; McDill, J.; Wong, G.K.; Li, J.; Li, X.; Deyholos, M.K.; Cronk, Q.C. Phylogenetic pinpointing of a paleopolyploidy event within the flax genus (Linum) using transcriptomics. Ann. Bot. 2014, 113, 753–761. [Google Scholar] [CrossRef]
- Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Carpenter, E.; Matasci, N.; Ayyampalayam, S.; Barker, M.S.; Burleigh, J.G.; Gitzendanner, M.A.; et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 2014, 111, E4859–E4868. [Google Scholar] [CrossRef]
- Mckain, M.R.; Wickett, N.J.; Zhang, Y.; Ayyampalayam, S.; Mccombie, W.R.; Chase, M.W.; Pires, J.C.; Depamphilis, C.W.; Leebensmack, J. Phylogenomic analysis of transcriptome data elucidates co-occurrence of a paleopolyploid event and the origin of bimodal karyotypes in Agavoideae (Asparagaceae). Am. J. Bot. 2012, 99, 397–406. [Google Scholar] [CrossRef]
- Wen, J.; Xiong, Z.; Nie, Z.L.; Mao, L.; Zhu, Y.; Kan, X.Z.; Ickert-Bond, S.M.; Gerrath, J.; Zimmer, E.A.; Fang, X.D. Transcriptome sequences resolve deep relationships of the grape family. PLoS ONE 2013, 8, e74394. [Google Scholar] [CrossRef]
- Brumfield, R.T.; Beerli, P.; Nickerson, D.A.; Edwards, S.V. The utility of single nucleotide polymorphisms in inferences of population history. Trends. Ecol. Evol. 2003, 18, 249–256. [Google Scholar] [CrossRef]
- Garvin, M.R.; Saitoh, K.; Gharrett, A.J. Application of single nucleotide polymorphisms to non-model species: A technical review. Mol. Ecol. Res. 2010, 10, 915–934. [Google Scholar] [CrossRef]
- Helyar, S.J.; Hemmer-Hansen, J.; Bekkevold, D.; Taylor, M.I.; Nielsen, E.E. Application of snps for population genetics of nonmodel organisms: New opportunities and challenges. Mol. Ecol. Res. 2011, 11, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Wang, S.; He, J.; Chen, W. De novo assembly and transcriptome characterization of Opisthopappus (Asteraceae) for population differentiation and adaption. Front. Genet. 2018, 9, 371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xiao, X.L.; Zong, J.Q.; Chen, J.B.; Li, J.J.; Guo, H.L. Comparative transcriptome analysis provides new insights into erect and prostrate growth in bermuda grass (Cynodon dactylon L.). Plant Physiol. Biochem. 2017, 2017, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Jordan, R.; Hoffmann, A.A.; Dillon, S.K.; Prober, S.M. Evidence of genomic adaptation to climate in Eucalyptus microcarpa: Implications for adaptive potential to projected climate change. Mol. Ecol. 2017, 26, 6002–6020. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.K.; Allendorf, F.W.; Holt, J.S.; Lodge, D.M.; Molofsky, J.; Baughman, S.; Cabin, R.J.; Cohen, J.E.; Ellstrand, N.C.; Mccauley, D.E.; et al. The population biology of invasive species. Annu. Rev. Ecol. Evol. Syst. 2001, 32, 305–332. [Google Scholar] [CrossRef]
- Lee, C.E. Evolutionary genetics of invasive species. Trends. Ecol. Evol. 2002, 17, 386–391. [Google Scholar] [CrossRef]
- Liu, P.; Que, Y.; Pan, Y.B. Highly polymorphic microsatellite DNA markers for sugarcane germplasm evaluation and variety identity testing. Sugar Tech. 2011, 13, 129–136. [Google Scholar] [CrossRef]
