Development of an In Vitro Method of Propagation for Artemisia tridentata subsp. tridentata to Support Genome Sequencing and Genotype-by-Environment Research
Abstract
1. Introduction
2. Results
2.1. Plant Materials
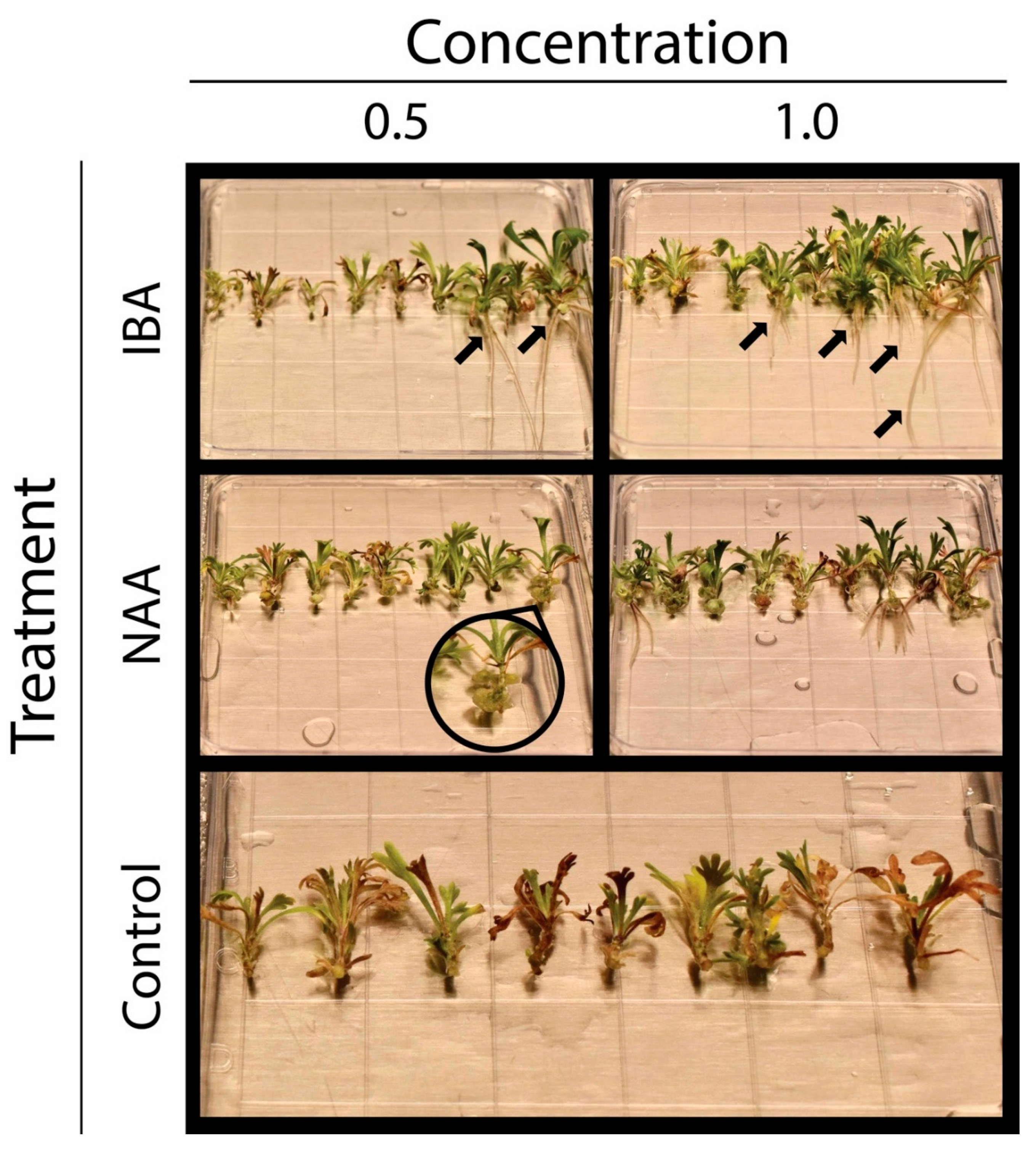
2.2. Effect of Growth Regulators on in vitro Calli Development on Shoot Tips
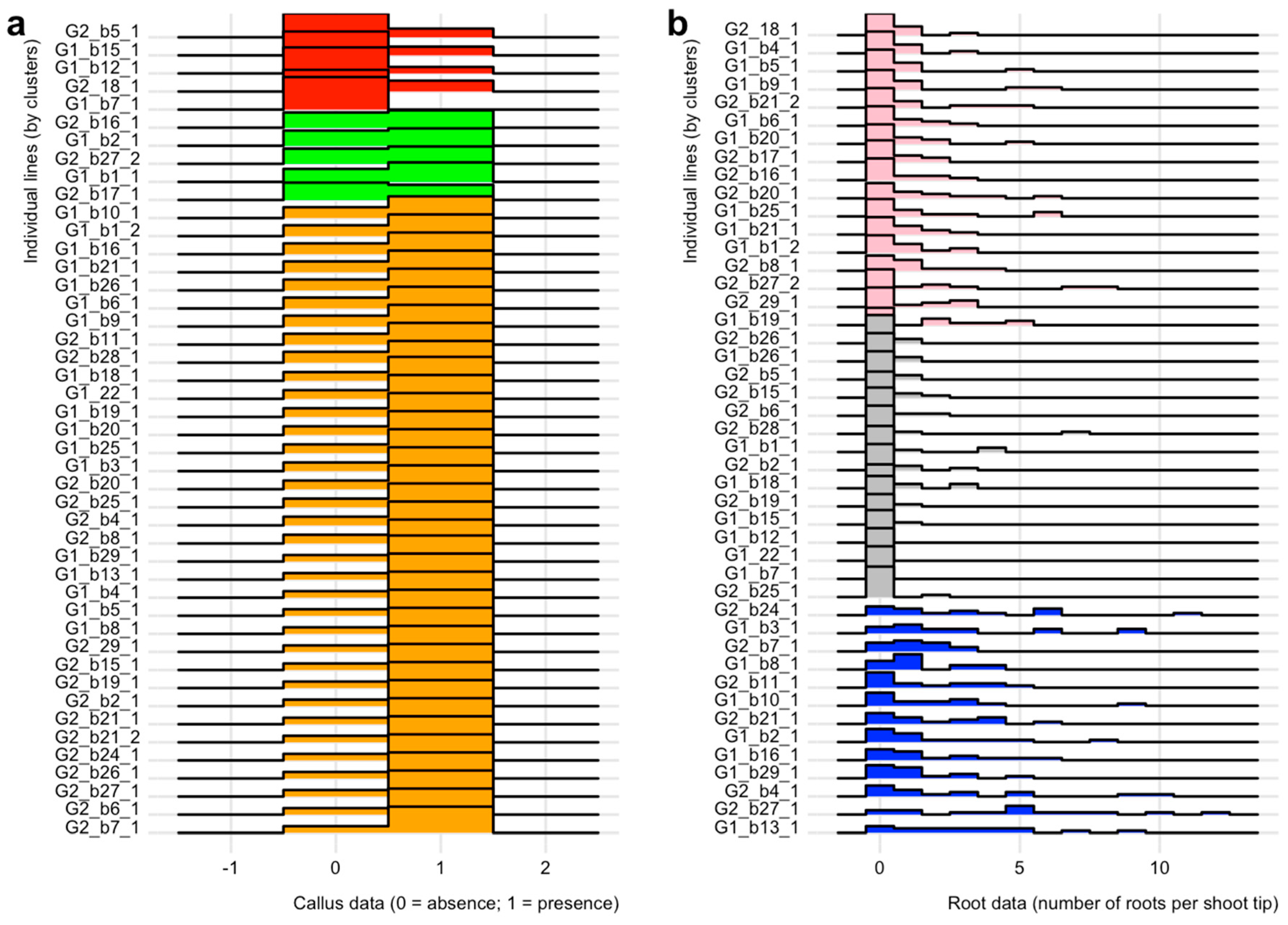
2.3. Effect of Growth Regulators on in vitro Rooting of Shoot Tips
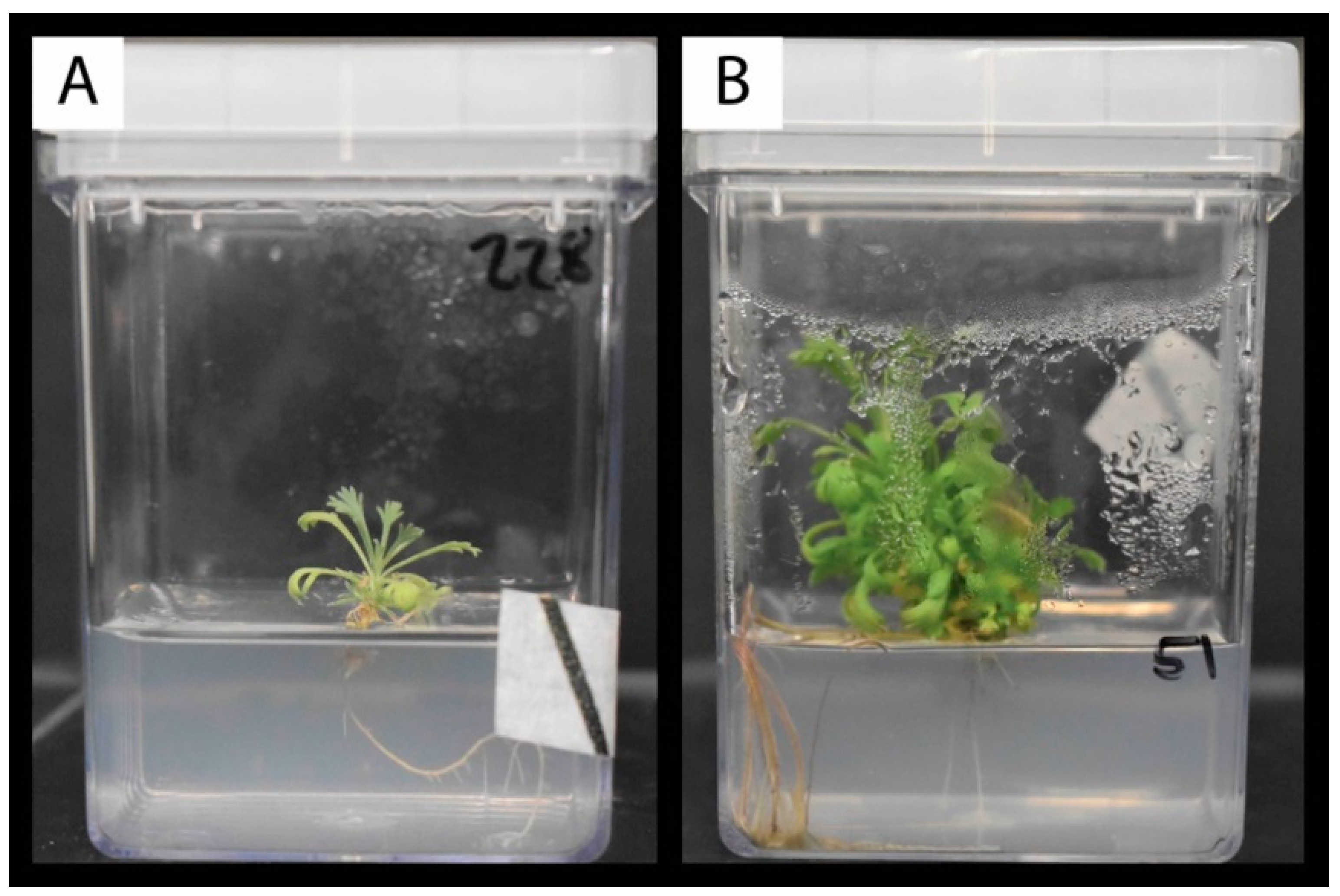
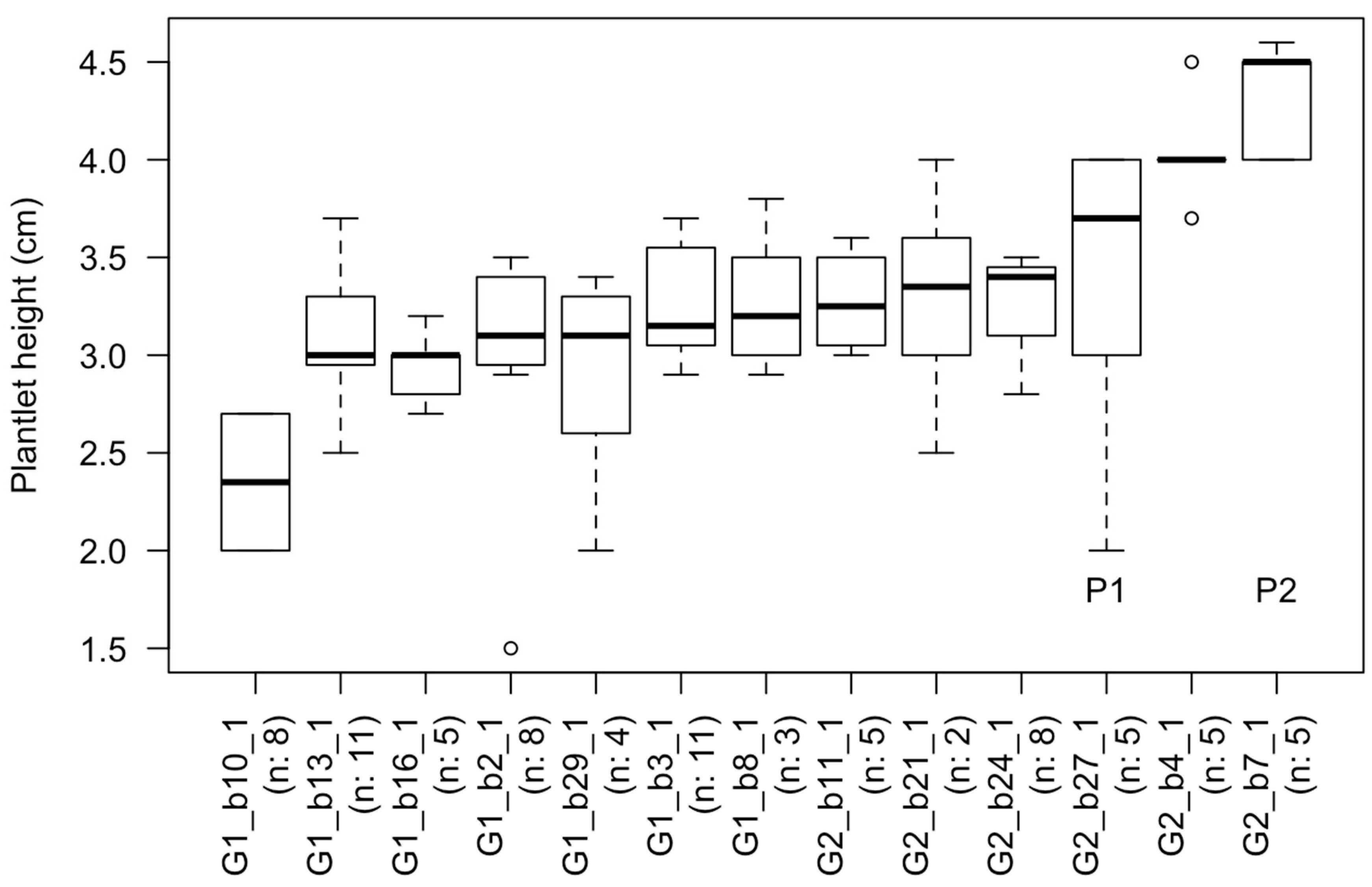
2.4. Survival and Plantlet Height of Rooted Shoot Tips Transplanted to Fresh Media
3. Discussion
3.1. Indirect Adventitious Rooting of Artemisia tridentata Shoot Tips
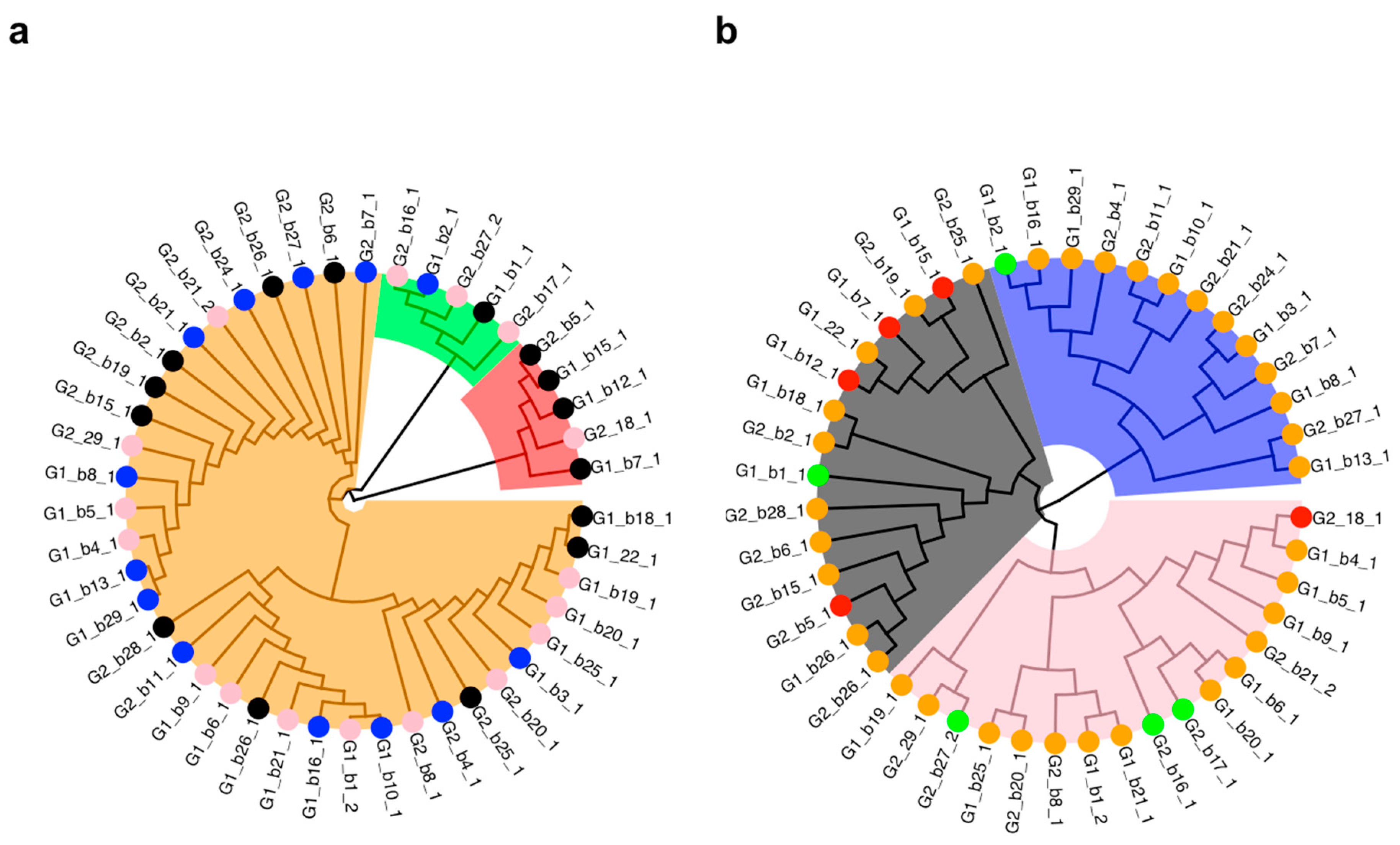
3.2. Basin Big Sagebrush Exhibits High Individual Rooting Plasticity
3.3. Growth Regulators Impact Rooting of Shoot Tips
3.4. Towards Selecting Individuals for in vitro Culture Propagation Program
3.5. Perspectives and Future Work
4. Materials and Methods
4.1. Plant Materials
4.2. Experimental Media Composition and Explant Preparation
4.3. Comparative Analyses on In Vitro Culture Data
4.4. Survival and Plantlet Height of Rooted Shoot Tips Transplanted to Fresh Media
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Williams, A.P.; Cook, E.R.; Smerdon, J.E.; Cook, B.I.; Abatzoglou, J.T.; Bolles, K.; Baek, S.H.; Badger, A.M.; Livneh, B. Large contribution from anthropogenic warming to an emerging North American megadrought. Science 2020, 368, 314–318. [Google Scholar] [CrossRef]
