Arabidopsis Response to Inhibitor of Cytokinin Degradation INCYDE: Modulations of Cytokinin Signaling and Plant Proteome
Abstract
1. Introduction
2. Results
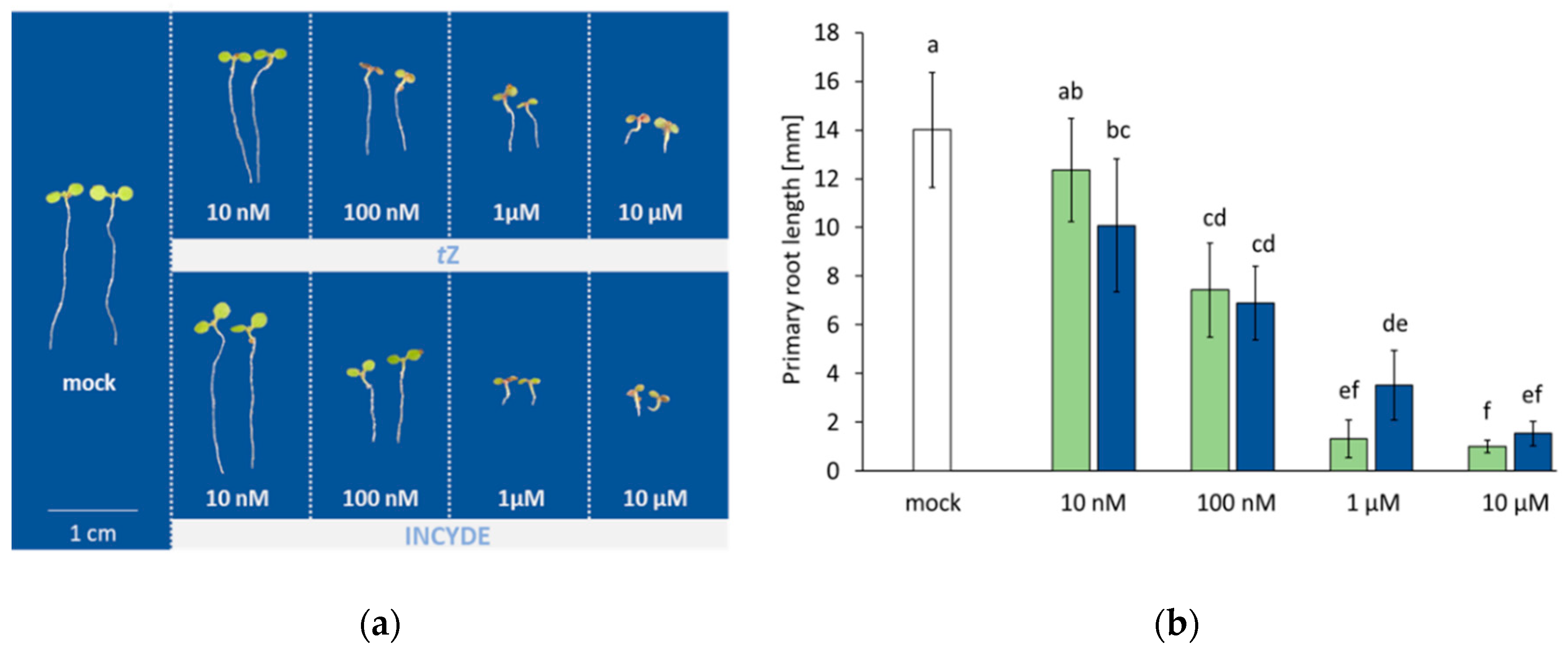
2.1. Root Growth in Response to tZ and INCYDE Treatment was not Significantly Different
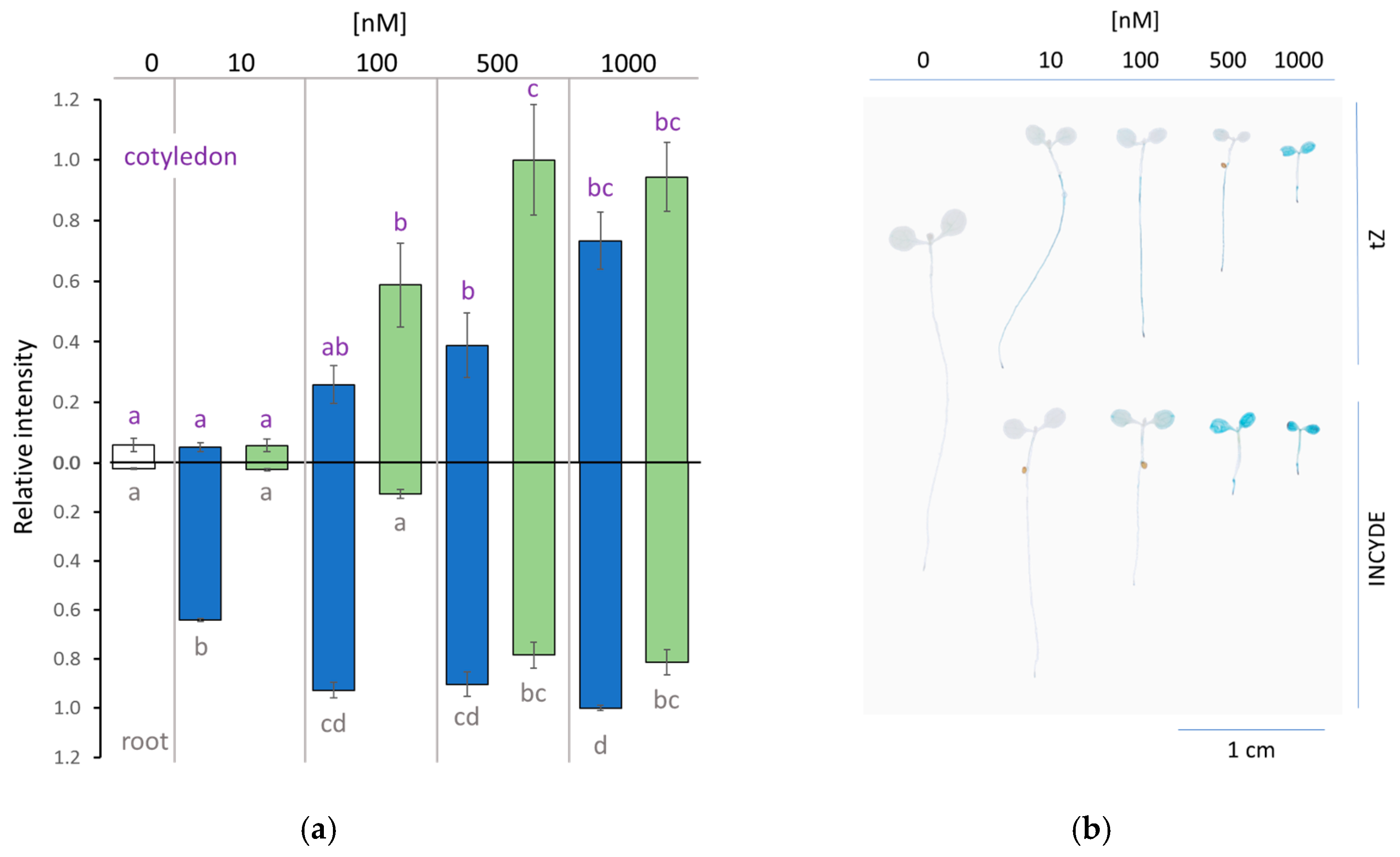
2.2. INCYDE Treatment Elicited Distinct Spatial Distribution of Cytokinin Signaling
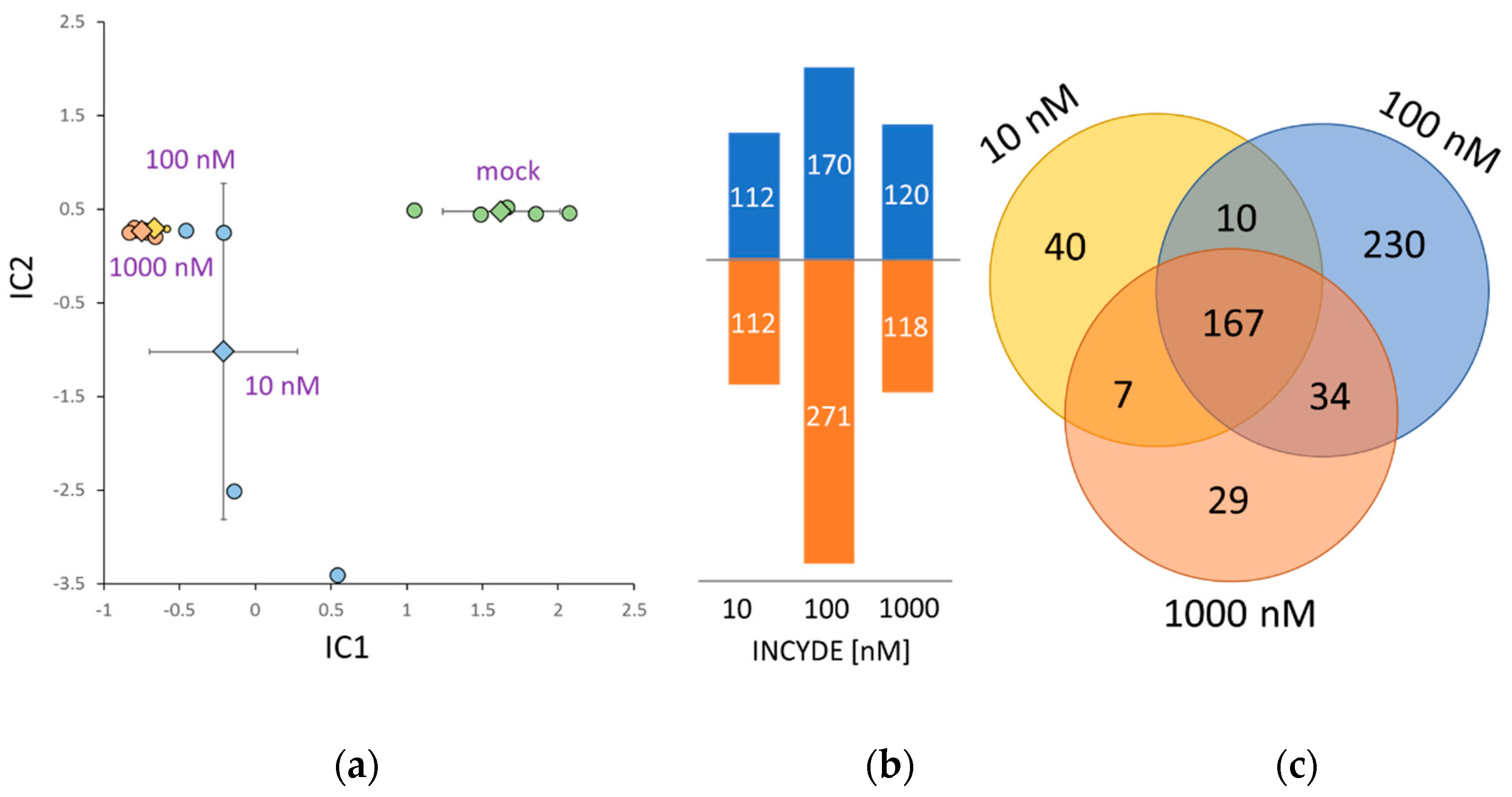
2.3. Early INCYDE Response Proteins of Arabidopsis Seedlings Highlight Similarity to Exogenous tZ Treatment
2.4. Correlation between INCYDE Concentration and Protein Abundance
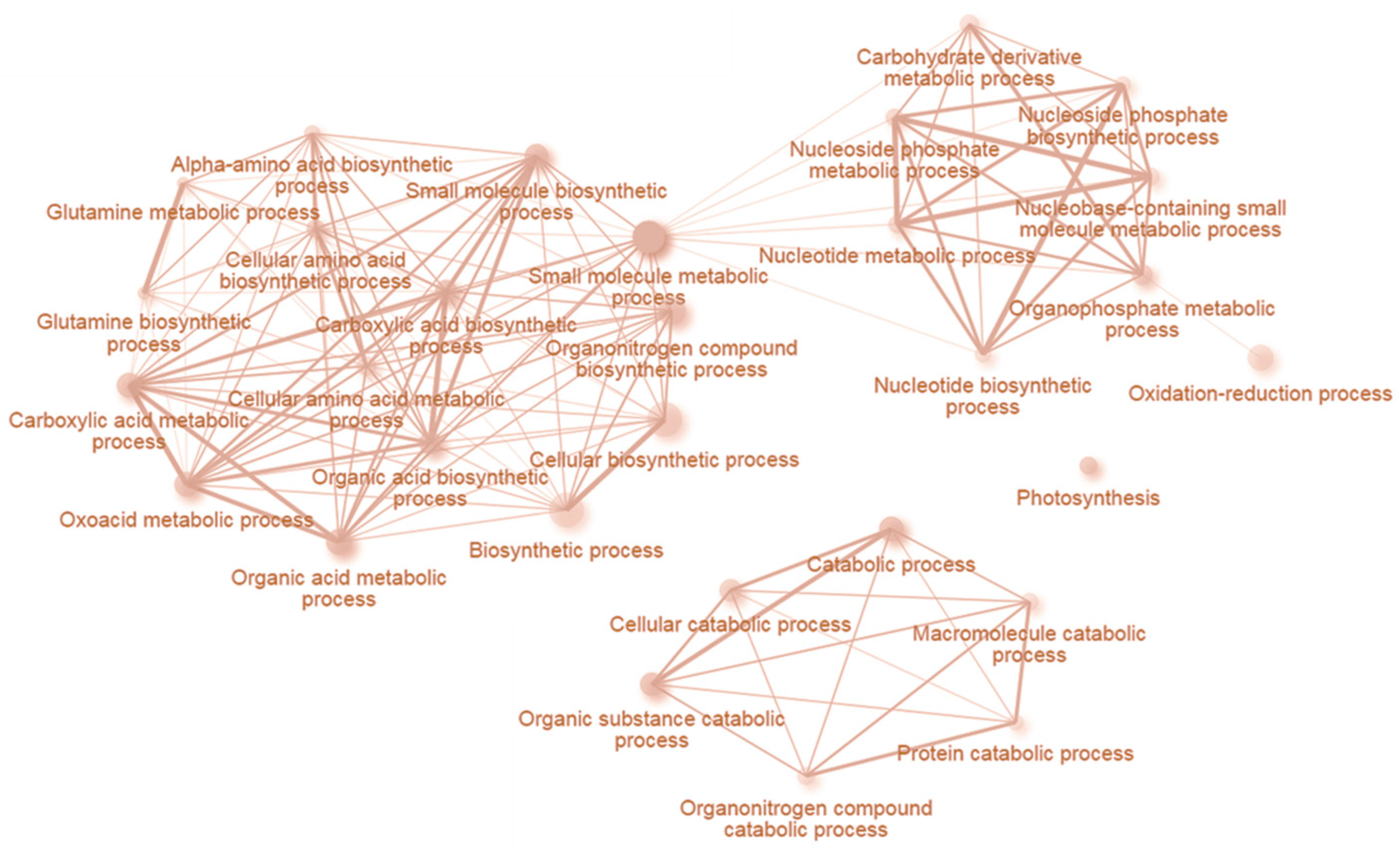
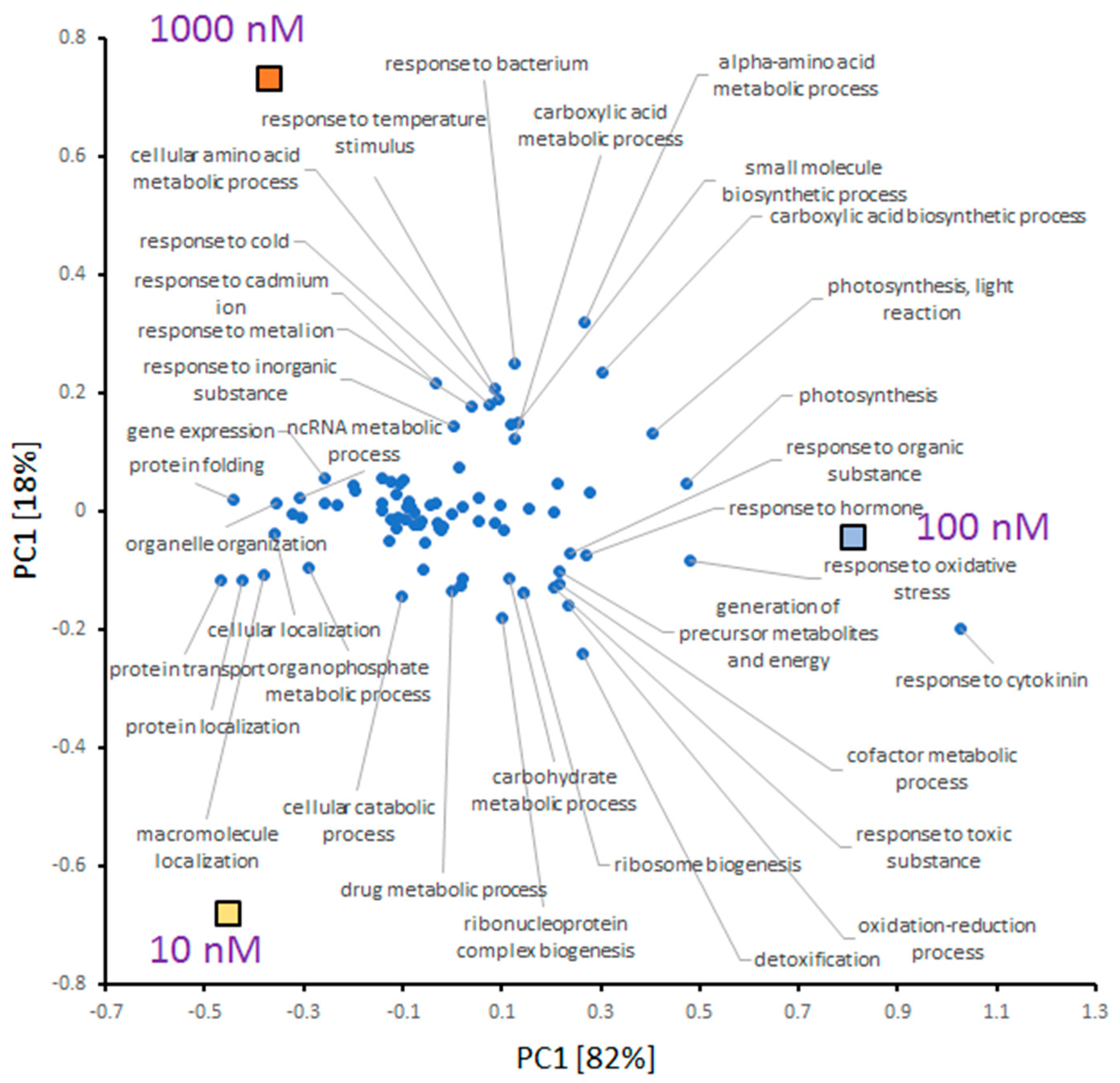
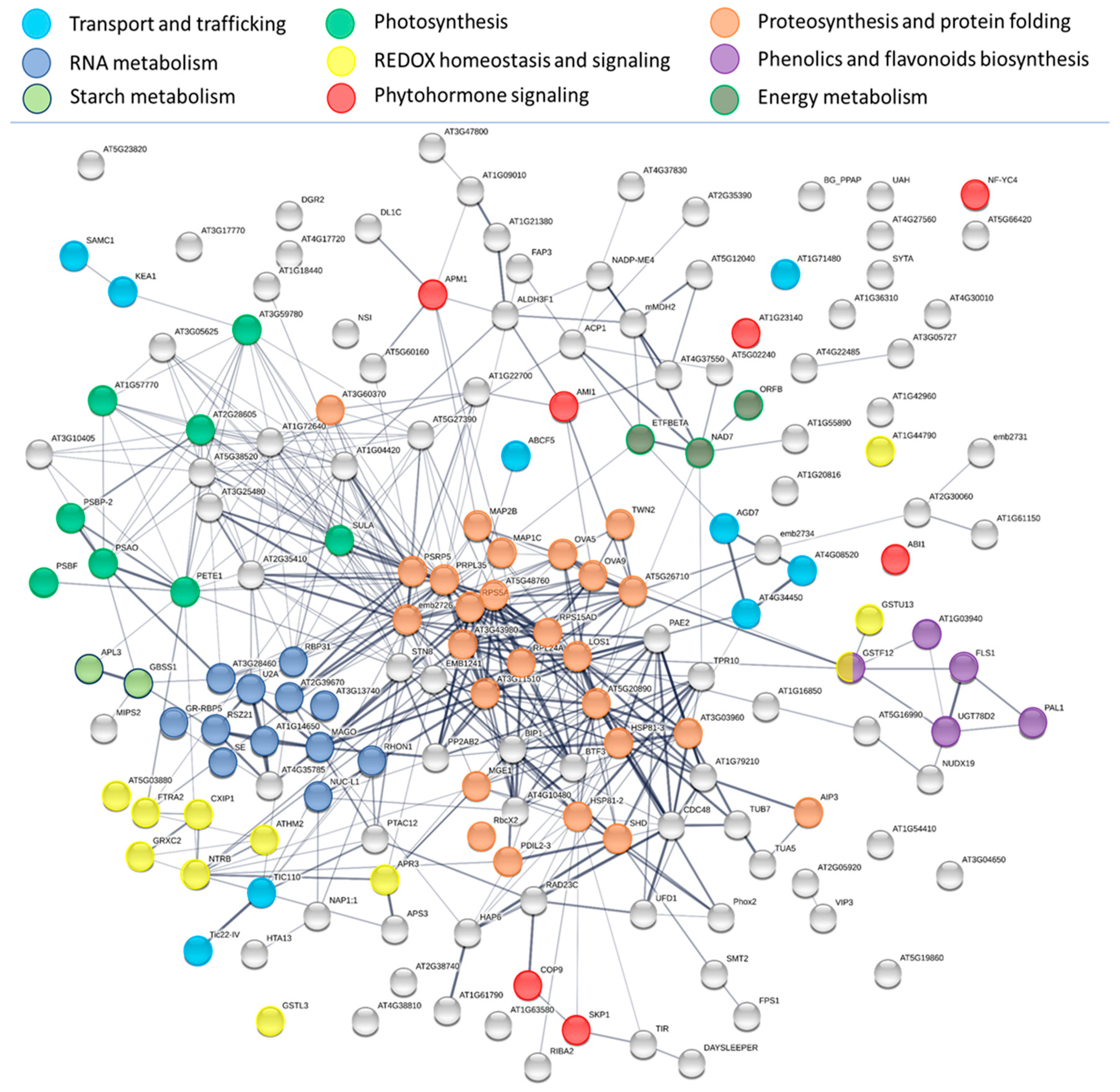
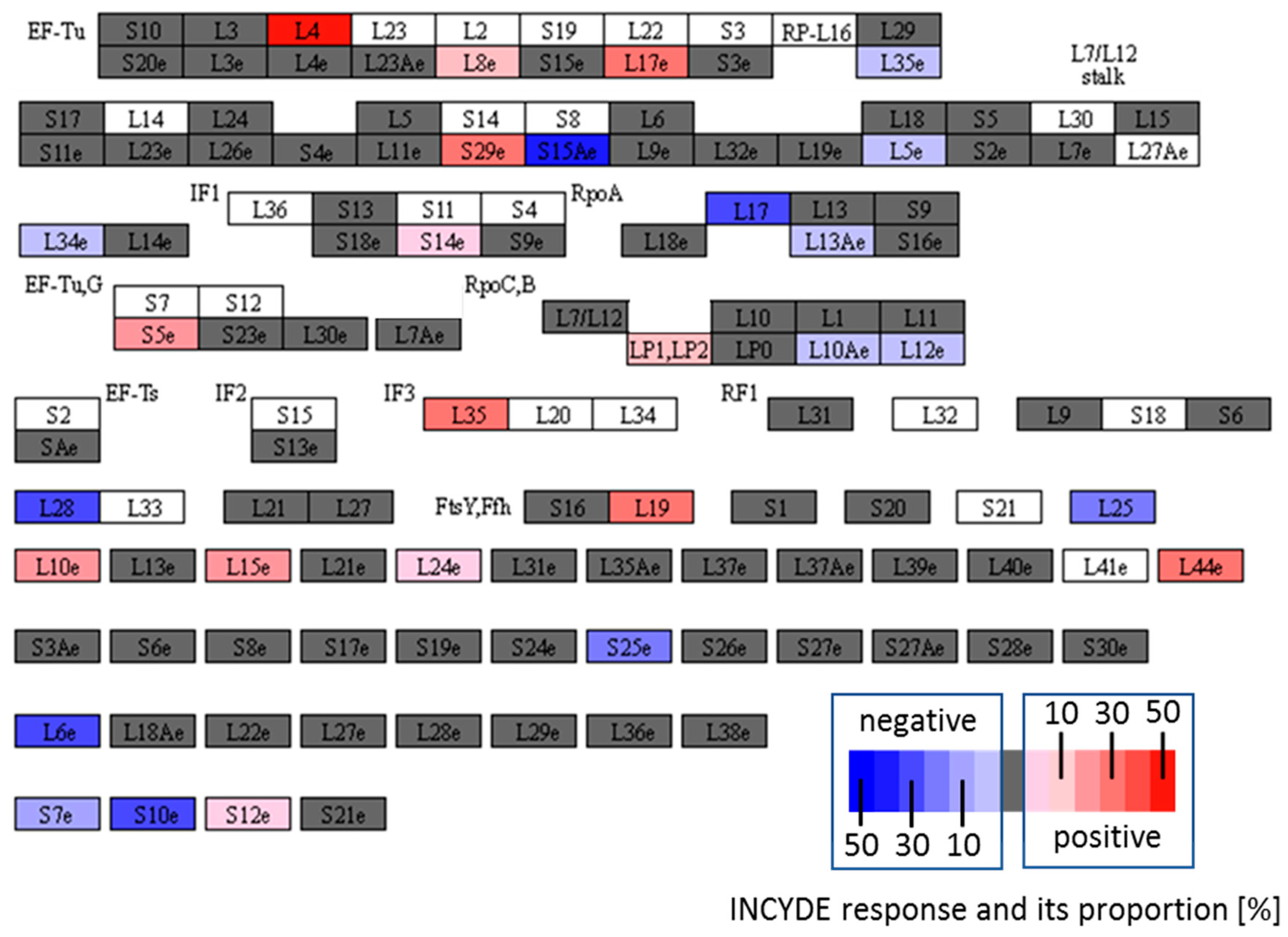
2.5. INCYDE Response Proteins Modulate Diverse Metabolic Processes
3. Discussion
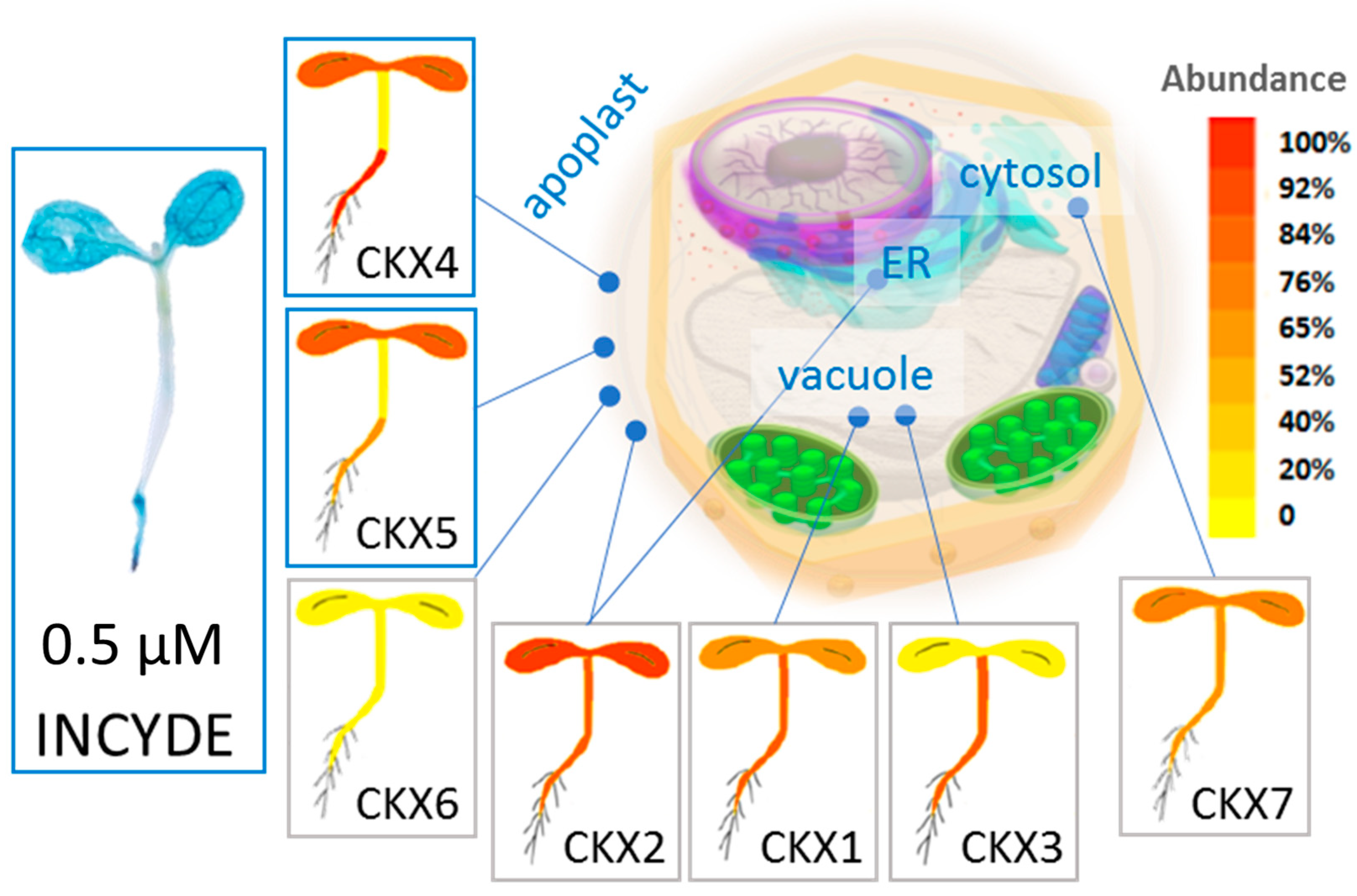
3.1. Cytokinin Dehydrogenase Isoforms Play a Key Role in the Contrasting Response between tZ and INCYDE
3.2. Cytokinin Response Targets Ribosomal Proteins
3.3. Positive Effect of INCYDE Could Coincide with Attenuated Stress Response
4. Materials and Methods
4.1. Plant Material
4.2. Histochemical Analysis
4.3. Proteome Analysis
4.4. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant. Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Pavlů, J.; Novák, J.; Koukalová, V.; Luklová, M.; Brzobohatý, B.; Černý, M. Cytokinin at the Crossroads of Abiotic Stress Signalling Pathways. Int. J. Mol. Sci. 2018, 19, 2450. [Google Scholar] [CrossRef]
- Zwack, P.J.; Rashotte, A.M. Interactions between cytokinin signalling and abiotic stress responses. J. Exp. Bot. 2015, 66, 4863–4871. [Google Scholar] [CrossRef]
- Joshi, S.; Choukimath, A.; Isenegger, D.; Panozzo, J.; Spangenberg, G.; Kant, S. Improved Wheat Growth and Yield by Delayed Leaf Senescence Using Developmentally Regulated Expression of a Cytokinin Biosynthesis Gene. Front. Plant Sci. 2019, 10, 1285. [Google Scholar] [CrossRef] [PubMed]