- Zhang, J.X.; Wang, M.L.; Guo, Z.P.; Guan, Y.Z.; Liu, J.Y.; Yan, X.B.; Guo, Y.X. Genetic Diversity and Population Structure of Bermuda grass [Cynodon dactylon (L.) Pers.] along Latitudinal Gradients and the Relationship with Polyploidy Level. Diversity 2019, 11, 135. [Google Scholar] [CrossRef]
- Janzen, D.H. Why mountain passes are higher in tropics. Am. Nat. 1967, 101, 233–249. [Google Scholar] [CrossRef]
- Ghalambor, C.K.; Huey, R.B.; Martin, P.R.; Tewksbury, J.J.; Wang, G. Are mountain passes higher in the tropics? Janzen’s hypothesis revisited. Integr. Comp. Biol. 2006, 46, 5–17. [Google Scholar] [CrossRef]
- Gazave, E.; Tassone, E.E.; Ilut, D.C.; Wingerson, M. Population genomic analysis reveals differential evolutionary histories and patterns of diversity across subgenomes and subpopulations of Brassica napus L. Front. Plant Sci. 2016, 7, 525. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.C.; Engqvist, G.M.; Karlsson, B. Comparison of rapeseed cultivars and resynthesized lines based on allozyme and RFLP markers. Theor. Appl. Genet. 1995, 91, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, F.; Xu, K.; Gao, G.; Chen, B.; Yan, G.; Wang, N.; Qiao, J.; Li, J.; Li, H.; et al. Assessing and broadening genetic diversity of a rapeseed germplasm collection. Breed. Sci. 2014, 64, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Hufford, M.B.; Lubinksy, P.; Pyhäjärvi, T.; Devengenzo, M.T.; Ellstrand, N.C.; Ross-Ibarra, J. The genomic signature of crop-wild introgression in maize. PLoS Genet. 2013, 9, e1003477. [Google Scholar] [CrossRef]
- Ebersbach, J.; Muellner-Riehl, A.N.; Michalak, I.; Tkach, N.; Hoffmann, M.H.; Roeser, M.; Sun, H.; Favre, A. In and out of the Qinghai-Tibet Plateau: Divergence time estimation and historical biogeography of the large arctic-alpine genus Saxifraga, L. J. Biogeogr. 2017, 44, 900–910. [Google Scholar] [CrossRef]
- Kim, C.; Deng, T.; Wen, J.; Nie, Z.L.; Sun, H. Systematics, biogeography, and character evolution of deutzia (hydrangeaceae) inferred from nuclear and chloroplast dna sequences. Mol. Phylogenetics Evol. 2015, 87, 91–104. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, J.; Deng, T.; Boufford, D.E. Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Divers. 2017, 39, 161. [Google Scholar] [CrossRef]
- Hajjar, R.; Hodgkin, T. The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 2007, 156, 1–13. [Google Scholar] [CrossRef]
- Kearns, R.; Zhou, Y.; Fukai, S.; Ye, C.; Loch, D.; Godwin, I.D.; Holton, T.; Innes, D.; Stirling, H.; Cao, N.; et al. Eco-turf: Water use efcient turfgrasses from Australian biodiversity. Acta Hortic. 2009, 829, 113–118. [Google Scholar] [CrossRef]
- Durka, W.; Michalski, S.G.; Berendzen, K.W.; Bossdorf, O.; Bucharova, A.; Hermann, J.M.; Hölzel, N.; Kollmann, J. Genetic differentiation within multiple common grassland plants supports seed transfer zones for ecological restoration. J. Appl. Ecol. 2017, 54, 116–126. [Google Scholar] [CrossRef]
- Michalski, S.G.; Durka, W.; Jentsch, A.; Kreyling, J.; Pompe, S.; Schweiger, O.; Willner, E.; Beierkuhnlein, C. Evidence for genetic differentiation and divergent selection in an autotetraploid forage grass (Arrhenatherum elatius). Theor. Appl. Genet. 2010, 120, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Jansson, R.; Davies, T.J. Global variation in diversification rates of flowering plants: Energy vs. climate change. Ecol. Lett. 2008, 11, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Svenning, J.C.; Borchsenius, F.; Bjorholm, S.; Balslev, H. High tropical net diversification drives the New World latitudinal gradient in palm (Arecaceae) species richness. J. Biogeogr. 2008, 35, 394–406. [Google Scholar] [CrossRef]