- McArthur, E. Ecology, Distribution, and Values of Sagebrush within the Intermountain Region. In Proceedings, Ecology and Management of Annual Rangelands; Monson, S.B., Kitchen, S.G., Eds.; General Technical Report; USDA Forest Service, Intermountain Research Station: Ogden, UT, USA, 1994; pp. 347–351. [Google Scholar]
- Green, J.S.; Flinders, J.T. Habitat and dietary relationships of the pygmy rabbit. Rangel. Ecol. Manag. 1980, 33, 136–142. [Google Scholar] [CrossRef]
- Prevéy, J.S.; Germino, M.J.; Huntly, N.J.; Inouye, R.S. Exotic plants increase and native plants decrease with loss of foundation species in sagebrush steppe. Plant Ecol. 2010, 207, 39–51. [Google Scholar] [CrossRef]
- Beetle, A.A. A Study of Sagebrush. The Section of Tridentatae of Artemisia; Bulletin; Wyoming Agricultural Experimental Station: Laramie, WY, USA, 1960; Volume 368, p. 83. [Google Scholar]
- Liang, Z.; Gupta, S.K.; Yeh, C.-T.; Zhang, Y.; Ngu, D.W.; Kumar, R.; Patil, H.T.; Mungra, K.D.; Yadav, D.V.; Rathore, A.; et al. Phenotypic data from inbred parents can improve genomic prediction in pearl millet hybrids. G3 (Bethesda) 2018, 8, 2513–2522. [Google Scholar] [CrossRef]
- Hoffman, A.M.; Smith, M.D. Thinking inside the box: Tissue culture for plant propagation in a key ecological species, Andropogon gerardii. Am. J. Plant Sci. 2018, 9, 1987–2003. [Google Scholar] [CrossRef][Green Version]
- McArthur, E.D.; Welch, B.L.; Sanderson, S.C. Natural and artificial hybridization between big sagebrush (Artemisia tridentata) subspecies. J. Hered. 1988, 79, 268–276. [Google Scholar] [CrossRef]
- McKey, D.; Elias, M.; Pujol, B.; Duputié, A. The evolutionary ecology of clonally propagated domesticated plants. New Phytol. 2010, 186, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Skirvin, R.M.; McPheeters, K.D.; Norton, M. Sources and frequency of somaclonal variation. HortScience 1994, 29, 1232–1237. [Google Scholar] [CrossRef]