- Zalewski, W.; Galuszka, P.; Gasparis, S.; Orczyk, W.; Nadolska-Orczyk, A. Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J. Exp. Bot. 2010, 61, 1839–1851. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Dobrá, J.; Černý, M.; Štorchová, H.; Dobrev, P.; Skalák, J.; Jedelský, P.L.; Lukšanová, H.; Gaudinová, A.; Pešek, B.; Malbecka, J.; et al. The impact of heat stress targeting on the hormonal and transcriptomic response in Arabidopsis. Plant Sci. 2015, 231, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Skalák, J.; Černý, M.; Jedelský, P.; Dobrá, J.; Ge, E.; Novák, J.; Hronková, M.; Dobrev, P.; Vanková, R.; Brzobohatý, B. Stimulation of ipt overexpression as a tool to elucidate the role of cytokinins in high temperature responses of Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, M.E.; Albacete, A.; Martínez-Andújar, C.; Acosta, M.; Romero-Aranda, R.; Dodd, I.C.; Lutts, S.; Pérez-Alfocea, F. Hormonal changes during salinity-induced leaf senescence in tomato (Solanum lycopersicum L.). J. Exp. Bot. 2008, 59, 3039–3050. [Google Scholar] [CrossRef]
- Iqbal, M.; Ashraf, M.; Jamil, A. Seed enhancement with cytokinins: Changes in growth and grain yield in salt stressed wheat plants. Plant Growth Regul. 2006, 50, 29–39. [Google Scholar] [CrossRef]
- Kariali, E.; Mohapatra, P.K. Hormonal regulation of tiller dynamics in differentially-tillering rice cultivars. Plant Growth Regul. 2007, 53, 215–223. [Google Scholar] [CrossRef]
- Koprna, R.; De Diego, N.; Dundálková, L.; Spíchal, L. Use of cytokinins as agrochemicals. Bioorgan. Med. Chem. 2016, 24, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Heyl, A.; Werner, T.; Schmülling, T. Cytokinin metabolism and signal transduction. In Annual Plant Reviews Online; John Wiley & Sons, Ltd.: Chichester, UK, 2018; pp. 93–123. [Google Scholar]
- Hallmark, H.T.; Černý, M.; Brzobohatý, B.; Rashotte, A.M. trans-Zeatin-N-glucosides have biological activity in Arabidopsis thaliana. PLoS ONE 2020, 15, e0232762. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-Deficient Transgenic Arabidopsis Plants Show Multiple Developmental Alterations Indicating Opposite Functions of Cytokinins in the Regulation of Shoot and Root Meristem Activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef]