- McFadden, I.R.; Sandel, B.; Tsirogiannis, C.; Moruetaholme, N.; Svenning, J.; Enquist, B.J.; Kraft, N.J.B. Temperature shapes opposing latitudinal gradients of plant taxonomic and phylogenetic β diversity. Ecol. Lett. 2019, 22, 1126–1135. [Google Scholar] [CrossRef]
- Lehnert, S.J.; DiBacco, C.; Van Wyngaarden, M.; Jeffery, N.W.; Ben Lowen, J.; Sylvester, E.V.A.; Wringe, B.F.; Stanley, R.R.E.; Hamilton, L.C.; Bradbury, I.R. Fine-scale temperature-associated genetic structure between inshore and offshore populations of sea scallop (Placopecten magellanicus). Heredity 2019, 122, 69. [Google Scholar] [CrossRef] [PubMed]
- Matz, M.V.; Treml, E.A.; Aglyamova, G.V.; Bay, L.K. Potential and limits for rapid genetic adaptation to warming in a Great Barrier Reef coral. PLoS Genet. 2018, 14, e1007220. [Google Scholar] [CrossRef] [PubMed]
- Rohde, K. Latitudinal gradients in species diversity: The search for the primary cause. Oikos 1992, 65, 514–527. [Google Scholar] [CrossRef]
- Jorde, P.E.; SoVik, G.; Westgaard, J.I.; Albretsen, J.; André, C.; Hvingel, C.; Johansen, T.; Sandvik, A.D.; Kingsley, M.; Jørstadet, K.E. Genetically distinct populations of northern shrimp, Pandalus borealis, in the North Atlantic: Adaptation to different temperatures as an isolation factor. Mol. Ecol. 2015, 24, 1742–1757. [Google Scholar] [CrossRef] [PubMed]
- Lowry, E.; Lester, S.E. The biogeography of plant reproduction: Potential determinants of species’ range sizes. J. Biogeogr. 2006, 33, 1975–1982. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.M.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.J.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. Ital. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequinversion3.0: Anintegrated software package for population genetics data analysis. Evol. Bioinform. 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]

| 5 °C | 20 °C | 35 °C | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ho | He | Nei’s | I | PIC | Ho | He | Nei’s | I | PIC | Ho | He | Nei’s | I | PIC | |
| 1 | 0.914 | 0.479 | 0.574 | 0.670 | 0.362 | 0.893 | 0.475 | 0.570 | 0.666 | 0.360 | 0.911 | 0.478 | 0.574 | 0.670 | 0.362 |
| 2 | 0.700 | 0.418 | 0.501 | 0.604 | 0.326 | 0.702 | 0.416 | 0.499 | 0.602 | 0.324 | 0.730 | 0.432 | 0.519 | 0.620 | 0.335 |
| 3 | 0.673 | 0.410 | 0.492 | 0.596 | 0.321 | 0.779 | 0.448 | 0.537 | 0.637 | 0.344 | 0.788 | 0.445 | 0.534 | 0.634 | 0.343 |
| 4 | 0.726 | 0.427 | 0.512 | 0.614 | 0.331 | 0.713 | 0.425 | 0.510 | 0.612 | 0.330 | 0.699 | 0.423 | 0.508 | 0.611 | 0.329 |
| 5 | 0.687 | 0.416 | 0.499 | 0.602 | 0.325 | 0.700 | 0.416 | 0.499 | 0.602 | 0.325 | 0.676 | 0.410 | 0.492 | 0.596 | 0.321 |
| 6 | 0.801 | 0.450 | 0.540 | 0.640 | 0.346 | 0.750 | 0.442 | 0.530 | 0.631 | 0.341 | 0.769 | 0.445 | 0.534 | 0.634 | 0.342 |
| 7 | 0.780 | 0.448 | 0.537 | 0.637 | 0.344 | 0.835 | 0.459 | 0.551 | 0.649 | 0.351 | 0.791 | 0.446 | 0.535 | 0.635 | 0.343 |