- Vyskot, B.; JáRa, Z. Clonal propagation of cacti through axillary buds in vitro. HortScience 1984, 59, 449–452. [Google Scholar] [CrossRef]
- Larkin, P.J.; Scowcroft, W.R. Somaclonal variation—A novel source of variability from cell cultures for plant improvement. Theoret. Appl. Genet. 1981, 60, 197–214. [Google Scholar] [CrossRef]
- McArthur, E.D.; Sanderson, S.C. Cytogeography and chromosome evolution of subgenus Tridentatae of Artemisia (Asteraceae). Am. J. Bot. 1999, 86, 1754–1775. [Google Scholar] [CrossRef] [PubMed]
- Richardson, B.A.; Page, J.T.; Baigain, P.; Sanderson, S.C.; Udall, J.A. Deep sequencing of amplicons reveals widespread intraspecific hybridization and multiple origins of polyploidy in big sagebrush (Artemisia tridentata; Asteraceae). Am. J. Bot. 2012, 99, 1962–1975. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, H.A.; DaMATTA, F.M.; Chaves, A.R.M.; Loureiro, M.E.; Ducatti, C. Drought tolerance is associated with rooting depth and stomatal control of water use in clones of Coffea canephora. Ann. Bot. 2005, 96, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Jinagool, W.; Rattanawong, R.; Sangsing, K.; Barigah, T.S.; Gay, F.; Cochard, H.; Kasemsap, P.; Herbette, S. Clonal variability for vulnerability to cavitation and other drought-related traits in Hevea brasiliensis. J. Plant Hydraul. 2015, 2, e001. [Google Scholar] [CrossRef]