- Frebort, I.; Kowalska, M.; Hluska, T.; Frebortova, J.; Galuszka, P. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2017, 45, D1064–D1074. [Google Scholar] [CrossRef]
- Galuszka, P.; Popelková, H.; Werner, T.; Frébortová, J.; Pospíšilová, H.; Mik, V.; Köllmer, I.; Schmülling, T.; Frébort, I. Biochemical characterization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. J. Plant Growth Regul. 2007, 26, 255–267. [Google Scholar] [CrossRef]
- Werner, T.; Holst, K.; Pörs, Y.; Guivarc’h, A.; Mustroph, A.; Chriqui, D.; Grimm, B.; Schmülling, T. Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J. Exp. Bot. 2008, 59, 2659–2672. [Google Scholar] [CrossRef]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef]
- Li, Y.; Song, G.; Gao, J.; Zhang, S.; Zhang, R.; Li, W.; Chen, M.; Liu, M.; Xia, X.; Risacher, T.; et al. Enhancement of grain number per spike by RNA interference of cytokinin oxidase 2 gene in bread wheat. Hereditas 2018, 155, 33. [Google Scholar] [CrossRef]
- Zatloukal, M.; Gemrotová, M.; Doležal, K.; Havlíček, L.; Spíchal, L.; Strnad, M. Novel potent inhibitors of A. thaliana cytokinin oxidase/dehydrogenase. Bioorganic Med. Chem. 2008, 16, 9268–9275. [Google Scholar] [CrossRef] [PubMed]
- Aremu, A.O.; Masondo, N.A.; Sunmonu, T.O.; Kulkarni, M.G.; Zatloukal, M.; Spichal, L.; Doležal, K.; Van Staden, J. A novel inhibitor of cytokinin degradation (INCYDE) influences the biochemical parameters and photosynthetic apparatus in NaCl-stressed tomato plants. Planta 2014, 240, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Prerostova, S.; Dobrev, P.I.; Kramna, B.; Gaudinova, A.; Knirsch, V.; Spichal, L.; Zatloukal, M.; Vankova, R. Heat Acclimation and Inhibition of Cytokinin Degradation Positively Affect Heat Stress Tolerance of Arabidopsis. Front. Plant Sci. 2020, 11, 87. [Google Scholar] [CrossRef] [PubMed]