| 8 | 0.791 | 0.448 | 0.538 | 0.638 | 0.345 | 0.816 | 0.456 | 0.548 | 0.647 | 0.350 | 0.840 | 0.461 | 0.553 | 0.651 | 0.352 |
| 9 | 0.783 | 0.447 | 0.537 | 0.636 | 0.344 | 0.771 | 0.445 | 0.534 | 0.634 | 0.343 | 0.714 | 0.421 | 0.505 | 0.607 | 0.327 |
| 10 | 0.837 | 0.458 | 0.549 | 0.648 | 0.350 | 0.915 | 0.480 | 0.640 | 0.672 | 0.364 | 0.771 | 0.446 | 0.535 | 0.635 | 0.343 |
| 11 | 0.784 | 0.447 | 0.537 | 0.636 | 0.344 | 0.709 | 0.432 | 0.518 | 0.620 | 0.335 | 0.776 | 0.444 | 0.533 | 0.633 | 0.342 |
| 12 | 0.683 | 0.414 | 0.497 | 0.600 | 0.324 | 0.726 | 0.430 | 0.516 | 0.618 | 0.334 | 0.691 | 0.425 | 0.510 | 0.613 | 0.331 |
| 14 | 0.815 | 0.455 | 0.546 | 0.645 | 0.348 | 0.831 | 0.458 | 0.550 | 0.648 | 0.350 | 0.865 | 0.467 | 0.560 | 0.657 | 0.355 |
| 15 | 0.683 | 0.414 | 0.496 | 0.600 | 0.323 | 0.618 | 0.398 | 0.478 | 0.583 | 0.314 | 0.653 | 0.406 | 0.487 | 0.591 | 0.319 |
| 16 | 0.820 | 0.449 | 0.539 | 0.638 | 0.345 | 0.711 | 0.423 | 0.508 | 0.610 | 0.329 | 0.695 | 0.425 | 0.510 | 0.612 | 0.330 |
| Overall | 0.543 | 0.353 | 0.357 | 0.526 | 0.280 | 0.528 | 0.349 | 0.353 | 0.521 | 0.277 | 0.539 | 0.356 | 0.360 | 0.529 | 0.282 |
| 5 °C | ||||||
| Source of Variation | df | SS | MS | Estimate of Variation | % | Fixation Index (Fst) |
| Among Populations | 15 | 44,715.134 | 2981.008933 | 4.11 | 5.56% | 0.041 * |
| Within Populations | 30 | 46,993.333 | 1566.444433 | 69.78 | 94.44% | |
| Total | 45 | 91,708.467 | 73.89 | |||
| 20 °C | ||||||
| Source of Variation | df | SS | MS | Estimate of Variation | % | Fixation Index (Fst) |
| Among Populations | 15 | 28,819.383 | 1921.2922 | 4.36 | 5.90% | 0.044 * |
| Within populations | 29 | 28,765.083 | 991.8994138 | 69.5 | 94.10% | |
| Total | 44 | 57,584.466 | 73.86 | |||
| 35 °C | ||||||
| Source of Variation | df | SS | MS | Estimate of Variation | % | Fixation Index (Fst) |
| Among Populations | 15 | 43,718.942 | 2914.596133 | 4.16 | 5.70% | 0.042 * |
| Within Populations | 30 | 46,420.667 | 1547.355567 | 68.85 | 94.30% | |
| Total | 45 | 90,139.609 | 73.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.-X.; Chen, M.-H.; Gan, L.; Zhang, C.-J.; Shen, Y.; Qian, J.; Han, M.-L.; Guo, Y.-X.; Yan, X.-B. Diversity Patterns of Bermuda Grass along Latitudinal Gradients at Different Temperatures in Southeastern China. Plants 2020, 9, 1778. https://doi.org/10.3390/plants9121778
Zhang J-X, Chen M-H, Gan L, Zhang C-J, Shen Y, Qian J, Han M-L, Guo Y-X, Yan X-B. Diversity Patterns of Bermuda Grass along Latitudinal Gradients at Different Temperatures in Southeastern China. Plants. 2020; 9(12):1778. https://doi.org/10.3390/plants9121778
Chicago/Turabian StyleZhang, Jing-Xue, Ming-Hui Chen, Lu Gan, Chuan-Jie Zhang, Yu Shen, Jin Qian, Meng-Li Han, Yu-Xia Guo, and Xue-Bing Yan. 2020. "Diversity Patterns of Bermuda Grass along Latitudinal Gradients at Different Temperatures in Southeastern China" Plants 9, no. 12: 1778. https://doi.org/10.3390/plants9121778
APA StyleZhang, J.-X., Chen, M.-H., Gan, L., Zhang, C.-J., Shen, Y., Qian, J., Han, M.-L., Guo, Y.-X., & Yan, X.-B. (2020). Diversity Patterns of Bermuda Grass along Latitudinal Gradients at Different Temperatures in Southeastern China. Plants, 9(12), 1778. https://doi.org/10.3390/plants9121778