- de Freitas Guedes, F.A.; Nobres, P.; Rodrigues Ferreira, D.C.; Menezes-Silva, P.E.; Ribeiro-Alves, M.; Correa, R.L.; DaMatta, F.M.; Alves-Ferreira, M. Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ. Exp. Bot. 2018, 147, 220–233. [Google Scholar] [CrossRef]
- Turi, C.E.; Axwik, K.E.; Murch, S.J. In vitro conservation, phytochemistry, and medicinal activity of Artemisia tridentata Nutt: Metabolomics as a hypothesis generating tool for plant tissue culture. Plant Growth Regul. 2014, 74, 239–250. [Google Scholar] [CrossRef]
- Alvarez-Cordero, E.; McKell, C.M. Stem cutting propagation of big sagebrush (Artemisia tridentata Nutt.). Rangel. Ecol. Manag. 1979, 32, 141–143. [Google Scholar] [CrossRef]
- Sujatha, G.; Kumari, B.R. Effect of phytohormones on micropropagation of Artemisia vulgaris L. Acta Physiol. Plant. 2007, 29, 189–195. [Google Scholar] [CrossRef]
- Alok, A.; Shukla, V.; Pala, Z.; Kumar, J.; Kudale, S.; Desai, N. In vitro regeneration and optimization of factors affecting Agrobacterium mediated transformation in Artemisia pallens, an important medicinal plant. Physiol. Mol. Bio. Plants 2016, 22, 261–269. [Google Scholar] [CrossRef]
- Dangash, A.; Ram, M.; Riturai, N.; Bharillya, A.; Misra, H.; Pandya, N.; Jain, D. In vitro selection and hormonal regulation in cell culture of Artemisia annua L. plant. JSM Cell Dev. Biol. 2015, 3, 1013. [Google Scholar]
- Barron, R. In Vitro Regeneration, Rooting, and Cloning of Artemisia tridentata. Master Thesis, Boise State University, Boise, Idaho, USA, 2020. [Google Scholar]
- GitHub. Available online: https://github.com/svenbuerki/Sagebrush_rooting_in_vitro_prop (accessed on 25 November 2020).
- Sagebrush Propagation. Available online: https://svenbuerki.github.io/Sagebrush_rooting_in_vitro_prop/ (accessed on 26 October 2020).