- Gemrotová, M.; Kulkarni, M.G.; Stirk, W.A.; Strnad, M.; Van Staden, J.; Spíchal, L. Seedlings of medicinal plants treated with either a cytokinin antagonist (PI-55) or an inhibitor of cytokinin degradation (INCYDE) are protected against the negative effects of cadmium. Plant Growth Regul. 2013, 71, 137–145. [Google Scholar] [CrossRef]
- Reusche, M.; Klásková, J.; Thole, K.; Truskina, J.; Novák, O.; Janz, D.; Strnad, M.; Spíchal, L.; Lipka, V.; Teichmann, T. Stabilization of cytokinin levels enhances arabidopsis resistance against Verticillium longisporum. Mol. Plant-Microbe Interact. 2013, 26, 850–860. [Google Scholar] [CrossRef]
- Růzǐčka, K.; Šimášková, M.; Duclercq, J.; Petrášek, J.; Zažímalová, E.; Simon, S.; Friml, J.; Van Montagu, M.C.E.; Benková, E. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 2009, 106, 4284–4289. [Google Scholar] [CrossRef]
- Deikman, J.; Hammer, P.E. Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol. 1995, 108, 47–57. [Google Scholar] [CrossRef]
- Dalal, J.; Lewis, D.R.; Tietz, O.; Brown, E.M.; Brown, C.S.; Palme, K.; Muday, G.K.; Sederoff, H.W. ROSY1, a novel regulator of gravitropic response is a stigmasterol binding protein. J. Plant Physiol. 2016, 196–197, 28–40. [Google Scholar] [CrossRef]
- Reinbothe, S.; Gray, J.; Rustgi, S.; Von Wettstein, D.; Reinbothe, C. Cell growth defect factor 1 is crucial for the plastid import of NADPH:protochlorophyllide oxidoreductase A in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 5838–5843. [Google Scholar] [CrossRef]
- Reboul, R.; Geserick, C.; Pabst, M.; Frey, B.; Wittmann, D.; Lütz-Meindl, U.; Léonard, R.; Tenhaken, R. Down-regulation of UDP-glucuronic acid biosynthesis leads to swollen plant cell walls and severe developmental defects associated with changes in pectic polysaccharides. J. Biol. Chem. 2011, 286, 39982–39992. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Berka, M.; Luklová, M.; Dufková, H.; Malých, V.; Novák, J.; Saiz-Fernández, I.; Rashotte, A.M.; Brzobohaty, B.; Cerny, M. Barley root proteome and metabolome in response to cytokinin and abiotic stimuli. Front. Plant Sci. 2020, 11, 1647. [Google Scholar] [CrossRef]
- Rodriguez, L.; Gonzalez-Guzman, M.; Diaz, M.; Rodrigues, A.; Izquierdo-Garcia, A.C.; Peirats-Llobet, M.; Fernandez, M.A.; Antoni, R.; Fernandez, D.; Marquez, J.A.; et al. C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in arabidopsis. Plant Cell 2014, 26, 4802–4820. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Galvão, R.M.; Li, M.; Burger, B.; Bugea, J.; Bolado, J.; Chory, J. Arabidopsis HEMERA/pTAC12 Initiates photomorphogenesis by phytochromes. Cell 2010, 141, 1230–1240. [Google Scholar] [CrossRef]
- Yang, Y.; Sage, T.L.; Liu, Y.; Ahmad, T.R.; Marshall, W.F.; Shiu, S.H.; Froehlich, J.E.; Imre, K.M.; Osteryoung, K.W. Clumped chloroplasts 1 is required for plastid separation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18530–18535. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. Von STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020. [Google Scholar] [CrossRef]
- Osugi, A.; Kojima, M.; Takebayashi, Y.; Ueda, N.; Kiba, T.; Sakakibara, H. Systemic transport of trans-zeatin and its precursor have differing roles in Arabidopsis shoots. Nat. Plants 2017, 3, 17112. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “electronic fluorescent pictograph” Browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Martinez-Seidel, F.; Beine-Golovchuk, O.; Hsieh, Y.C.; Kopka, J. Systematic review of plant ribosome heterogeneity and specialization. Front. Plant Sci. 2020, 11, 948. [Google Scholar] [CrossRef]
- Černý, M.; Kuklová, A.; Hoehenwarter, W.; Fragner, L.; Novák, O.; Rotková, G.; Jedelský, P.L.P.L.; Žáková, K.K.; Šmehilová, M.; Strnad, M.; et al. Proteome and metabolome profiling of cytokinin action in Arabidopsis identifying both distinct and similar responses to cytokinin down- and up-regulation. J. Exp. Bot. 2013, 64, 4193–4206. [Google Scholar] [CrossRef] [PubMed]
- Karunadasa, S.S.; Kurepa, J.; Shull, T.E.; Smalle, J.A. Cytokinin-induced protein synthesis suppresses growth and osmotic stress tolerance. New Phytol. 2020, 227, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, M.L.F.; Pezza, A.; Biarc, J.; Burlingame, A.L.; Casati, P. Plant L10 Ribosomal Proteins Have Different Roles during Development and Translation under Ultraviolet-B Stress. Plant Physiol. 