- de Klerk, G.-J.; van der Krieken, W.; de Jong, J.C. Review the formation of adventitious roots: New concepts, new possibilities. In Vitro Cell Dev. Biol.-Plant 1999, 35, 189–199. [Google Scholar] [CrossRef]
- Yu, J.; Liu, W.; Liu, J.; Qin, P.; Xu, L. Auxin control of root organogenesis from callus in tissue culture. Front. Plant Sci. 2017, 8, 1385. [Google Scholar] [CrossRef] [PubMed]
- Mok, D.W.; Mok, M.C. Cytokinin Metabolism and Action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Sugimoto, K.; Iwase, A. Plant callus: Mechanisms of induction and repression. Plant Cell 2013, 25, 3159–3173. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Iwase, A.; Rymen, B.; Lambolez, A.; Kojima, M.; Takebayashi, Y.; Heyman, J.; Watanabe, S.; Seo, M.; De Veylder, L.; et al. Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 2017, 175, 1158–1174. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, 4. [Google Scholar] [CrossRef]
- Gopu, C.; Chakilam, C.S.; Chirumamilla, P.; Vankudoth, S.; Taduri, S. Rapid in vitro adventitious rooting and proliferation by leaf and nodal cultures of Momordica cymbalaria Fenzl. J. Appl. Biol. Biotechnol. 2020, 80, 103–107. [Google Scholar] [CrossRef]
- Druege, U.; Zerche, S.; Kadner, R. Nitrogen- and storage-affected carbohydrate partitioning in high-light-adapted Pelargonium cuttings in relation to survival and adventitious root formation under low light. Ann. Bot. 2004, 94, 831–842. [Google Scholar] [CrossRef]
- Ruedell, C.M.; de Almeida, M.R.; Schwambach, J.; Posenato, C.F.; Fett-Neto, A.G. Pre and post-severance effects of light quality on carbohydrate dynamics and microcutting adventitious rooting of two Eucalyptus species of contrasting recalcitrance. Plant Growth Regul. 2013, 69, 235–245. [Google Scholar] [CrossRef]
- Geiss, G.; Gutierrez, L.; Bellini, C. Adventitious root formation: New insights and perspectives. Ann. Plant Rev. Online 2018, 15, 127–156. [Google Scholar] [CrossRef]
- Karron, J.D.; Marshall, D.L. Fitness consequences of multiple paternity in wild radish, Raphanus sativus. Evolution 1990, 44, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Kibbler, H.; Johnston, M.E.; Williams, R.R. Adventitious root formation in cuttings of Backhousia citriodora F. Muell: 1. Plant genotype, juvenility and characteristics of cuttings. Sci. Hortic. 2004, 102, 133–143. [Google Scholar] [CrossRef]
- Metaxas, D.J.; Syros, T.D.; Yupsanis, T.; Economou, A.S. Peroxidases during adventitious rooting in cuttings of Arbutus unedo and Taxus baccata as affected by plant genotype and growth regulator treatment. Plant Growth Regul. 2004, 44, 257–266. [Google Scholar] [CrossRef]
- Sorin, C.; Negroni, L.; Balliau, T.; Corti, H.; Jacquemot, M.P.; Davanture, M.; Sandberg, G.; Zivy, M.; Bellini, C. Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol. 2006, 140, 349–364. [Google Scholar] [CrossRef]
- Mishra, P.; Roggen, A.; Ljung, K.; Albani, M.C. Natural variation in adventitious rooting in the Alpine perennial Arabis alpina. Plants 2020, 9, 184. [Google Scholar] [CrossRef]
- Garcia, S.; Canela, M.Á.; Garnatje, T.; McArthur, E.D.; Pellicer, J.; Sanderson, S.C.; Vallès, J. Evolutionary and ecological implications of genome size in the North American endemic sagebrushes and allies (Artemisia, Asteraceae). Biol. J. Linn. Soc. 2008, 94, 631–649. [Google Scholar] [CrossRef]