2010, 153, 1878–1894. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Skirycz, A.; Vandenbroucke, K.; Clauw, P.; Maleux, K.; De Meyer, B.; Dhondt, S.; Pucci, A.; Gonzalez, N.; Hoeberichts, F.; Tognetti, V.B.; et al. Survival and growth of Arabidopsis plants given limited water are not equal. Nat. Biotechnol. 2011, 29, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.-J.; Zhang, F.-J.; Zhang, G.-Z.; Jiang, X.-Y.; Yu, H.-M.; Hou, B.-K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.P.; Christopher, D.A. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol. Genet. Genom. 2008, 280, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Fridborg, I.; Johansson, A.; Lagensjö, J.; Leelarasamee, N.; Floková, K.; Tarkowská, D.; Meijer, J.; Bejai, S. ML3: A novel regulator of herbivory-induced responses in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 935–948. [Google Scholar] [CrossRef]
- Černý, M.; Novák, J.; Habánová, H.; Cerna, H.; Brzobohatý, B. Role of the proteome in phytohormonal signaling. Biochim. Biophys. Acta 2016, 1864, 1003–1015. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Béziat, C.; Kleine-Vehn, J.; Feraru, E. Histochemical staining of β-glucuronidase and its spatial quantification. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2017; Volume 1497, pp. 73–80. [Google Scholar]
- Nolte, H.; MacVicar, T.D.; Tellkamp, F.; Krüger, M. Instant Clue: A Software Suite for Interactive Data Visualization and Analysis. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Mierswa, I.; Wurst, M.; Klinkenberg, R.; Scholz, M.; Euler, T. YALE. In Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining—KDD ’06, Philadelphia, PA, USA, 20–23 August 2006; Association for Computing Machinery (ACM): New York, NY, USA, 2006; p. 935. [Google Scholar]

| Accession | Process | Name | r | p-Value |
|---|---|---|---|---|
| AT4G20260 | Calmodulin signaling | Plasma membrane-associated cation-binding protein 1 | −0.66 | 1.5 × 10−3 |
| AT3G55800 | Carbohydrate metabolism | Sedoheptulose-1,7-bisphosphatase | −0.72 | 3.4 × 10−4 |
| AT2G31390 | Carbohydrate metabolism | Fructokinase-1 | −0.73 | 2.7 × 10−4 |
| AT1G65960 | GABA biosynthesis | Glutamate decarboxylase 2 | 0.66 | 1.5 × 10−3 |
| AT5G26000 | Glucosinolate degradation | Myrosinase 1 | 0.65 | 1.7 × 10−3 |
| AT5G56010 | Chaperon, defense response | Heat shock protein 90-3 | 0.70 | 5.9 × 10−4 |
| AT4G37000 | Chlorophyll biosynthesis | Red chlorophyll catabolite reductase | 0.65 | 1.9 × 10−3 |
| AT1G63680 | Chloroplast biogenesis | UDP-N-acetylmuramoyl-l-alanyl-d-glutamate-2,6-diaminopimelate ligase | 0.77 | 8.1 × 10−5 |
| AT1G08520 | Chloroplast biogenesis | Magnesium-chelatase subunit | 0.68 | 9.6 × 10−4 |
| AT2G04030 | Chloroplast biogenesis | Heat shock protein 90-5 | 0.66 | 1.5 × 10−3 |
| AT3G18420 | Chloroplast biogenesis | protein SLOW GREEN 1 | −0.65 | 2.1 × 10−3 |
| AT1G72150 | Membrane-trafficking | Patellin-1 | 0.80 | 1.9 × 10−5 |
| AT5G21060 | Methionine metabolism | Homoserine dehydrogenase | 0.70 | 6.6 × 10−4 |
| AT1G20220 | Nucleic acid binding | Alba DNA/RNA-binding protein | −0.65 | 1.7 × 10−3 |
| AT5G60360 | Nutrition | Thiol protease aleurain | 0.70 | 6.7 × 10−4 |
| AT1G53310 | Nutrition, anaplerotic reaction | Phosphoenolpyruvate carboxylase 1 | 0.68 | 9.6 × 10−4 |
| AT5G43830 | Other | DUF3700 domain-containing protein | 0.67 | 1.3 × 10−3 |
| AT5G14910 | Other | Heavy metal transport/detoxification superfamily protein | −0.67 | 1.3 × 10−3 |
| AT5G44130 | Other | Fasciclin-like arabinogalactan protein 13 | 0.79 | 3.8 × 10−5 |
| AT1G48600 | Phospholipid metabolism | Phosphomethylethanolamine N-methyltransferase | 0.67 | 1.2 × 10−3 |
| AT5G13120 | Photosynthesis | Photosynthetic NDH subunit of lumenal location 5 | 0.67 | 1.2 × 10−3 |
| AT1G34000 | Photosynthesis | Light-harvesting complex-like protein OHP2 | −0.68 | 9.9 × 10−4 |
| AT4G38630 | Protein degradation | 26S proteasome non-ATPase regulatory subunit 4 | 0.73 | 2.3 × 10−4 |
| AT1G07320 | Proteosynthesis | 50S ribosomal protein L4, chloroplastic | 0.72 | 3.9 × 10−4 |
| AT1G09620 | Proteosynthesis | Leucine--tRNA ligase | 0.66 | 1.4 × 10−3 |