- Torrell, M.; Vallès, J. Genome size in 21 Artemisia L. species (Asteraceae, Anthemideae): Systematic, evolutionary, and ecological implications. Genome 2001, 44, 231–238. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, L.; Liao, Z.; Wang, S.; Yan, T.; Shi, P.; Liu, M.; Fu, X.; Pan, Q.; Wang, Y.; et al. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol. Plant 2018, 11, 776–788. [Google Scholar] [CrossRef]
- Chaney, L.; Richardson, B.A.; Germino, M.J. Climate drives adaptive genetic responses associated with survival in big sagebrush (Artemisia tridentata). Evol. Appl. 2017, 10, 313–322. [Google Scholar] [CrossRef]
- McAndrews, J.H.; Wright, H.E. Modern pollen rain across the Wyoming basins and the northern Great Plains (U.S.A.). Rev. Palaeobot. Palynol. 1969, 9, 17–43. [Google Scholar] [CrossRef]
- Laursen, S.C.; Reiners, W.A.; Kelly, R.D.; Gerow, K.G. Pollen dispersal by Artemisia tridentata (Asteraceae). Int. J. Biometeorol. 2007, 51, 465. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth wand bioassays with Tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Gamborg, O.L. Aromatic metabolism in plants II. Enzymes of the skikimate pathway in suspension cultures of plant cells. Can. J. Biochem. 1966, 44, 791–799. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: http://www.R-project.org/ (accessed on 20 November 2020).
- Wilke, C.O. ggridges: Ridgeline Plots in ggplot2. R Package Version 0.5.2. 2020. Available online: https://wilkelab.org/ggridges (accessed on 20 November 2020).