| AT2G40290 | Proteosynthesis | Translation initiation factor eIF-2a | 0.65 | 1.9 × 10−3 |
| AT4G29060 | Proteosynthesis | Polyprotein of EF-Ts | 0.67 | 1.2 × 10−3 |
| AT2G19870 | RNA metabolism | tRNA/rRNA methyltransferase | 0.93 | 4.4 × 10−9 |
| AT3G55460 | RNA metabolism | Serine/arginine-rich SC35-like splicing factor | 0.80 | 2.1 × 10−5 |
| AT1G80670 | RNA metabolism | Protein RAE1 (RNA export factor 1) | −0.65 | 2.1 × 10−3 |
| AT3G57150 | RNA metabolism | H/ACA ribonucleoprotein complex subunit 4 | 0.67 | 1.3 × 10−3 |
| AT4G09000 | Signaling | 14-3-3-like protein GF14 chi | −0.65 | 2.1 × 10−3 |
| AT3G44750 | Transcription | Histone deacetylase HDT1 | 0.72 | 3.3 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berková, V.; Kameniarová, M.; Ondrisková, V.; Berka, M.; Menšíková, S.; Kopecká, R.; Luklová, M.; Novák, J.; Spíchal, L.; Rashotte, A.M.; et al. Arabidopsis Response to Inhibitor of Cytokinin Degradation INCYDE: Modulations of Cytokinin Signaling and Plant Proteome. Plants 2020, 9, 1563. https://doi.org/10.3390/plants9111563
Berková V, Kameniarová M, Ondrisková V, Berka M, Menšíková S, Kopecká R, Luklová M, Novák J, Spíchal L, Rashotte AM, et al. Arabidopsis Response to Inhibitor of Cytokinin Degradation INCYDE: Modulations of Cytokinin Signaling and Plant Proteome. Plants. 2020; 9(11):1563. https://doi.org/10.3390/plants9111563
Chicago/Turabian StyleBerková, Veronika, Michaela Kameniarová, Vladěna Ondrisková, Miroslav Berka, Simona Menšíková, Romana Kopecká, Markéta Luklová, Jan Novák, Lukáš Spíchal, Aaron M. Rashotte, and et al. 2020. "Arabidopsis Response to Inhibitor of Cytokinin Degradation INCYDE: Modulations of Cytokinin Signaling and Plant Proteome" Plants 9, no. 11: 1563. https://doi.org/10.3390/plants9111563
APA StyleBerková, V., Kameniarová, M., Ondrisková, V., Berka, M., Menšíková, S., Kopecká, R., Luklová, M., Novák, J., Spíchal, L., Rashotte, A. M., Brzobohatý, B., & Černý, M. (2020). Arabidopsis Response to Inhibitor of Cytokinin Degradation INCYDE: Modulations of Cytokinin Signaling and Plant Proteome. Plants, 9(11), 1563. https://doi.org/10.3390/plants9111563