| Growth Regulator | Concentration (mg/L) | Response (%) |
|---|---|---|
| Control | - | 2.9 ± 1.5 b |
| IBA | 0.5 | 75.5 ± 7.5 a |
| IBA | 1 | 84.4 ± 7.9 a |
| NAA | 0.5 | 81.5 ± 7.8 a |
| NAA | 1 | 87.4 ± 8.04 a |
| Growth Regulator | Concentration (mg/L) | Response (%) | Av. No. of Roots |
|---|---|---|---|
| Control | - | 8.9 ± 2.5 b | 0.13 ± 0.04 c |
| IBA | 0.5 | 47.4 ± 5.9 a | 1.37 ± 0.20 ab |
| IBA | 1 | 60.0 ± 6.6 a | 1.92± 0.26 a |
| NAA | 0.5 | 40.7 ± 5.5 a | 0.74 ± 0.12 b |
| NAA | 1 | 40.0 ± 5.4 a | 1.00 ± 0.15 b |
| Cluster | Growth Regulator | Concentration (mg/L) | Response (%) | Av. No. of Roots |
|---|---|---|---|---|
| grey | Control | - | 0 ± 0 | 0 ± 0 |
| grey | IBA | 0.5 | 11.1 ± 4.2 | 0.22 ± 0.10 |
| grey | IBA | 1 | 20.0 ± 7.1 | 0.44 ± 0.21 |
| grey | NAA | 0.5 | 13.3 ± 6.3 | 0.16 ± 0.08 |
| grey | NAA | 1 | 13.3 ± 5.4 | 0.22 ± 0.13 |
| pink | Control | - | 0 ± 0 | 0 ± 0 |
| pink | IBA | 0.5 | 49.0 ± 8.1 | 1.16 ± 0.19 |
| pink | IBA | 1 | 74.5 ± 6.1 | 1.82 ± 0.32 |
| pink | NAA | 0.5 | 39.2 ± 6.5 | 0.63 ± 0.12 |
| pink | NAA | 1 | 43.1 ± 4.7 | 0.86 ± 0.2 |
| blue | Control | - | 30.8 ± 8.8 | 0.62 ± 0.23 |
| blue | IBA | 0.5 | 87.2 ± 4.7 | 2.97 ± 0.37 |
| blue | IBA | 1 | 87.2 ± 6.0 | 3.59 ± 0.45 |
| blue | NAA | 0.5 | 74.4 ± 9.4 | 1.74 ± 0.37 |
| blue | NAA | 1 | 66.7 ± 6.5 | 2.28 ± 0.33 |
| Cluster | N. Individual Lines | N. Shoot Tips | Growth Regulator | Concentration (mg/L) | Survival (%) | Height (cm) |
|---|---|---|---|---|---|---|
| grey | 0 | 0 | Control | - | NA | NA |
| grey | 5 | 5 | IBA | 0.5 | 40.00 | 1.2+/−1.64 |
| grey | 4 | 6 | IBA | 1 | 33.33 | 0.9+/−1.59 |
| grey | 4 | 6 | NAA | 0.5 | 66.67 | 2.27+/−1.94 |
| grey | 8 | 10 | NAA | 1 | 40.00 | 1.32+/−1.71 |
| TOTAL | 12 | 27 | ||||
| pink | 0 | 0 | Control | - | NA | NA |
| pink | 14 | 24 | IBA | 0.5 | 37.50 | 1.04+/−1.39 |
| pink | 16 | 35 | IBA | 1 | 60.00 | 1.82+/−1.59 |
| pink | 13 | 19 | NAA | 0.5 | 42.11 | 1.28+/−1.59 |
| pink | 15 | 26 | NAA | 1 | 53.85 | 1.56+/−1.57 |
| TOTAL | 16 | 104 | ||||
| blue | 8 | 12 | Control | - | 58.33 | 1.97+/−1.8 |
| blue | 13 | 32 | IBA | 0.5 | 65.62 | 2.27+/−1.7 |
| blue | 13 | 34 | IBA | 1 | 64.71 | 1.95+/−1.54 |
| blue | 12 | 27 | NAA | 0.5 | 37.04 | 1.21+/−1.64 |
| blue | 13 | 29 | NAA | 1 | 68.97 | 2.29+/−1.64 |
| TOTAL | 13 | 134 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barron, R.; Martinez, P.; Serpe, M.; Buerki, S. Development of an In Vitro Method of Propagation for Artemisia tridentata subsp. tridentata to Support Genome Sequencing and Genotype-by-Environment Research. Plants 2020, 9, 1717. https://doi.org/10.3390/plants9121717
Barron R, Martinez P, Serpe M, Buerki S. Development of an In Vitro Method of Propagation for Artemisia tridentata subsp. tridentata to Support Genome Sequencing and Genotype-by-Environment Research. Plants. 2020; 9(12):1717. https://doi.org/10.3390/plants9121717
Chicago/Turabian StyleBarron, Rachael, Peggy Martinez, Marcelo Serpe, and Sven Buerki. 2020. "Development of an In Vitro Method of Propagation for Artemisia tridentata subsp. tridentata to Support Genome Sequencing and Genotype-by-Environment Research" Plants 9, no. 12: 1717. https://doi.org/10.3390/plants9121717
APA StyleBarron, R., Martinez, P., Serpe, M., & Buerki, S. (2020). Development of an In Vitro Method of Propagation for Artemisia tridentata subsp. tridentata to Support Genome Sequencing and Genotype-by-Environment Research. Plants, 9(12), 1717. https://doi.org/10.3390/plants9121717







